Abstract
Recent studies have revealed that long noncoding RNAs (lncRNAs) are connected with pathogenesis of neurodegenerative diseases. Additionally, glucocorticoids have fundamental regulatory roles on the immune system, and act as potent therapeutic compounds for autoimmune and inflammatory diseases. The long noncoding RNA growth arrest-specific 5 (GAS5) which accumulates inside the cells in response to cellular starvation/growth arrest, acts as a potent repressor of the glucocorticoid receptor (GR) through its glucocorticoid response element (GRE). The aim of the present study was to investigate the role of lncRNA GAS5 and its downstream target Nuclear Receptor Subfamily 3 Group C Member 1(NR3C1) in the pathogenesis of multiple sclerosis (MS), and to define the role of GAS5 in the regulation of NR3C1 expression. Quantitative polymerase chain reaction was performed for investigating the expression of GAS5 and NR3C1 in MS patients and healthy subjects. We found that GAS5 levels were up-regulated in the MS patients, blood compared with healthy subjects in correlation with NR3C1 expression. Our findings suggest that GAS5 may play on important role in the molecular etiology and treatment of MS.
Key Words: Multiple sclerosis, GAS5, NR3C1
Multiple sclerosis (MS) is a multifocal chronic progressive inflammatory demy-elinating disease of the central nervous system (CNS) that leads to severe neurological disability through axonal degeneration and gliosis (1). Recent studies have reported that human genome encodes many long non-coding RNAs (lncRNAs) (2). LncRNAs participate in multiple biological mechanisms and regulate gene expression via transcriptional, post-transcriptional, and epigenetic regulation. Most of lncRNAs have tissue-specific expression pattern with precise temporal and spatial regulation in the CNS. Dysregulation in lncRNAs expression has been shown to be associated with different types of neurodegenerative disorders such as MS, Alzheimer's, Parkinson's and Huntington's diseases. However, the exact mechanisms of such contribution have remained unclear.The growth arrest-specific 5 (GAS5), a small nucleolar RNA (snoRNA) host gene, is accumulated in cells showing growth arrest upon lack of nutrients or growth factors. This lncRNA has been detected for the first time through screening of potential tumor suppressor genes expressed at high levels during growth arrest (3). GAS5 has multiple functions at cellular level such as cell cycle arrest control at the G0/G1 phase, alteration of protein synthesis, and modulation of apoptosis (4). This gene encodes 10 box C/D snoRNAs within 11 introns and has been considered as a member of the 5′-terminal oligopyrimidine tract (5′ TOP) gene family. The nutrient's levels alter cellular growth and survival through affecting gene transcription.
The transcription of GAS5 is increased during growth arrest due to the presence of anti-translation inhibitors, and serum starvation. Such alteration in GAS5 expression prepares cells for apoptosis via inhibition of glucocorticoid-mediated induction of numerous responsive genes. GAS5 RNA, despite of little protein-coding potential, is spliced, polyadenylated, and associated with ribosomes. GAS5 has been recently reported to act as a starvation- or growth arrest-linked riborepressor with a central role in the glucocorticoid receptor (GR) regulation. GAS5 binds to the GR DNA-binding domain and interferes with glucocorticoid response element (GRE) (5, 6).
Glucocorticoids modulate gene transcription, and have roles on cell growth, energy expenditure, and cell survival as well. They are vastly used in the treatment of inflammatory and autoimmune diseases. Upon binding with GRs, they translocate into the nucleus, and participate in the transcriptional activity of target genes by binding to specific promoter DNA sequences. They have interaction with transcription factors, and different signaling pathways. In this current study, we examined the expression of GAS5 in correlation with one of downstream targets called Nuclear Receptor Subfamily 3 Group C Member 1 (NR3C1) which encodes for a glucocorticoid receptor.
