Abstract
This study aims to investigate the role of miR-106b-5p in cervical cancer by performing a comprehensive analysis on its expression and identifying its putative molecular targets and pathways based on The Cancer Genome Atlas (TCGA) dataset, Gene Expression Omnibus (GEO) dataset, and literature review. Significant upregulation of miR-106b-5p in cervical cancer is confirmed by meta-analysis with the data from TCGA, GEO, and literature. Moreover, the expression of miR-106b-5p is significantly correlated with the number of metastatic lymph nodes. Our bioinformatics analyses show that miR-106b could promote cervical cancer progression by modulating the expression of GSK3B, VEGFA, and PTK2 genes. Importantly, these three genes play a crucial role in PI3K-Akt signaling, focal adhesion, and cancer. Both the expression of miR-106b-5p and key genes are upregulated in cervical cancer. Several explanations could be implemented for this upregulation. However, the specific mechanism needs to be investigated further.
Subject terms: Oncogenes, Oncogenes
Introduction
At present, with an increase in morbidity and mortality, cancer has become the leading cause of death and a significant public health problem. There are over 500,000 novel cases and approximately 274,000 deaths estimated for cervical cancer (CC) each year all over the world1. In 2015, the number of new cases and deaths in China was 4,292,000 and 2,814,000, respectively2. CC is the fourth most common cancer in women in the world, and incidence and mortality are still rising3. Although the relationship between persistent high-risk human papillomavirus (HPV) infection and CC has been confirmed4, the specific molecular cellular network mechanism is still unclear.
Genetic mutations lead to cancer by affecting gene expression and protein function in the cells. However, the dysregulation of microRNA (miRNA) expression is detected in a variety of tumors and is considered to be a significant contributor to the development of cancer in recent years5,6. miRNAs are small single-stranded non-coding RNAs that specifically silence gene expression and alter cell or organism phenotypes. Previous studies have confirmed that miRNAs participate in proliferation, apoptosis, morphogenesis7, antiviral defense8, and tumorigenesis9.
Recently, growing evidence reveals that miR-106b-5p plays a critical role in various cancers. Huang and Hu10 showed that the upregulation of miR-106b can be observed in the endometrium and knockdown of miR-106b suppresses proliferation and promotes apoptosis. Shi et al.11 reported that upregulation of miR-106b-5p exhibited a promoting role in hepatocellular carcinoma (HCC) cell properties and migration, whereas downregulation exhibited an opposite effect. Lu et al.12 and Xiang et al.13 submitted that overexpression of miR-106b-5p could promote the proliferation and increase the number of metastatic colonies, whereas inhibition would induce cell cycle arrest, suppress cell proliferation, and promote cell apoptosis in renal cell carcinoma.
For CC, miR-106b, the pre-miRNA of miR-106b-5p, also has a pivotal role in occurrence and development. While constructing a miRNA-mRNA network for CC, Ma et al.14 found that miR-106b was one of the key nodes in the network. Overexpression of miR-106b promoted the migration of HeLa and SiHa cells significantly while inhibition displayed an opposite phenomenon15. However, few studies concern the mechanisms of miR-106b for CC at present. Cheng et al.15 found that DAB2 is identified as a direct target of miR-106b and it is inhibited by TGF-β1 partly through miR-106b and is involved in TGF-β1-induced CC cell migration. Piao et al.16 reported that miR-106 overexpression and DAB2 knockdown induced epithelial to mesenchymal transition (EMT) of CC cells cultured on substrate. As miR-106b plays an essential role in CC, its molecular mechanisms need to be further studied.
The purpose of this study is to investigate the role of miR-106b-5p in CC by performing comprehensive research on its expression and identify its putative molecular targets and pathways based on The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), and literature review.
Results
Clinical significance of miR-106b-5p
As the expression data of mature miR-106b-5p are absent in TCGA, a comparison of miR-106b between CC and healthy samples is provided. The expression level of miR-106b is higher in CC (Fig. 1) and significantly associated with the number of metastatic lymph nodes (Cor = 24.510, P = 0.006). However, there is no significant correlation in tumor purity, race, pathological M/N/T stage, number of years of birth, histological type, race, radiotherapy, or overall survival (Table 1).
Fig. 1. Expression of miR-106b-5p in cervical cancer from TCGA.
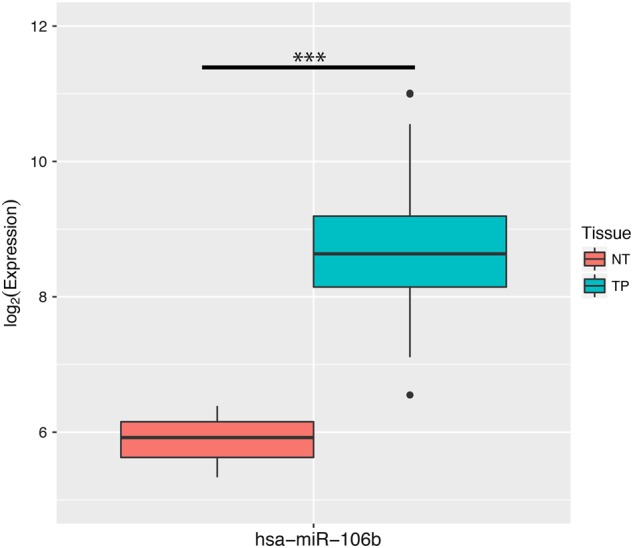
NT normal tissue, TP primary tumor. There are three samples in the NT group and 309 samples in the TP group. Student's t-test is used for the statistical analysis
Table 1.
