Abstract
Multiple Sclerosis patients' clinical symptoms do not correlate strongly with structural assessment done with traditional magnetic resonance images. However, its diagnosis and evaluation of the disease's progression are based on a combination of this imaging analysis complemented with clinical examination. Therefore, other biomarkers are necessary to better understand the disease. In this paper, we capitalize on machine learning techniques to classify relapsing-remitting multiple sclerosis patients and healthy volunteers based on machine learning techniques, and to identify relevant brain areas and connectivity measures for characterizing patients. To this end, we acquired magnetic resonance imaging data from relapsing-remitting multiple sclerosis patients and healthy subjects. Fractional anisotropy maps, structural and functional connectivity were extracted from the scans. Each of them were used as separate input features to construct support vector machine classifiers. A fourth input feature was created by combining structural and functional connectivity. Patients were divided in two groups according to their degree of disability and, together with the control group, three group pairs were formed for comparison. Twelve separate classifiers were built from the combination of these four input features and three group pairs. The classifiers were able to distinguish between patients and healthy subjects, reaching accuracy levels as high as 89% ± 2%. In contrast, the performance was noticeably lower when comparing the two groups of patients with different levels of disability, reaching levels below 63% ± 5%. The brain regions that contributed the most to the classification were the right occipital, left frontal orbital, medial frontal cortices and lingual gyrus. The developed classifiers based on MRI data were able to distinguish multiple sclerosis patients and healthy subjects reliably. Moreover, the resulting classification models identified brain regions, and functional and structural connections relevant for better understanding of the disease.
Abbreviations: MS, Multiple Sclerosis; RRMS, Relapsing-Remitting Multiple Sclerosis; MRI, Magntenic Resonance Imaging; rsfMRI, resting state functional Magnetic Resonance Imaging; DTI, Diffusion Tensor Imaging; FA, Fractional Anisotropy; SVM, Support Vector Machine
Keywords: Resting state, fMRI, DTI, SVM, Multiple sclerosis, Classification
Graphical abstract
Highlights
-
•
Classifiers based on functional and diffusion imaging data identified multiple sclerosis patients.
-
•
These classifiers allowed to identify brain regions relevant for the disease.
-
•
Functional imaging data was more relevant than diffusion data for the classification.
1. Introduction
Multiple sclerosis (MS) is a complex disease that causes inflammation, demyelination, axonal degeneration and neuronal loss of the central nervous system (CNS; Budde et al., 2009; Loizou et al., 2013), with axonal damage leading to global and regional atrophy from the onset of the disease (Barkhof and Filippi, 2009). Its diagnosis and progression evaluation are based on a clinical examination complemented with structural assessment using Magnetic Resonance Imaging (MRI; Droby et al., 2016; Filippi and Agosta, 2010; Loizou et al., 2013; Thompson et al., 2017). The location and load of leasions caused by MS are of special importance. These lesions are seen as hyperintense regions in T2 weighted images (Budde et al., 2009; Mollison et al., 2017; Richiardi et al., 2012; Thompson et al., 2017). However, traditional imaging findings do not correlate strongly with patients' clinical symptoms. This discrepancy is known as the clinico-radiological paradox in MS (Filippi and Agosta, 2010; Fu et al., 1998; Mollison et al., 2017; Richiardi et al., 2012). Redundancy of pathways in the CNS or its restoration capacity and functional adaptation may explain this paradox (Barkhof and Filippi, 2009; Rocca et al., 2005). Lesions in T2 weighted images might even include areas with normal function or caused by reasons different from MS (Barkhof and Filippi, 2009). Additionally, conventional processing of structural MR images has been unable to detect some types of damages to brain tissue (Filippi and Agosta, 2010; Fu et al., 1998; Lin et al., 2005; Richiardi et al., 2012).
To better understand the clinico-radiological paradox in MS, other MR sequences have been used to characterize the disease. For instance, diffusion tensor imaging (DTI) and resting state functional MRI (rsfMRI) have been explored to provide new insights about this disease (Filippi et al., 2013; Rocca et al., 2012; Roosendaal et al., 2009; Sbardella et al., 2017). DTI is sensitive to white matter pathology in MS, as radial diffusivity has been shown to increase in response to demyelination and axial diffusivity decreases with axonal damage (Budde et al., 2009). rsfMRI has the ability to show a maladaptive role of cortical functional changes (Filippi and Rocca, 2011) due to CNS injuries during acute relapse and in clinically stable patients (Rocca et al., 2005).
DTI and rsfMRI generate a large amount of data per scan, which can be difficult to analyze and to take full advantage of the available information. Therefore, it would be beneficial to have a method that can automatically process such information in the context of MS.