Materials and methods
Study design and eligibility criteria
Blood samples were collected from 50 healthy volunteers (27 female and 23 male, mean age: 35.3 ± 2.1 years) and 50 relapsing remitting (RR) MS patients in early stage of the disease (31 female and 19 male, mean age: 36.2 ± 2.9 years, age of onset: 31.36 ± 2.4 years, duration of disease: 6.1 ± 3.1 years, expanded disability status scale EDSS: 2.72±2.3). Exclusion criteria were pregnancy, steroid therapy or infection within one month prior to sample collection, existence of other autoimmune diseases, malignancies or chronic infectious diseases. Inclusion criteria for RR-MS patients were diagnosis of RRMS according to the revised McDonald’s criteria, and stable phase of the disease (7). Exclusion criteria for RR-MS were clinical or radiological relapse within one month from sample collection. We also excluded those with smoking history. No significant difference was seen in body mass index between cases and healthy controls.
Blood sampling
Whole blood samples were collected from participants after acquiring informed consent form. Ethics approval for the study was acquired from the local ethics committee of Shahid Beheshti University of Medical Sciences (No. 3275). Blood samples were collected from MS patients in Iranian MS society clinic and Imam Hossein Hospital in Tehran. Total RNA was extracted using Geneall Hybrid-RTM blood RNA extraction Kit (cat No. 305-101) according to the manufacturer’s instructions. Extracted RNA was subjected to complementary DNA (cDNA) synthesis using Geneall cDNA synthesis kit according to the manufacturer’s instructions. Total RNA was treated with DNase I for 30 min at 42 °C. Allele ID 7 software (Premier Biosoft, PaloAlto, USA) was used to design the specific probes and primers. HPRT1 as a housekeeping gene was used for normalization of the gene expression level of samples. The sequence of probes and primers are presented in Table 1.
Table 1.
The sequence of probes and primers
| Gene name | Primer and probe length (bp) | Primer and probe sequences | Product length (bp) |
|---|---|---|---|
| HPRT1 | 18 | F: AGCCTAAGATGAGAGTTC | 88 |
| 21 | R: CACAGAACTAGAACATTGATA | ||
| 24 | FAM -CATCTGGAGTCCTATTGACATCGC- TAMRA |
||
| GAS5 | 20 | F: CTGCTTGAAAGGGTCTTGCC | 98 |
| 21 | R: GGAGGCTGAGGATCACTTGAG | ||
| 24 | FAM- ACCCAAGCTAGAGTGCAGTGGCCT- TAMRA | ||
| NR3C1 | 22 | F: AGAGGAGGAGCTACTGTGAAGG | 78 |
| 21 | R: TCGCTGCTTGGAGTCTGATTG | ||
| 24 | FAM -TGCGTCTTCACCCTCACTGGCTGT- TAMRA |
Quantitative real-time PCR (TaqMan®)
Expression levels of GAS5 and its target NR3C1 were assessed by real-time RT-PCR TaqMan® analysis using the Corbett Rotor gene 6000 instrument (Corbett Life Science). The reverse transcription reaction was carried out with the high capacity RNA-to-cDNA kit (Geneall) and the real-time PCR was performed in duplicate using the TaqMan® gene expression assay according to the manufacturer’s instructions. The thermal cycling conditions for TaqMan assays were as follows: 10 min at 95 °C followed by 40 cycles at 95 °C for 15 s and 60 °C for 45 s. The expression levels of GAS5 and NR3C1 were evaluated using the comparative Ct method (2-ΔΔCt method). Ct values were corrected based on PCR efficiencies using LinReg PCR. The GAS5 and NR3C1 expression values were normalized using the housekeeping gene. Quality control was done using negative control in every reaction.
Statistical analysis
SPSS Version 18 (Chicago, IL, USA) statistical package was applied for statistical analysis. Independent t test was done to compare the differences between the results of two groups. Pearson correlation coefficient was used for the evaluation of the correlation between variables. P values less than 0.05 were regarded as significant. Spearman rank order correlation test was considered to evaluate the possible correlation between relative expression levels of genes and clinical variables as well as between the levels of gene expression in patients.
Results
Demographic data of participants
Clinical characteristics, disease duration, and demographic characteristics of RRMS patients and healthy controls are presented in Table 2. Participants were classified into three different groups based on their age ranges (<30, 30–40, and>40 years), and sex (male or female). Separate computations were done for the total numbers in both groups. All patients were treated with interferon (IFN)-β for at least two years (intramuscular injection of 20 μg of CinnoVex three-times a week), and were recognized as IFN-β responders (9).
Table 2.