Correlations between expression of miR-106b-5p and clinical outcomes
| Item | Method | Cor. | P value | FDR |
|---|---|---|---|---|
| Number of lymph nodes | Kruskal–Wallis Test | 24.510 | 0.006a | 0.070 |
| Tumor purity | Spearman Correlation | 0.107 | 0.078 | 0.430 |
| Race | Kruskal–Wallis Test | 7.214 | 0.125 | 0.458 |
| Pathology M stage | Wilcox Test | 0.025 | 0.193 | 0.530 |
| Years to birth | Spearman Correlation | 0.063 | 0.300 | 0.553 |
| Histological type | Kruskal–Wallis Test | 6.047 | 0.302 | 0.553 |
| Ethnicity | Wilcox Test | −0.015 | 0.386 | 0.606 |
| Radiation therapy | Wilcox Test | 0.011 | 0.545 | 0.750 |
| Pathology N stage | Wilcox Test | −0.006 | 0.683 | 0.758 |
| Pathology T stage | Kruskal–Wallis Test | 2.256 | 0.689 | 0.758 |
| Overall survival | Cox Regression Test | 0.045 | 0.792 | 0.792 |
aSignificant difference
Meta-analysis of miR-106b-5p expression
Meta-analysis based on TCGA and GEO
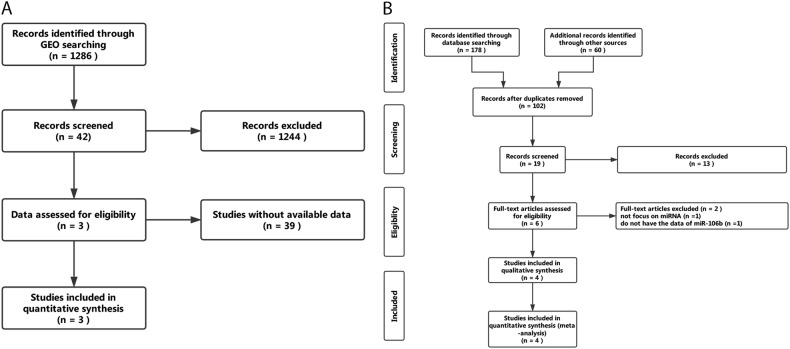
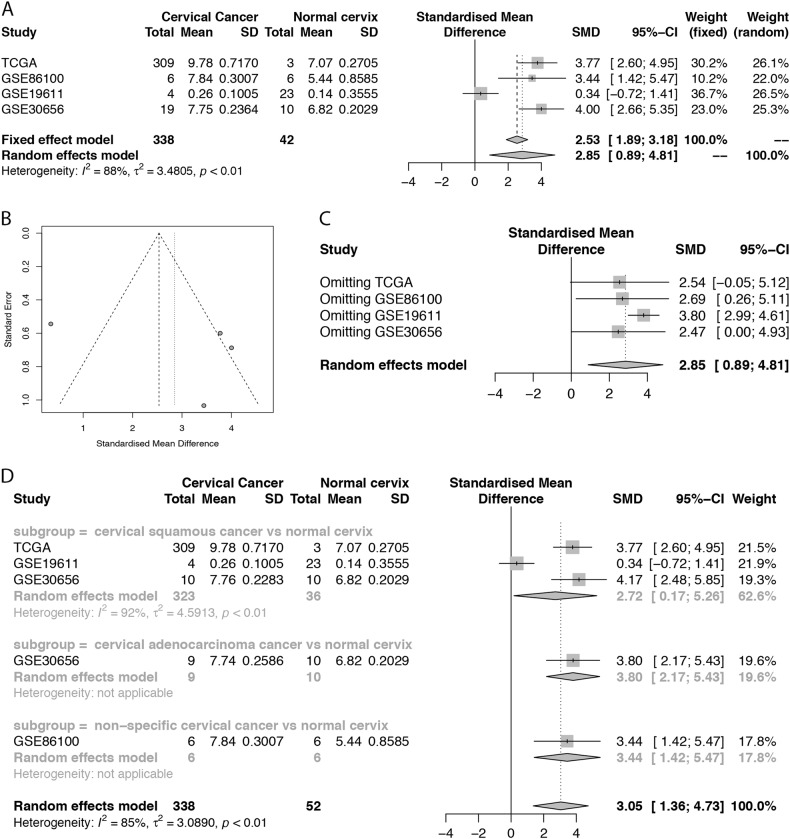
A total of 1286 microarrays were obtained from GEO. After careful screening, the three microarrays, GSE86100, GSE19611, and GSE30656, meet the criteria and are included in the analysis (Fig. 2a). The forest plot presents an overall standard mean difference (SMD) of 2.85 (95% confidence interval (CI): 0.89–4.81) with P = 0.0045 and I2 = 88% (random effect used), suggesting that miR-106b-5p is upregulated in CC (Fig. 3a).
Fig. 2.
Searching workflow for the expression of miR-106b-5p between cervical cancer and non-cancerous tissue. a Searching strategy in GEO; bSearching strategy in literature review
Fig. 3. Meta-analysis of miR-106b-5p between healthy and cancerous cervical tissue based on TCGA and GEO.
a Forest plot of SMD. The expression of miR-106b-5p is significantly higher in cervical cancer tissue; b Funnel plot for four studies that are marked as circles. No significant publication bias is detected (P = 0.5187); c Influence analysis for four studies. No study had an impact on the overall SMD estimation. d Subgroup forest plot based on cancer type. As I2 value is still relatively high, the cancer subtype is not the only source of heterogeneity
A funnel plot of miR-106b-5p expression (Fig. 3b) reveals that no significant publication bias is detected by Egger’s test (P = 0.5187). Sensitivity analysis shows that similar results are obtained for the fixed effects models except for a lower difference (SMD: 2.53, 95%CI: 1.89–3.18, P < 0.0001).
The influence analysis (Fig. 3c) shows that no study had an impact on the overall SMD estimation because the point estimate for any of the studies is not outside the combined analysis CI and there is no significant statistical change.
Except for a decrease in I2 values, similar results are obtained in the subgroup analysis of cancer subtypes (Fig. 3d). The results show that the cancer subtype is not the only source of heterogeneity as the I2 value is still relatively high, but miR-106b-5p continues to be highly expressed in CC tissues.
Meta-analysis based on literature review
The workflow for searching is presented in Fig. 2b. Finally, four studies14,15,17,18 that met the criteria were selected. Consistent with the result of the meta-analysis, a common pattern of upregulation for miR-106b-5p in CC was reported across the included studies (Table 2).
Table 2.
Overview of the four studies selected in the literature review
| Author | Year | Country | Cancer (n) | Normal (n) | Result | Detection methods |
|---|---|---|---|---|---|---|
| Cheng et al. | 2016 | China | 19 | 19 | Upregulated | qRT-PCR |
| Gao et al. | 2016 | China | 30 | 26 | Upregulated | qRT-PCR |
| Ma et al. | 2012 | China | 8 | 8 | Upregulated | qRT-PCR |
| Liu et al. | 2016 | China | 10 | 10 | Upregulated | qRT-PCR |
Bioinformatics analyses of miR-106b-5p
Screening of candidate genes
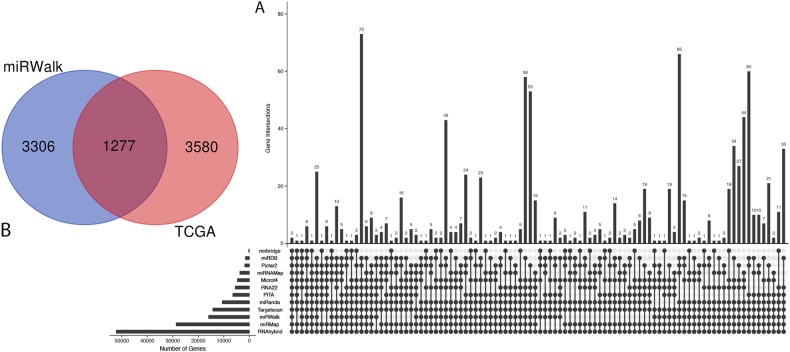
By analyzing the data from Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) with the criterion of log|FC| > 1 and FDR < 0.05, 4857 differentially expressed genes (DEGs) were selected, including 4619 upregulated genes and 238 downregulated genes. Meanwhile, predicting using 12 databases in miRWalk, 10,073 target genes that were overlapping in at least five databases were found (Fig. 4a). After merging DEGs and the predicted target genes, 1277 candidate genes were collected (Fig. 4b).
Fig. 4. Predication of miR-106b-5p target genes and candidate genes screening.