Demographic information of RRMS patients and healthy controls
| Variables | MS patients | Controls |
|---|---|---|
| Female/male [no. (%)] | 31(62%)/19(38%) | 27(54%)/23(46%) |
| Age (mean ± SD, year) | 36.2 ± 2.9 | 35.3 ± 2.1 |
| Age at onset (mean ± SD, year) | 31.41 ± 2.8 | - |
| Duration (mean ± SD, Year) | 4.58 ± 3.2 | - |
| EDSS (mean ± SD) | 3.07 ± 2.7 | - |
| EDSS: expanded disability status scale. | ||
GAS5 and NR3C1 expression levels
The expression level of NR3C1 has been compared between MS patients and control group based on age, and sex of the participants. Obtained results are presented in Table 3. Statistical analysis showed significant up-regulation of NR3C1 gene in MS patients compared with healthy controls (P=0.03), and in male subgroup compared with the corresponding age and sex-matched healthy subjects (P = 0.001).
Table 3.
NR3C1 expression levels in MS patients compared with control group, based on age and sex of the participants
| NR3C1 expression | Controls no. | MS patients no. | Expression ratio | Standard error | P value | 95% confidence interval | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 50 | 50 | 1.9751 | 0.34 | 0.03 | [0.44, 1.97] | |||||