a The number of overlapped genes across 12 databases; 10,073 target genes which overlapped at least five databases are obtained. b Venn plot for the integration between DEGs and predicted target genes of miR-106b-5p
Gene ontology enrichment analysis
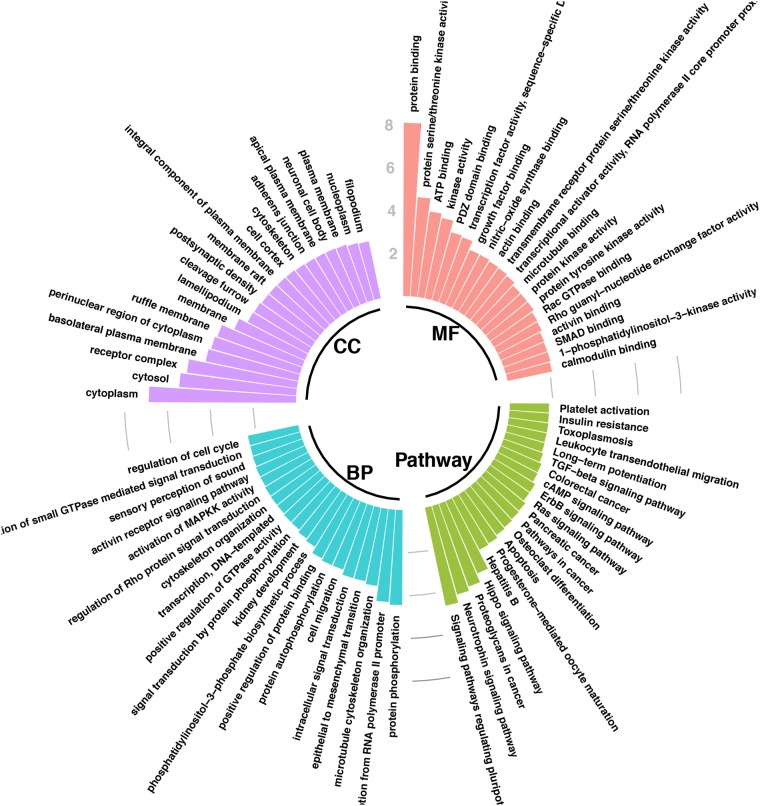
The DAVID database was used for Gene Ontology (GO) analysis of the 1277 genes (Fig. 5). Using the criterion of P < 0.001, the results showed that while cellular component (CeC), target genes are mainly involved in “cytoplasm”, “cytosol”, “receptor complex”, “basolateral plasma membrane”, “perinuclear region of cytoplasm”, “ruffle membrane”, “membrane”, “lamellipodium”, “cleavage furrow”, “postsynaptic density”, “membrane raft”, “integral component of plasma membrane”, and “cell cortex”. In terms of biological process (BP), the target genes mainly participate in “protein phosphorylation”, “positive regulation of transcription from RNA polymerase II promoter”, “microtubule cytoskeleton organization”, “epithelial to mesenchymal transition”, “intracellular signal transduction”, “cell migration”, “protein autophosphorylation”, and “positive regulation of protein binding”. With regard to MF, these genes are mainly enriched in “protein binding”, “protein serine/threonine kinase activity”, “ATP binding”, “kinase activity”, “PDZ domain binding”, and “transcription factor activity, sequence-specific DNA binding”.
Fig. 5. The top 20 items of cellular component (CeC), biological process (BP) pathways, molecular function (MF), and pathways in Gene Ontology (GO) and pathway enrichment analysis for candidate target genes of miR-106b-5p in CC.
Values are expressed as −log10 (P-value)
Protein–protein interaction network analysis
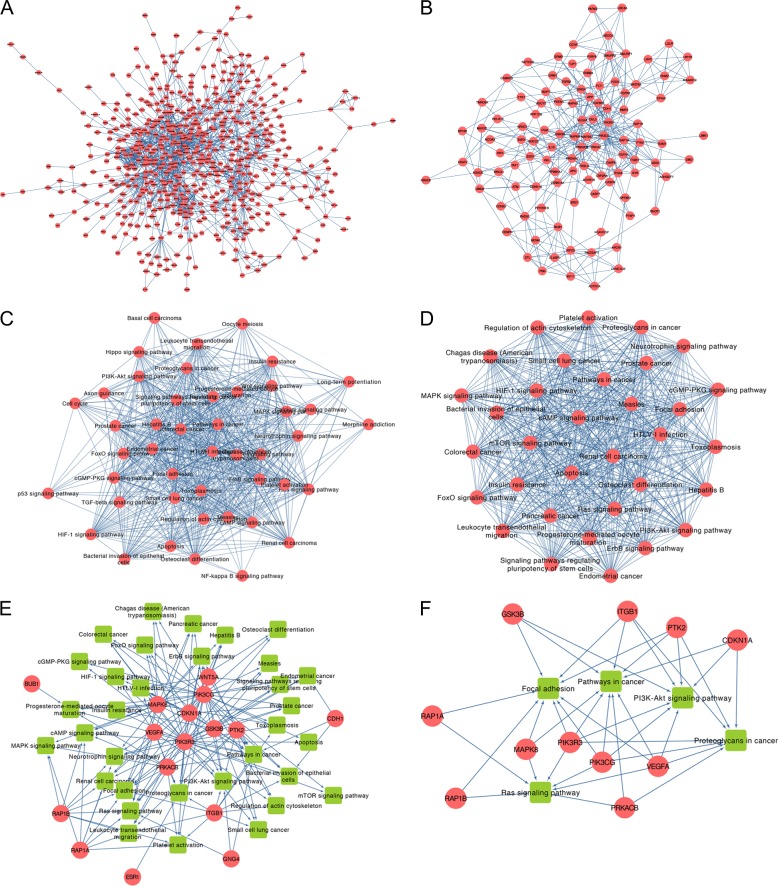
The original network contains 668 nodes and 1779 edges. After cleaning the isolated genes, the main network containing 579 nodes and 1716 edges was obtained (Fig. 6a). By extracting the nodes with degree and betweenness which are higher than average, a subnetwork that contains 110 nodes and 404 edges was gained (Fig. 6b; Table 3).
Fig. 6. Protein–protein analysis (PPI) analysis of candidate genes and pathway crosstalk and gene-pathway analysis of hub genes.
a PPI network of candidate genes; b Subnetwork of PPI for main nodes extracted according to degree and betweenness being higher than average; c Pathway crosstalk analysis of hub genes. The thickness of lines between nodes are represented by the average value of Jaccard coefficient (JC) and overlapping coefficient (OC); d Subnetwork of pathway crosstalk extracted by MCODE; e Comprehensive gene-pathway network constructed by mapping the hub genes to the subnetwork. The arrow direction between gene and pathway is determined by KEGG. Red circle: genes; green square: pathway. f Subnetwork of gene-pathway collected according to the criteria that node’s degree > average
Table 3.