| Male | 23 | 19 | 3.2544 | 0.404 | 0.001 | [0.98, 3.05] | |||||
| Female | 27 | 31 | 1.3278 | 0.325 | 0.538 | [-0.57, 1.31] | |||||
| <30 years | Male | 6 | 4 | 1.7151 | 0.295 | 0.476 | [-2.17, 3.91] | ||||
| Female | 3 | 7 | 2.3501 | 4.65 | 0.183 | [-9.87, 12.5] | |||||
| 30–40 years | Male | 8 | 5 | 5.5470 | 0.91 | 0.042 | [0.253, 0.75] | ||||
| Female | 5 | 10 | 1.5884 | 1.14 | 0.679 | [-2.27, 3.85] | |||||
| >40 years | Male | 9 | 10 | 3.2386 | -1.01 | 0.038 | [0.027, 4.2] | ||||
| Female | 19 | 14 | 1.0221 | 0.39 | 0.788 | [-1.43, 1.27] | |||||
GAS5 mRNA level was not significantly different in MS patients compared with healthy subjects. The results of the GAS5 expression level in MS patients in comparison with control group are shown in Table 4.
Table 4.
GAS5 expression levels in MS patients compared with control group, based on age and sex of the participants
| GAS5 expression | Controls no. |
MS
patients no. |
Expression ratio | Standard error | P value | 95% confidence interval | |
|---|---|---|---|---|---|---|---|
| Total | 50 | 50 | 0.6687 | 0.08 | 0.061 | [-1.19, 0.08] | |
| Male | 23 | 19 | 0.8945 | -0.227 | 0.561 | [-1.25, 0.74] | |