Characteristics of main genes
| Gene | Degree | Betweenness |
|---|---|---|
| Average value | 5.928 | 2064.408 |
| PIK3CG | 46 | 41488.54899 |
| PIK3R3 | 41 | 25199.73317 |
| APP | 35 | 32767.23403 |
| MAPK8 | 33 | 35071.18269 |
| PRKACB | 29 | 27367.34287 |
| PTK2 | 28 | 25355.409 |
| ITGB1 | 28 | 17088.50374 |
| SMURF1 | 27 | 7546.059618 |
| SMURF2 | 27 | 6748.334913 |
| BUB1 | 26 | 11066.28571 |
| GNG4 | 26 | 10653.80228 |
| H2AFV | 25 | 20305.78635 |
| WNT5A | 25 | 13709.55435 |
| VEGFA | 23 | 20760.79426 |
| KIF23 | 21 | 10253.88184 |
| RAD51 | 21 | 17892.93805 |
| KIF11 | 21 | 5135.989346 |
| CCNF | 20 | 5540.628454 |
| RAP1A | 19 | 7521.72323 |
| PPP2R1B | 19 | 14143.0877 |
| BRCA1 | 19 | 6029.273042 |
| NEDD4 | 19 | 3192.727805 |
| CLASP1 | 19 | 7496.485807 |
| SYK | 19 | 11386.15814 |
| GSK3B | 19 | 18438.62749 |
| DNM2 | 19 | 5911.698768 |
| ADCY2 | 18 | 7560.286406 |
| ATM | 18 | 9420.324388 |
| CDKN1A | 18 | 18240.66944 |
| RHOT1 | 18 | 16492.35297 |
| HIP1R | 17 | 3517.329081 |
| RAP1B | 17 | 5215.72323 |
| MCM4 | 17 | 5808.281935 |
| UBE4A | 17 | 2304 |
| PARK2 | 17 | 2366.125584 |
| CDH1 | 17 | 14376.3563 |
| UBE2I | 17 | 10854.31473 |
| SYNJ2 | 17 | 5273.61775 |
| IL10 | 16 | 11158.43666 |
| LRP2 | 16 | 3590.998058 |
| KIAA0319 | 15 | 3475.143506 |
| RACGAP1 | 15 | 5861.489126 |
| ACTR1A | 15 | 3744.522591 |
| PRDM10 | 15 | 14008.2127 |
| FASLG | 15 | 5210.263877 |
| BMP2 | 15 | 5337.049932 |
| LDLR | 15 | 2191.70904 |
| HDAC9 | 15 | 10403.97449 |
| GRM5 | 14 | 3250.915796 |
| CASP8 | 14 | 5874.766056 |
| ESR1 | 14 | 11585.62988 |
| NFATC2 | 14 | 4879.928683 |
| PBK | 14 | 4821.131434 |
| VAV2 | 14 | 8716.774939 |
| GRM1 | 14 | 3250.915796 |
| FGFR1OP | 13 | 6727.611655 |
| DTL | 13 | 2540.356126 |
| TGFBR1 | 13 | 4022.943631 |
| CAMK2D | 13 | 5055.739472 |
| CASP9 | 12 | 4919.739038 |
| P2RY6 | 12 | 2304 |
| YWHAZ | 11 | 4040.292887 |
| MAP3K7 | 11 | 6201.810194 |
| FLT1 | 11 | 6634.16899 |
| PPP1CB | 11 | 2005.113067 |
| NCOA3 | 11 | 7457.814126 |
| FGFR2 | 11 | 3030.464668 |
| ITGA2 | 10 | 3529.023629 |
| NOTCH2 | 10 | 10947.19567 |
| CENPN | 10 | 1873.156984 |
| FGFR1 | 10 | 8007.32537 |
| IRF4 | 10 | 5202.998605 |
| PTBP1 | 10 | 6730.910039 |
| MEF2C | 10 | 4165.497288 |
| ARHGEF7 | 9 | 3147.468396 |
| TIAM1 | 9 | 4058.542014 |
| ERBB4 | 9 | 4519.484052 |
| ARCN1 | 9 | 2088.364196 |
| CASP7 | 9 | 5818.243304 |
| PGR | 9 | 4778.496029 |
| SPTBN1 | 9 | 10172.1474 |
| DMD | 9 | 2456.15705 |
| FZD3 | 9 | 2260.879218 |
| MAPK9 | 9 | 2693.578423 |
| DYNC1LI2 | 9 | 2495.446083 |
| NR3C1 | 9 | 4868.398944 |
| DVL3 | 9 | 3925.165711 |
| HDAC8 | 9 | 2385.834093 |
| APC | 9 | 4483.983598 |
| TNRC6A | 8 | 2766.776691 |
| RPS6KA1 | 8 | 2691.499073 |
| TJP1 | 8 | 4534.026739 |
| ACVR1B | 8 | 1933.546424 |
| TGFB2 | 8 | 2116.806735 |
| SP100 | 8 | 2757.903141 |
| POLR1E | 7 | 6748.802196 |
| ETS1 | 7 | 4087.280316 |
| CSNK1A1 | 7 | 2157.303406 |
| PSEN1 | 7 | 3233.020264 |
| TAF1 | 7 | 4214.786588 |
| ERC1 | 7 | 3537.432599 |
| CCNE2 | 7 | 1750.370853 |
| REL | 7 | 2794.403409 |
| MAPT | 7 | 2044.275299 |
| LIMK1 | 7 | 1750.999958 |
| CSF2RA | 7 | 1782.306466 |
| E2F2 | 7 | 4585.295895 |
Twelve topological algorithms were applied and the top 20 genes of each method for the subnetwork of PPI were extracted. These selected genes that appeared at least twice are conserved as hub genes (Table 4).
Table 4.
Hub genes identified from top 20 of 12 topological algorithms
| Rank | Gene | Counts |
|---|---|---|
| 1 | APP | 9 |
| 2 | MAPK8 | 9 |
| 3 | PIK3CG | 9 |
| 4 | PIK3R3 | 9 |
| 5 | VEGFA | 9 |
| 6 | ITGB1 | 8 |
| 7 | PRKACB | 8 |
| 8 | PTK2 | 8 |
| 9 | GNG4 | 6 |
| 10 | GSK3B | 6 |
| 11 | PRDM10 | 6 |
| 12 | WNT5A | 6 |
| 13 | CDKN1A | 5 |
| 14 | RAD51 | 5 |
| 15 | SMURF1 | 5 |
| 16 | SMURF2 | 5 |
| 17 | CCNF | 4 |
| 18 | CDH1 | 4 |
| 19 | ESR1 | 4 |
| 20 | H2AFV | 4 |
| 21 | NEDD4 | 4 |
| 22 | TRIM36 | 4 |
| 23 | BUB1 | 3 |
| 24 | EHHADH | 3 |
| 25 | FBXL5 | 3 |
| 26 | RAP1A | 3 |
| 27 | RAP1B | 3 |
| 28 | RHOT1 | 3 |
| 29 | AGFG1 | 2 |
| 30 | ASB13 | 2 |
| 31 | FASLG | 2 |
| 32 | FLT1 | 2 |
| 33 | KBTBD8 | 2 |
| 34 | KIAA0319 | 2 |
| 35 | KIF11 | 2 |
| 36 | KIF23 | 2 |
| 37 | KLHL20 | 2 |
| 38 | KLHL5 | 2 |
| 39 | LDLR | 2 |
| 40 | LRP2 | 2 |
| 41 | MKRN1 | 2 |
| 42 | PACSIN1 | 2 |
| 43 | PARK2 | 2 |
| 44 | PCYT1B | 2 |
| 45 | PGR | 2 |
| 46 | RLIM | 2 |
| 47 | RNF213 | 2 |
| 48 | SPSB4 | 2 |
| 49 | UBE4A | 2 |
Pathway enrichment and crosstalk analysis
Pathway enrichment analysis was also performed by the DAVID database. The results indicate that the pathways of “Signaling pathways regulating pluripotency of stem cells”, “Neurotrophin signaling pathway”, “Proteoglycans in cancer”, and “Hippo signaling pathway” are significantly enriched.
For pathway crosstalk analysis, 45 out of 47 pathways that contain more than two genes met the crosstalk analysis criteria and were selected to construct the network (Fig. 6c). The thickness of the edges indicates measurements of the average value of OC and JC. By using MCODE, a major cluster with 33 nodes and 523 edges was identified from the initial network (Fig. 6d).
Comprehensive gene-pathway analysis
After mapping the hub genes into the subnetwork of pathways guided by KEGG, a potential gene-pathway network including 33 essential pathways and 16 hub genes were constructed (Fig. 6e). This network shows that “PIK3R3” and “PIK3CG” participate in most of the pathways. Furthermore, “pathways in cancer” and “focal adhesion” and “proteoglycans in cancer” rank as the top three pathways according to the genes that they involved.
To screen the main factors (including genes and pathways) in the gene-pathway network, those nodes with degree > average were selected (Fig. 6f). It was found that 11 genes (PIK3R3, PIK3CG, MAPK8, GSK3B, ITGB1, CDKN1A, PTK2, VEGFA, PRKACB, RAP1B, and RAP1A) with five pathways (pathways in cancer, focal adhesion, proteoglycans in cancer, PI3K-Akt signaling pathway, and Ras signaling pathway) are involved and considered more likely to play a role as influential agents.
Identification of key genes and pathways
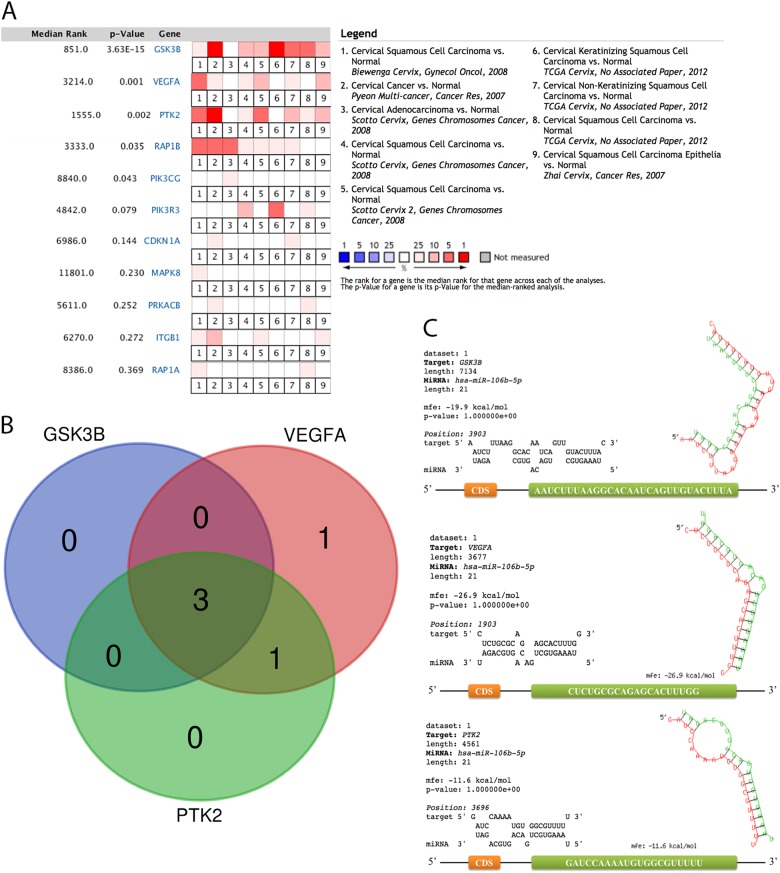
The expression of the main genes was investigated with meta-analysis through nine studies in Oncomine. The results show that all of the main genes are upregulated in CC, among which five genes (GSK3B, VEGFA, PTK2, RAP1B, and PIK3CG) gain significant differences (Fig. 7a). However, the influence analysis indicates that RAP1B and PIK3CG obtain a discrepant result with the meta-analysis for the altering of significance (Table 5). Hence, only GSK3B, VEGFA, and PTK2 are considered to be key genes.
Fig. 7.
The expression of main genes of cervical cancer across nine studies (a), Venn plot for the interaction between key genes and their pathways (b), and the binding sequence and location between miR-106b-5p and each critical gene (c). Orange square: coding sequence (CDS). Green square: binding site
Table 5.
Influence analysis of 5 key genes
| Omitting study | PGSK3B | PVEGFA | PPTK2 | PRAP1B | PPIK3CG |
|---|---|---|---|---|---|
| Omitting Study 1 | 2.16*10-6 | 0.023 | 9.22*10-4 | 0.067 | 0.165 |
| Omitting Study 2 | 1.88*10-4 | 0.023 | 9.22*10-4 | 0.067 | 0.489 |
| Omitting Study 3 | 2.16*10-6 | 7.55*10-4 | 9.51*10-4 | 0.067 | 0.367 |
| Omitting Study 4 | 2.16*10-6 | 0.022 | 9.51*10-4 | 0.023 | 0.165 |
| Omitting Study 5 | 1.88*10-4 | 0.023 | 9.22*10-4 | 0.067 | 0.367 |
| Omitting Study 6 | 1.88*10-4 | 7.55*10-4 | 9.51*10-4 | 0.023 | 0.165 |
| Omitting Study 7 | 1.88*10-4 | 7.55*10-4 | 2.97*10-5 | 0.055 | 0.367 |
| Omitting Study 8 | 1.90*10-4 | 7.55*10-4 | 9.51*10-4 | 0.023 | 0.165 |
| Omitting Study 9 | 2.16*10-6 | 0.023 | 9.22*10-4 | 0.023 | 0.367 |
Study 1: Cervical Squamous Cell Carcinoma vs. Normal .Biewenga Cervix, Gynecol Oncol, 2008
Study 2: Cervical Cancer vs. Normal. Pyeon Multi-cancer, Cancer Res, 2007
Study 3: Cervical Adenocarcinoma vs. Normal. Scotto Cervix, Genes Chromosomes Cancer, 2008
Study 4: Cervical Squamous Cell Carcinoma vs. Normal. Scotto Cervix, Genes Chromosomes Cancer, 2008
Study 5: Cervical Squamous Cell Carcinoma vs. Normal. Scotto Cervix 2, Genes Chromosomes Cancer, 2008
Study 6: Cervical Keratinizing Squamous Cell Carcinoma vs. Normal. TCGA Cervix, No Associated Paper, 2012
Study 7: Cervical Non-Keratinizing Squamous Cell Carcinoma vs. Normal. TCGA Cervix, No Associated Paper, 2012
Study 8: Cervical Squamous Cell Carcinoma vs. Normal. TCGA Cervix, No Associated Paper, 2012
Study 9: Cervical Squamous Cell Carcinoma Epithelia vs. Normal. Zhai Cervix, Cancer Res, 2007.
PGene: The p-Value for a gene and is the p-Value for the median-ranked analysis
We merged the pathways that each essential gene participates in order to discover the most important pathways. Three pathways, PI3K-Akt signaling pathway, focal adhesion, and pathways in cancer, which all of the three key genes involved in, were identified (Fig. 7b).
Location and characteristic of the binding site
As presented in Fig. 7c, all of the binding sites for miR-106b-5p are located in 3′UTR of GSK3B, VEGFA, and PTK2. Furthermore, when inspecting the sequence of binding sites, adenine (A) and uracil (U) are occupied in most of the sequence.
Discussion
This study confirms that the expression of miR-106b-5p is significantly upregulated in CC and its expression is highly correlated with the number of metastatic lymph nodes. By using bioinformatics analyses, miR-106b-5p is found to be a key issue in the progression of CC by interacting with three key genes and pathways.
Our results show that miR-106b-5p promotes the progression of CC by targeting GSK3B, VEGFA, and PTK2. GSK3B is dysregulated in a variety of tumor tissues including CC19–24, and it participates in the development of CC caused by HPV16 infection by regulating Wnt signaling/β-catenin pathway23,24. In addition, GSK3B exerts antiproliferative effects by promoting APC-dependent phosphorylation and thus promotes protease-mediated degradation of β-catenin which is a transcription factor that positively regulates Myc and cyclin D1 expression21. When referring to VEGFA, its overexpression in CC can enhance the growth and invasion of tumor cells25. More importantly, VEGFA can promote the proliferation and migration of CC cells by activating the PI3K/Akt/mTOR pathway26. In addition, the expression and stability of VEGFA have a close relationship with cellular hypoxia27 and serum VEGFA level can be used as a biomarker for prognosis evaluation28. In terms of PTK2, it is a cytoplasmic protein tyrosine kinase and has an expression in various solid tumors such as ovarian cancer, gastric cancer, and bladder cancer29. It is found that PTK2 is associated with the sensitivity of colon cancer cells to DNA damage therapy30. However, there are few reports of PTK2 in CC.