| Female | 27 | 31 | 0.5459 | 0.411 | 0.061 | [-1.84, 0.16] | |
| <30 years | Male | 6 | 4 | 0.9636 | -0.01 | 0.476 | [-4.16, 3.73] |
| Female | 3 | 7 | 0.1920 | 5.13 | 0.067 | [-16.7, 10.3] | |
| 30–40 years |
Male | 8 | 5 | 1.7716 | 0.33 | 0.171 | [-0.56, 2.77] |
| Female | 5 | 10 | 0.6850 | 1.11 | 0.206 | [-3.05, 1.79] | |
| >40 years | Male | 9 | 10 | 0.5529 | -0.96 | 0.156 | [-2.93, 0.72] |
| Female | 19 | 14 | 0.5677 | 0.16 | 0.226 | [-2.17, 0.74] | |
Correlation between GAS5 and NR3C1 expres-sion levels and clinical characteristics of patients
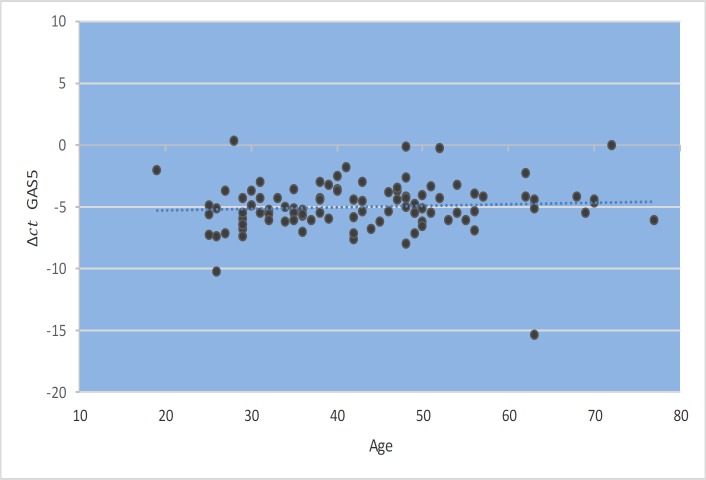
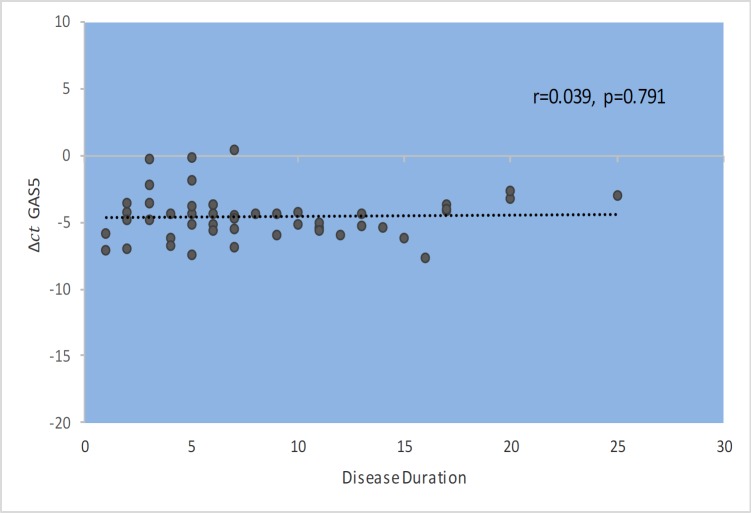
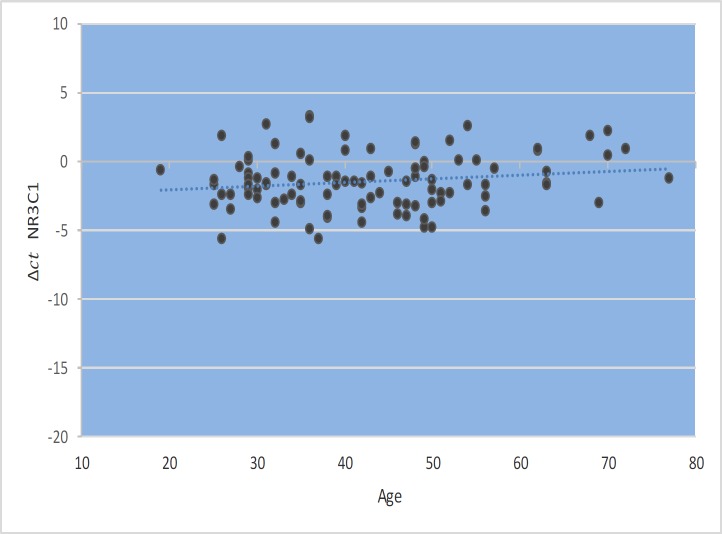
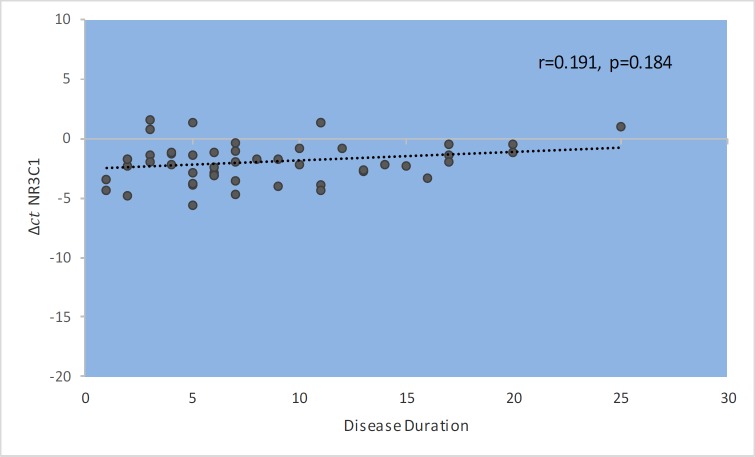
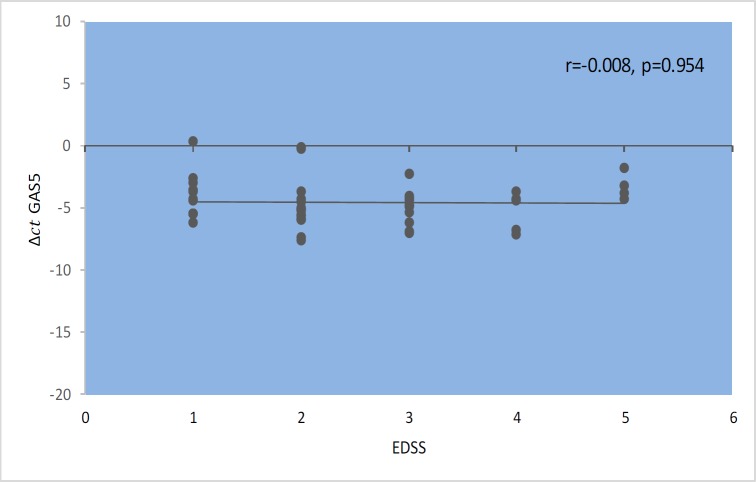
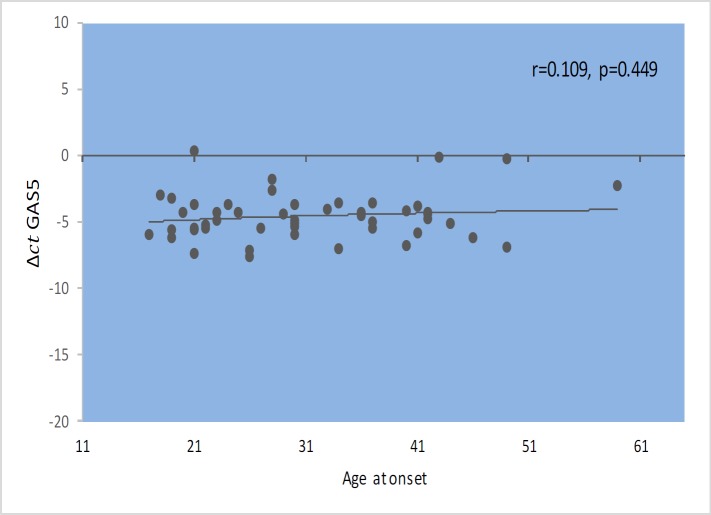
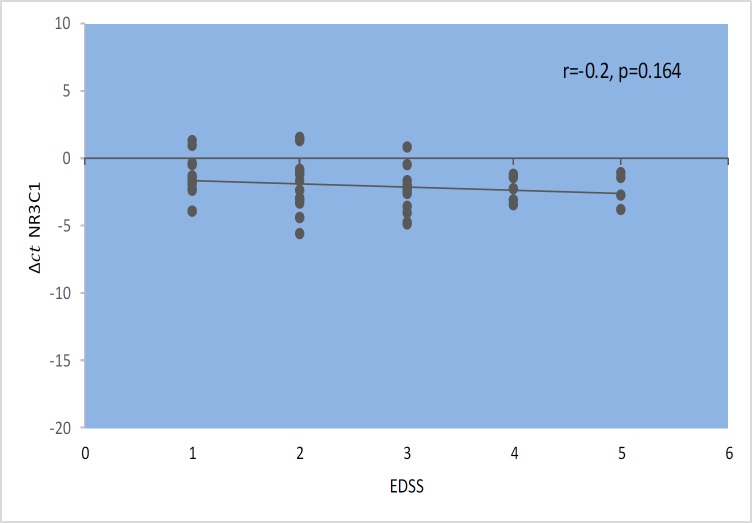
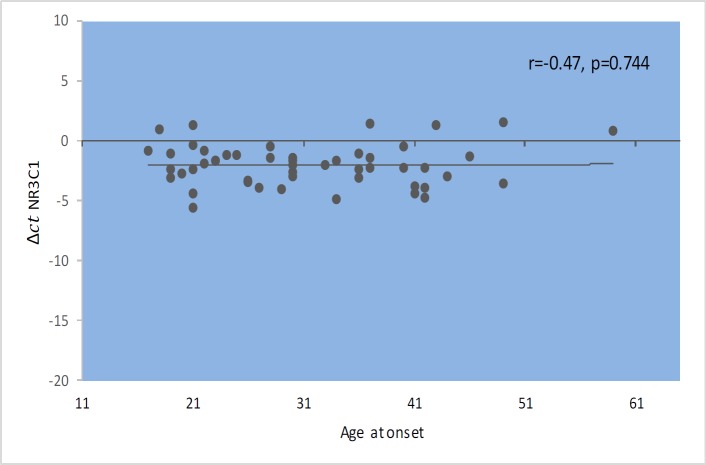
GAS5 gene expression level did not show significant correlations with EDSS, disease duration, or age at onset (Figures 1–4). Also, there were no significant correlations between NR3C1 expression level and disease duration, or age at onset (Figures 5-8).
Fig. 1.
Spearman correlation between GAS5 expression and age.
Fig. 4.
Spearman correlation between GAS5 expression and disease duration in patients.
Fig. 5.
Spearman correlation between NR3C1 expression and age.
Fig. 8.
Spearman correlation between NR3C1 expression and disease duration in patients
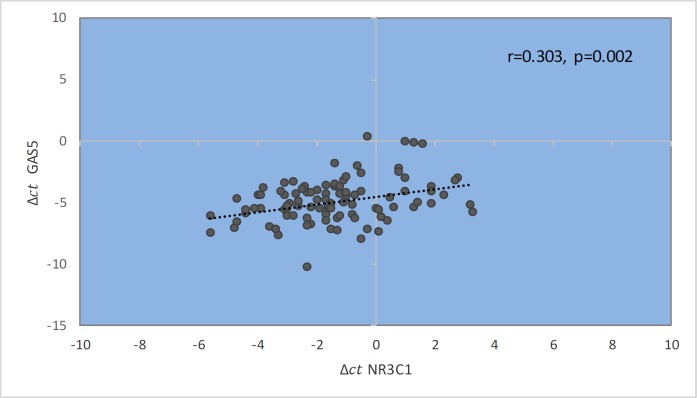
Correlation between expression levels of GAS5 and NR3C1 genes
Our results have shown a significant correlation between GAS5 and NR3C1 expression levels (Fig.9).
Fig. 9.
Spearman correlation between expression levels of GAS5 and NR3C1.
Discussion
Recent studies have shown that lncRNAs are involved in the development and progress of human diseases such as neurodegenerative diseases (10, 11). Certain circulating lncRNAs, such as inflammation regulators, and immune response, have been identified as useful biomarkers for neurodegenerative disorders as well as pathway prediction and determination (12, 13).
Fig. 2.
Spearman Correlation between GAS5 expression and EDSS in patients .
Fig. 3.
Spearman correlation between GAS5 expression and age at onset in patients.
Fig. 6.
Spearman correlation between NR3C1 expression and EDSS in patients.
Fig. 7.
Spearman correlation between NR3C1 expression and age at onset in patients
GAS5 is a lncRNA involved in the regulation of cell cycle with central role in normal growth arrest in T-cell and non‑transformed lymphocytes (14). GAS5 has a conserved 5'‑terminal oligopyrimidine tract (5'‑TOP) that may explain its cellular accumulation upon growth arrest in a range of species (15). Recent studies hawe reported the role of GAS5 in many types of cancer (16, 17). Functional studies have shown that GAS5 inhibition may suppress cell apoptosis, whereas overexpression increases cell apoptosis and reduces the rate of progression through the cell cycle. It hase been concluded that modulated GAS5 is required for normal growth arrest (18).