The results of pathway analyses reveal that all of the three key genes are involved in the pathway of PI3K-Akt signaling pathway, focal adhesion, and Pathways in cancer. With regard to PI3K-Akt signaling pathway, its imbalance in expression is associated with a variety of tumors, including cervical, endometrial, and non-small cell lung cancers31–33. It is activated during the G1/S transition of the cell cycle and regulates several key cell cycle regulators34. Tumors associated with HPV infection are closely related to the PI3K/Akt pathway. Activation of this pathway contributes to genetic instability, dysregulation, apoptosis resistance, and altered metabolic properties ultimately leading to the malignant transformation of infected cells. Concerning focal adhesion, this pathway is involved in the invasion and metastasis of many kinds of tumors and is related to the medicine resistance of certain tumors35,36. The decreased expression of focal adhesion kinase (FAK), a key gene involved in the pathway, can inhibit the invasion and migration of CC cells37. Another study reports that targeted FAK therapy makes pancreatic cancer cells more sensitive to immunotherapy38.
It has been recognized that miRNA negatively regulates gene expression by guiding the RNA-induced silencing complex (RISC) to silence its target mRNA through degradation or translation repression39. However, it is interesting to observe that both the expression of miR-106b-5p and key genes are upregulated in CC with the binding sites located in 3′UTR of mRNAs. According to previous studies40–44, the mechanisms of miRNA-mediated gene upregulation include (1) the presence of cellular state (G0 or G0-like state) and/or specific factors (AGO1, AGO2, GW182, and FXR1); (2) miRNA directly binds to 5′UTR of RNA and increases its association with 40S and polysome formation or enhances their translation by alleviating their TOP-mediated translational repression during amino acid starvation, respectively; (3) miRNA prevents tristetraprolin (TTP) binding to the AU-rich element (ARE) sites of mRNA and inhibits its degradation by ARE-mediated mRNA decay (AMD) pathway; (4) derepression from miRNA-mediated downregulation in response to cell stresses by HuR (an RNA binding protein that interacts with ARE in 3′UTR of the mRNA). As none of the binding sites are located in 5′UTR in our results, and cells are considered active in cancer tissue, the mechanisms of upregulation for cellular state or binding site in 5′UTR may be less possible.
AREs are found in the 3′UTR of mRNAs that code for proto-oncogenes, nuclear transcription factors, and cytokines45. They can be classified into three types: (1) having dispersed AUUUA motifs within or near U-rich regions; (2) having overlapping AUUUA motifs within or near U-rich regions; (3) a much less well-defined class having a U-rich region but no AUUUA repeats46. Our results reveal that A and U are occupied in most of the binding sequence. To our knowledge, it has been verified that TTP has interactions with GSK3B47 and VEGFA48 whereas there are no reports on PTK2. It is possible that miR-106b-5p prevents TTP from binding to the mRNAs in 3′UTR and therefore regulate their expression, but the specific mechanism needs to be further investigated. In addition, it has been confirmed that HuR can bind to VEGF49 and reverse the repression effect by miR-200b50. This also could be a contributing factor to the upregulation of VEGFA and be adopted for the other key genes.
However, several limitations exist in the present study: (1) Significant heterogeneity can be observed in the meta-analysis. This high heterogeneity may result from population, race, stage, and the type of CC (squamous or adenocarcinoma). Hence, more data from large-scale clinical trials are needed to evaluate the source of the heterogeneity. (2) Parts of genes are removed due to the selection criteria. However, it is possible that these genes might also impact the progression of CC and they also need to be analyzed. (3) As this study is an in silico research, a further experiment is needed for validation. (4) Upregulations of both miR-106-5p and key genes are identified. Despite the fact that several possibilities are analyzed, the specific mechanisms still need to be studied and verified further.
Conclusions
In summary, significant upregulation of miR-106b-5p in CC is confirmed by meta-analysis with the data from TCGA and GEO, and the expression of miR-106b-5p is significantly correlated with the number of metastatic lymph nodes. Furthermore, miR-106b-5p promotes the progression of CC by targeting three key genes (GSK3B, VEGFA, PTK2) through three crucial pathways (PI3K-Akt signaling pathway, focal adhesion, and pathways in cancer). miR-106b-5p might upregulate the key genes by preventing TTP from binding to the mRNAs in 3’UTR with/without the effect of derepression of HuR. However, the specific mechanism needs to be further investigated.
Materials and methods
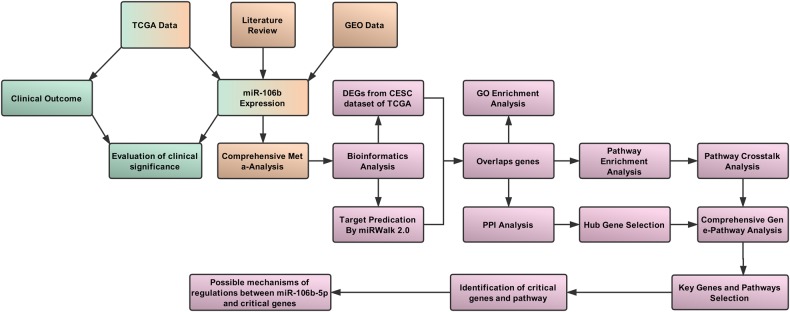
The workflow of this study is presented in Fig. 8 and this study is performed according to guidelines of MIAME51 and Meta-Analysis of Gene Expression Microarray Datasets52. Firstly, the clinical significance of miR-106b-5p in CC is assessed according to the CESC data from TCGA. Secondly, the expression data of miR-106b-5p from TCGA, GEO, and literature are synthesized by meta-analysis. Thirdly, DEGs from TCGA are screened and 12 databases are used to predict target genes of miR-106b-5p. The overlap genes between DEGs from TCGA and predicted target genes are explored by bioinformatic analyses.
Fig. 8. Work flow of the clinical significance evaluation and comprehensive analysis for miR-106b-5p in cervical cancer.
The modules of clinical value evaluation (green), meta-analysis (brown), and bioinformatics analyses (pink) are included
Correlations between the expression of miR-106b-5p and clinical outcomes
To identify the clinical significance of miR-106b-5p in CC, we adopt LinkedOmics53 which is constructed based on TCGA with 309 cancer samples and three normal samples as controls in CESC dataset to explore the correlations between the expression of miR-106b-5p and the clinical outcomes, including the number of metastatic lymph nodes, tumor purity, race, pathology M/N/T stage, years to birth, histological type, ethnicity, radiation therapy, and overall survival.
Meta-analysis of miR-106b-5p expression based on TCGA/GEO and literature
A systematic searching in GEO for the miR-106b-5p expression between CC and healthy tissue was performed on 30 March 2018 with the terms and eligibility criteria that are presented in Tables 5 and 6. For adjusting the data to the normal distribution to reduce variation, Log2 scale transformation was applied to all of the expression data. The meta-analysis was conducted by R Version 3.4.154 and meta package55. We pooled SMD using the Mantel–Haenszel formula (fixed-effect model) or the DerSimonian–Laird formula (random effects model) and assessed the heterogeneity using I2 statistics. Random-effect models (P < 0.05 or I2 > 50%) are used when the heterogeneity is considered significant. Otherwise a fixed-effects model would be used.
Table 6.
Searching terms used in GEO and literature review
| (1) Microarray searching | |
| #1 | microRNA OR miRNA OR micro RNA noncoding RNA OR ncRNA OR small RNA |
| #2 | Cervical OR cervix |
| #3 | Cancer OR carcinoma OR tumor OR neoplasia OR neoplasm OR malignant OR malignancy |
| #4 | #1 AND #2 AND #3 |
| (2) Literature search | |
| #1 | microRNA OR miRNA OR micro RNA noncoding RNA OR ncRNA OR small RNA |
| #2 | 106b OR 106b-5p |
| #3 | Cervical OR cervix |
| #4 | Cancer OR carcinoma OR tumor OR neoplasia OR neoplasm OR malignant OR malignancy |
| #5 | #1 AND #2 AND #3 AND #4 |
Funnel plot with Egger’s test56 was utilized to evaluate the publication bias. P < 0.1 was considered to be significant asymmetry for the funnel plot.