On the other hand, glucocorticoids are commonly applied in inflammatory, autoimmune disorders, and in the prevention of rejection in transplanted patients (19, 20). Their efficacy is different between individuals, and the side effects have been shown in recent reports (21). Glucocorticoids regulate gene expression of target cells through binding with the NR3C1 encoded GR. The NR3C1 gene has a C-terminal ligand-binding domain (LBD), an N-terminal transcriptional regulatory region, and a central DNA binding domain (DBD) (22). GR autoregulation by a negative feedback mechanism has been identified through the binding of the activated receptor to intragenic sequences called GRE-like elements contained in GR. Along with these findings; interaction of GAS5 with the activated GR suppresses its transcriptional activity through preventing its association with GRE targets (6).
In the present study, correlation analyses between GAS5 lncRNA expression levels and clinical data of MS patients revealed no significant correlation between its expression levels with age, age at onset, EDSS, and disease duration. Also, we found correlations between the expression levels of NR3C1 and clinical data of RRMS patients. However, expression ratio in MS patients hase increased by 1.9 times, and this increase was significantly different between two groups (P=0.03). The results of our study show an increase in the NR3C1 expression by about 3.2 times in affected males compared with the control group (P=0.001). Moreover, this upward trend increases the expression of the gene in a manner consistent with age. The peak in the range between 30 and 40 years in men is up to 5.5 fold, which is statistically significant (P =0.042). In addition, we found higher expression level of GAS5 in male MS patients in the same age range compared with male controls, although the difference was not significant. Such finding might be related to higher threshold for initiation of MS in males. On the opposite side, the increase in the expression of this gene in women with a disease was lower than that of men.
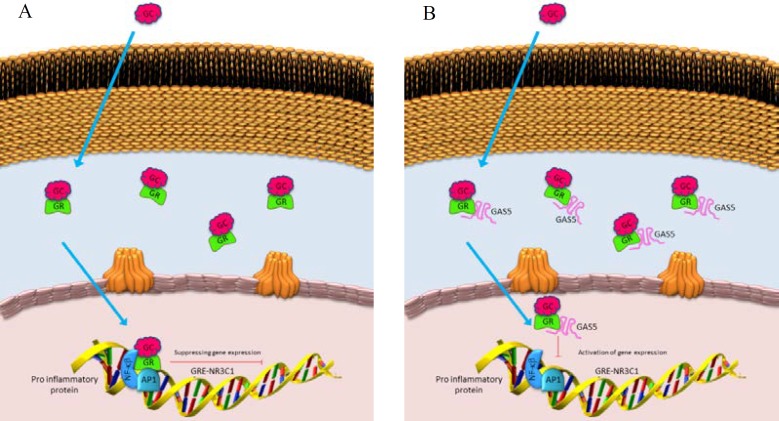
It can be concluded that GAS5 and its downstream target, NR3C1, might participate in a complex multigene interaction network which regulates the expression of several targets in the pathways that may be deregulated in MS. It has been recently reported that the nonsense-mediated degradation (NMD) pathway can regulate the function of GAS5 in mammalian cells (22). Interestingly, the ability of GAS5 in binding to GR will result in the inhibition of its ligand dependent association with DNA. Along with this action, GR binds the promoters of various glucocorticoid responsive genes, including inflammatory pathway genes (Figure 10 A and B). The mechanism of GR anti-inflammatory effects is through its interaction with other transcription factors such as activator protein 1 (AP1) or nuclear factor κB, thereby trans-repressing their transcriptional activity, which leads to inhibition of pro-inflammatory transcription factors. It has been shown that poor responders to glucicorticoids have higher levels of GAS5 and NR3C1 in comparison with good responders (23, 24). It seems that abnormal expression levels of GAS5 may modify glococorticoid effectiveness by interfering with the mechanism of GR autoregulation. The present study provides a novel finding that NR3C1 gene expression in the peripheral blood mononuclear cells is modulated by GAS5. We found a significant correlation between GAS5 and NR3C1 expression levels (figure 5). Our results showed that there is a direct and moderate relation between the expression of these two genes, so that by increasing the expression of each, the expression of the other one also increases, which is quite significant (r= 0.3, P = 0.002).
Fig. 10.
Mechanism of GAS5 (A) and NRC3C1 (B) contribution in proinflammatory pathway genes expression.