To detect the robustness of the pool results, sensitivity analysis was performed by alternating analysis model. In addition, to further evaluate the impact of individual studies on the overall effect estimates, influence analysis was performed and the combined estimates were recalculated by omitting one study at a time.
Furthermore, a full-scale search of miR-106b-5p expression in eight electronic databases (PubMed, Chinese VIP, CNKI, Wanfang Database, Embase, Web of Science, Science Direct, and Wiley Online Library) accompanied with manual searching by screening the references cited in the acquired articles was conducted on 30 March 2018. The searching terms and eligible criteria are also shown in Tables 5 and 6. The data of the publications will be extracted and analyzed, including the author, year of publication, country, number of cancer and control samples, regulations, and testing methods.
Screening of candidate genes for miR-106b-5p
The CESC data were analyzed by the limma package57 to identify DEGs. We determined the significance of the difference in gene expression as Log2 |fold change (FC) | > 1 and False Discovery Rate (FDR) < 0.05. Furthermore, the miR-106b-5p targeted genes are predicted by 12 databases (Microt4, miRWalk, mir-bridge, miRanda, miRDB, miRMap, Pictar2, PITA, miRNAMap, RNAhybrid, RNA22, and Targetscan) in miRWALK version 2.058. To increase prediction accuracy, the genes that are overlapping in at least five databases were selected. Finally, the overlap genes between DEGs and predicted genes were analyzed by upsetR59 and Venn Plot.
GO and pathway enrichment analysis
GO and pathway enrichment analyses were performed by DAVID60 on the overlapping genes. The significantly enriched biological items for CeC, BP, and molecular functions (MF) were identified as P < 0.01.
Protein-protein interaction network analysis
The STRING database61 was used to construct an interaction network between the overlapped genes. To obtain precise results, those nodes in the network would be removed: (a) interaction score < 0.7; (b) not connected to the major network; (3) the value of degree and betweenness is less than average.
CytoHubba62 was used to explore the important nodes by 12 topological algorithms, including Degree, Edge Percolated Component (EPC), Maximum Neighborhood Component (MNC), Density of Maximum Neighborhood Component (DMNC), Maximal Clique Centrality (MCC), and centralities based on shortest paths, such as Bottleneck (BN), EcCentricity, Closeness, Radiality, Betweenness, ClusteringCoefficient, and Stress. The top 20 genes in each topological algorithm were extracted and the duplication of each gene was also calculated. The genes with less than 2 in repetitiveness are excluded for ensuring the genes are closely linked to CC and the rest are considered as hub genes.
Pathway enrichment and crosstalk analysis
For pathway enrichment, the predicted targets were mapped using the Kyoto Gene and Genome Encyclopedia (KEGG) database by online analysis of DAVID. Significant pathways were considered as P < 0.05.
The obtained pathways were recruited for further crosstalk analysis to explore their interactions. The method is based on the assumption that if two pathways share a proportion of genes, they are considered crosstalk63. As described in the previous study64, in order to better measure the overlap between any two pathways, JC (Jaccard coefficient) = | (A ∩ B) / (A ∪ B) | and OC (overlapping coefficient) = (|A ∩ B|) / (min(| A |, | B |)) were adopted, where A and B are the genes contained in the two pathways. As pathways with too few genes may have insufficient biological information, we excluded pathways containing fewer than three genes. Likewise, pairs of pathways with less than two overlapping genes were also removed. After obtaining the network of pathway crosstalk, the plug-in app Molecular Complex Detection (MCODE) in Cytoscape was used to screen the hub subnetwork which had the score >4.
Comprehensive gene-pathway analysis
The hub genes are mapped into the subnetwork of crosstalk to further explore the mechanism by KEGG. To screen the main genes and pathways, the nodes with degree > average are collected for constituting a subnetwork.
Identification of key genes and pathways
To further identify the key genes, we evaluated the expression of main genes between CC and healthy samples with meta-analysis in Oncomine65. P < 0.05 is considered as a significant difference. Moreover, influence analysis was also conducted to access the pool estimates. The pathways that all of the key genes participate in were determined as crucial pathways.
Possible mechanisms of regulations between miR-106b-5p and key genes
The regulation between miR-106b-5p and the key genes is recognized by their expression. To explore the possible mechanisms of the regulation, we collected the sequence of miR-106b-5p and the three key genes from miRbase66 and NCBI–nucleotide to predict their binding site by RNAhybrid67 with the criteria that mRNA has perfect nucleotide pairing between the second and eighth positions of the 5′ end of miRNA sequences. Furthermore, the character of the binding sequence was also investigated.
Electronic supplementary material
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by: M.V. Niklison Chirou
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/10/2019
Due to a technical error, content intended for publication in Volume 4 (2018) published in Volume 5 (2019). The content has been moved into the correct volume, and the citation information was updated accordingly.
Electronic supplementary material
The online version of this article (10.1038/s41420-018-0096-8) contains supplementary material, which is available to authorized users.
References
- 1.Torre LA, et al. Global Cancer Statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Cancer Statistics in China. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Petry KU. HPV and cervical cancer. Scand. J. Clin. Lab. Invest. 2014;74:59–62. doi: 10.3109/00365513.2014.936683. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, Xu H, Qi M, Yan S, Tian X. miRNA dysregulation and the risk of metastasis and invasion in papillary thyroid cancer: a systematic review and meta-analysis. Oncotarget. 2018;9:5473–5479. doi: 10.18632/oncotarget.16681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos JMO, Gil da Costa RM, Medeiros R. Dysregulation of cellular microRNAs by human oncogenic viruses – Implications for tumorigenesis. Biochim. Biophys. Acta. 2018;1861:95–105. doi: 10.1016/j.bbagrm.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN. Small RNAs: classification, biogenesis, and function. Mol. Cells. 2005;19:1–15. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 8.Lecellier CH. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 9.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 10.Huang C, Hu G. Shikonin suppresses proliferation and induces apoptosis in endometrioid endometrial cancer cells via modulating miR-106b/PTEN/AKT/mTOR signaling pathway. Biosci. Rep. 2018;0:BSR20171546. doi: 10.1042/BSR20171546. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Shi DM, Bian XY, Qin CD, Wu WZ. miR-106b-5p promotes stem cell-like properties of hepatocellular carcinoma cells by targeting PTEN via PI3K/Akt pathway. Onco. Targets Ther. 2018;11:571–585. doi: 10.2147/OTT.S152611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, et al. miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget. 2017;8:21461–21471. doi: 10.18632/oncotarget.15591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang W, et al. miR-106b-5p targets tumor suppressor gene SETD2 to inactive its function in clear cell renal cell carcinoma. Oncotarget. 2015;6:4066–4079. doi: 10.18632/oncotarget.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma D, Zhang YY, Guo YL, Li ZJ, Geng L. Profiling of microRNA-mRNA reveals roles of microRNAs in cervical cancer. Chin. Med. J. (Engl.). 2012;125:4270–4276. [PubMed] [Google Scholar]