This clinical study prepared the foundation for decoding the molecular pathways, and genetic mechanisms underlying the pathology of MS disease. For this reason, assessing lncRNAs as important regulators of gene expression will help to identify the gene targets of GAS5 in the context of MS. LncRNAs including GAS5 are multifaceted and have complex functional responsibilities which are often dictated by the cell type and molecular targets. This is the first study that investigated and correlated levels of circulating lncRNA GAS5 with downstream target NR3C1 in MS. In conclusion, we proposed the altered expression of GAS5 as a pathologic event in MS which might lead to alteration of the NR3C1 gene function or expression. The design may be easily adapted to larger series and in patients with other chronic inflammatory, and autoimmune diseases. GAS5 can be considered as a candidate marker of glucocorticoid resistance in such disorders. In addition, large association studies are needed to assess the effects of GAS5 and NR3C1 functional polymorphisms in conferring risk to MS. These results can be expanded to verify the model that by combining GAS5 levels with other molecular markers such as NR3C1 levels may predict onset of MS.
Conflict of interest
The authors declared no conflict of interest.
References
- 1.Azimi T, Ghafouri-Fard S, Davood Omrani M, et al. Vaccinia Related Kinase 2 (VRK2) expression in neurological disorders: schizophrenia, epilepsy and multiple sclerosis. Mult Scler Relat Disord. 2018;19:15–9. doi: 10.1016/j.msard.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Eftekharian MM, Ghafouri-Fard S, Soudyab M, et al. Expression Analysis of Long Non-coding RNAs in the Blood of Multiple Sclerosis Patients. J Mol Neurosci. 2017;63:333–41. doi: 10.1007/s12031-017-0982-1. [DOI] [PubMed] [Google Scholar]
- 3.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–93. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 4.Pickard MR, Williams GT. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes (Basel) 2015;6:484–99. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mourtada-Maarabouni M, Williams GT. Growth arrest on inhibition of nonsense-mediated decay is mediated by noncoding RNA GAS5. Biomed Res Int. 2013;2013:358015. doi: 10.1155/2013/358015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayad A, Ghafouri-Fard S, Omrani MD, et al. Myxovirus resistance protein A (MxA) polymorphism is associated with IFNbeta response in Iranian multiple sclerosis patients. Neurol Sci. 2017;38:1093–9. doi: 10.1007/s10072-017-2935-4. [DOI] [PubMed] [Google Scholar]
- 9.Taheri M, Ghafouri-Fard S, Solgi G, et al. Determination of cytokine levels in multiple sclerosis patients and their relevance with patients' response to Cinnovex. Cytokine. 2017;96:138–43. doi: 10.1016/j.cyto.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Yan I, Haga H, et al. Long noncoding RNA in liver diseases. Hepatology. 2014;60:744–53. doi: 10.1002/hep.27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang EB, Yin DD, Sun M, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu L, Ye H, Huang G, et al. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691–702. doi: 10.1007/s13277-015-4111-x. [DOI] [PubMed] [Google Scholar]
- 13.Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 14.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84–8. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 16.Glover AR, Zhao JT, Ip JC, et al. Long noncoding RNA profiles of adrenocortical cancer can be used to predict recurrence. Endocr Relat Cancer. 2015;22:99–109. doi: 10.1530/ERC-14-0457. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953–8. doi: 10.3892/ol.2015.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mourtada-Maarabouni M, Pickard MR, Hedge VL, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 19.Riccardi C, Bruscoli S, Migliorati G. Molecular mechanisms of immunomodulatory activity of glucocorticoids. Pharmacol Res. 2002;45:361–8. doi: 10.1006/phrs.2002.0969. [DOI] [PubMed] [Google Scholar]
- 20.Inaba H, Pui CH. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol. 2010;11:1096–106. doi: 10.1016/S1470-2045(10)70114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maranville JC, Baxter SS, Torres JM, et al. Inter-ethnic differences in lymphocyte sensitivity to glucocorticoids reflect variation in transcriptional response. Pharmacogenomics J. 2013;13:121–9. doi: 10.1038/tpj.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Cidlowski JA. The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids. 2005;70:407–17. doi: 10.1016/j.steroids.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucafo M, De Iudicibus S, Di Silvestre A, et al. Long noncoding RNA GAS5: a novel marker involved in glucocorticoid response. Curr Mol Med. 2015;15:94–9. doi: 10.2174/1566524015666150114122354. [DOI] [PubMed] [Google Scholar]