- 15.Cheng Y, et al. MicroRNA-106b is involved in transforming growth factor β1-induced cell migration by targeting disabled homolog 2 in cervical carcinoma. J. Exp. Clin. Cancer Res. 2016;35:1–11. doi: 10.1186/s13046-016-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piao J, et al. Substrate stiffness affects epithelial-mesenchymal transition of cervical cancer cells through miR-106b and its target protein DAB2. Int. J. Oncol. 2017;50:2033–2042. doi: 10.3892/ijo.2017.3978. [DOI] [PubMed] [Google Scholar]
- 17.Gao D, Zhang Y, Zhu M, Liu S, Wang X. MiRNA expression profiles of HPV-infected patients with cervical cancer in the Uyghur population in China. PLoS ONE. 2016;11:1–12. doi: 10.1371/journal.pone.0164701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, et al. MicroRNA-27b up-regulated by human papillomavirus 16 E7 promotes proliferation and suppresses apoptosis by targeting polo-like kinase2 in cervical cancer. Oncotarget. 2016;7:19666–19679. doi: 10.18632/oncotarget.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer L, et al. Expression profiling of stem cell-related genes in neoadjuvant-treated gastric cancer: a NOTCH2, GSK3B and β-catenin gene signature predicts survival. PLoS ONE. 2012;7:1–9. doi: 10.1371/journal.pone.0044566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espinosa I, et al. Gene expression analysis identifies two groups of ovarian high-grade serous carcinomas with different prognosis. Mod. Pathol. 2011;24:846–854. doi: 10.1038/modpathol.2011.12. [DOI] [PubMed] [Google Scholar]
- 21.Grassilli E, et al. Inhibition of GSK3B bypass drug resistance of p53-null colon carcinomas by enabling necroptosis in response to chemotherapy. Clin. Cancer Res. 2013;19:3820–3831. doi: 10.1158/1078-0432.CCR-12-3289. [DOI] [PubMed] [Google Scholar]
- 22.Gürsel DB, et al. Tight regulation between cell survival and programmed cell death in GBM stem-like cells by EGFR/GSK3b/PP2A signaling. J. Neurooncol. 2015;121:19–29. doi: 10.1007/s11060-014-1602-3. [DOI] [PubMed] [Google Scholar]
- 23.Ma C, et al. GSK3β mediates the carcinogenic effect of HPV16 in cervical cancer. Sci. Rep. 2015;5:2–10. doi: 10.1038/srep16555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rath G, Jawanjal P, Salhan S, Nalliah M, Dhawan I. Clinical significance of inactivated glycogen synthase kinase 3β in HPV-associated cervical cancer: relationship with Wnt/β-Catenin pathway activation. Am. J. Reprod. Immunol. 2015;73:460–478. doi: 10.1111/aji.12346. [DOI] [PubMed] [Google Scholar]
- 25.V B, G C. The angiogenic switch in carcinogenesis. Semin. Cancer Biol. 2009;19:329–337. doi: 10.1016/j.semcancer.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, Zhang C, Dong P, Guo Y, Mu N. Molecular regulation of cervical cancer growth and invasion by VEGFa. Tumor Biol. 2014;35:11587–11593. doi: 10.1007/s13277-014-2463-2. [DOI] [PubMed] [Google Scholar]
- 27.Dibbens JA, et al. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol. Biol. Cell. 1999;10:907–919. doi: 10.1091/mbc.10.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du K, Gong H, Gong Z. Influence of serum VEGF levels on therapeutic outcome and diagnosis / prognostic value in patients with cervical. Cancer. 2014;15:8793–8796. doi: 10.7314/apjcp.2014.15.20.8793. [DOI] [PubMed] [Google Scholar]
- 29.Kong, D., Chen, F. & Sima, N. Focal adhesion kinases crucially regulate TGFβ-induced migration and invasion of bladder cancer cells via Src kinase and E-cadherin. Onco. Targets. Ther. 10, 1783–1792 (2017). [DOI] [PMC free article] [PubMed]
- 30.Ma K, et al. PTK2-mediated degradation of ATG3 impedes cancer cells susceptible to DNA damage treatment. Autophagy. 2017;13:579–591. doi: 10.1080/15548627.2016.1272742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann. Med. 2014;46:372–383. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 32.Owonikoko TK, Khuri FR. Am. Soc. Clin. Oncol. Educ. Book. 2013. Targeting the PI3K/AKT/mTOR pathway: biomarkers of success and tribulation; pp. 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai C, et al. Anti-proliferative effect of RCE-4 from Reineckia carnea on human cervical cancer HeLa cells by inhibiting the PI3K/Akt/mTOR signaling pathway and NF-κB activation. Naunyn Schmiedebergs Arch. Pharmacol. 2016;389:573–584. doi: 10.1007/s00210-016-1217-7. [DOI] [PubMed] [Google Scholar]
- 34.Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003;2:336–342. doi: 10.4161/cc.2.4.433. [DOI] [PubMed] [Google Scholar]
- 35.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin. Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Gari HH, DeGala GD, Ray R, Lucia MS, Lambert JR. PRL-3 engages the focal adhesion pathway in triple-negative breast cancer cells to alter actin structure and substrate adhesion properties critical for cell migration and invasion. Cancer Lett. 2016;380:505–512. doi: 10.1016/j.canlet.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao Z, et al. MicroRNA-7 inhibits metastasis and invasion through targeting focal adhesion kinase in cervical cancer. Int. J. Clin. Exp. Med. 2015;8:480–487. [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 2016;22:851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta. 2010;1803:1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Orang, A. V., Safaralizadeh, R. & Kazemzadeh-Bavili, M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genomics2014, 1–15 (2014). [DOI] [PMC free article] [PubMed]
- 41.Catalanotto, C., Cogoni, C. & Zardo, G. MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 17, 1712–1729 (2016). [DOI] [PMC free article] [PubMed]
- 42.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 43.Truesdell SS, et al. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep. 2012;2:0–11. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens K. When microRNAs activate translation. Nat. Methods. 2008;5:122–123. doi: 10.1038/nmeth0208-122a. [DOI] [Google Scholar]
- 45.Chen CYA, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/S0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 46.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao H, Lin R. Phosphorylation of recombinant tristetraprolin in vitro. Protein J. 2008;27:163–169. doi: 10.1007/s10930-007-9119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Essafi-Benkhadir K, Pouysségur J, Pagès G. Implication of the ERK pathway on the post-transcriptional regulation of VEGF mRNA stability. Methods Mol. Biol. 2010;661:451–469. doi: 10.1007/978-1-60761-795-2_28. [DOI] [PubMed] [Google Scholar]
- 49.Kurosu T, et al. HuR keeps an angiogenic switch on by stabilising mRNA of VEGF and COX-2 in tumour endothelium. Br. J. Cancer. 2011;104:819–829. doi: 10.1038/bjc.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang SH, et al. Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-a expression and angiogenesis. J. Biol. Chem. 2013;288:4908–4921. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 52.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5:1320–1332. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R. Core Team. R: A Language and Environment for Statistical Computing, Vol. 1 (R Foundation for Statistical Computing, 2017).
- 55.Schwarzer G. meta: An R package for meta-analysis. R. News. 2007;7:40–45. [Google Scholar]
- 56.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dweep H, Gretz N. MiRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 59.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiao X, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szklarczyk D, et al. STRINGv10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chin CH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8:S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jia P, Kao CF, Kuo PH, Zhao Z. A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC Syst. Biol. 2011;5:S12. doi: 10.1186/1752-0509-5-S3-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Y, Pan Z, Hu Y, Zhang L, Wang J. Network and pathway-based analyses of genes associated with Parkinson’s disease. Mol. Neurobiol. 2017;54:4452–4465. doi: 10.1007/s12035-016-9998-8. [DOI] [PubMed] [Google Scholar]
- 65.Rhodes DR, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozomara A, Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rehmsmeier, M., Steffen, P., Höchsmann, M., Giegerich, R. & Ho, M. Fast and effective prediction of microRNA / target duplexes. RNA10, 1507–1517(2004). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.