Machine learning approaches, in particular support vector machine (SVM), allow to classify data by generating a separation model (Vapnik, 1998). This model has relative weights associated to each input feature that represent their importance for distinguishing between groups, thus contributing to the characterization of the data (Griva et al., 2009). This way, instead of analyzing predefined areas, there is an objective evaluation of the whole brain, which may highlight relevant areas that might not have been otherwise considered. Furthermore, SVM is a multivariate approach that, unlike the traditional statistical parametric mapping approaches, combines information from multiple features for the purpose of classification (Griva et al., 2009).
Machine leaning approaches have been developed to differentiate MS from other pathologies (Eshaghi et al., 2016; Muthuraman et al., 2016), as well as to distinguish sub-categories within the disease (Bendfeldt et al., 2012; Kocevar et al., 2016). Some of these approaches have applied DTI (e.g., Stamile et al., 2015) and rsfMRI in patients with MS (e.g., Richiardi et al., 2012; Zhong et al., 2017). One of these studies (e.g., Richiardi et al., 2012) successfully classified MS patients and healthy controls with sensitivities and specificities above 80%. The authors identified the right middle temporal pole as the most important area for distinguishing between the groups. To the best of our knowledge, machine learning approaches have not yet been applied to combined DTI and rsfMRI data in patients with MS, but only in separate analyses.
In this study, we aim to develop classifiers based on DTI and fMRI data that are able to identify areas of the brain that may help to better characterize the disease. We hypothesize that classifiers based on multimodal data extracted from the combination of DTI and fMRI will lead to a reliable classification of relapsing-remitting multiple sclerosis (RRMS) patients from healthy subjects.
2. Materials and methods
2.1. Subjects and image acquisition
The study was conducted in accordance with the regulations of the Scientific Ethics Committee, Faculty of Medicine, Pontificia Universidad Católica de Chile. Patients and control subjects gave written informed consent for using their clinical data for research purposes. In this prospective study, we included 107 RRMS patients and a control group of 50 subjects without any clinical symptoms of any brain disease. Both, patient and control groups, were in the same age distribution, had at least 13 years of education and did not have any MR-incompatible implants in their body.
Patients did not have any major medical or psychiatric comorbidity other than RRMS. They were diagnosed with MS according to McDonald's Criteria 2010 (Polman et al., 2011) early from symptom onset (median diagnostic delay of 3 months, range 0–180 months). They began disease modifying treatment as soon as they were diagnosed. 97% of the patient group was receiving treatment.
All subjects underwent MR scans, and an expanded disability status scale (EDSS) evaluation was made for each patient. The MR scans consisted of T1 weighted, DTI and rsfMRI on a clinical Philips 3 T Ingenia MR Scanner (Best, The Netherlands) between November 2016 and August 2017. During the rsfMRI acquisition, subjects were asked to lie supine with eyes closed, and not think of anything in particular. Table 1 summarizes MRI acquisition parameters.
Table 1.
MRI acquisition parameters.
| MRI acquisition parameters | T1W-3D | DTI | rsfMRI |
|---|---|---|---|
| TR (ms) | 7.8 | 8834 | 2500 |
| TE (ms) | 3.6 | 92 | 35 |
| Matrix (mm) | 240 × 240 | 100 × 102 | 80 × 80 |
| Field of view (mm) | 240 × 240 × 164 | 224 × 224 × 140 | 220 × 220 × 132 |
| Acquisition resolution (mm3) | 1.00/1.00/1.00 | 2.24/2.20/2.00 | 2.75/2.75/3.00 |
| Reconstructed resolution (mm3) | 0.50/0.50/0.50 | 2.00/2.00/2.00 | 2.75/2.75/3.00 |
| Flip angle (°) | 8 | 90 | 82 |
| Inversion time (ms) | 977 | – | – |
| Number of signal averages | 1 | 2 | 1 |
| Bandwidth (Hz) | 191.5 | 26.5 | 35.9 |
| SENSE factor | 2.5 | 2 | 1.8 |
| Slices | 327 | 70 | 40 |
| Acquisition time | 4 min 8 s | 5 min 23 s | 8 min 27 s |
| Dynamic scan volumes | – | – | 200 |
| b value (mm2/s) | – | 1000 | – |
| Number of directions | – | 15 | – |
MRI: Magnetic Resonance Imaging; T1W-3D: T1 weighted 3D image; DTI: Diffusion Tensor Imaging; rsfMRI: resting state functional Magnetic Resonance Imaging; TR: repetition time; TE: echo time; SENSE: sensitivity encoding.
2.2. Image processing
We obtained fractional anisotropy (FA) maps from the DTI scans using DSI Studio (http://dsi-studio.labsolver.org). These maps were manually centered to the anterior commissure to avoid the spatial correction process to get suboptimally trapped in a local minimum. The images were then coregistered with the T1 weighted images and then normalized to the MNI space using SPM12 (http://www.fil.ion.ucl.ac.uk/spm/software/spm12/). All resulting images were visually inspected to verify proper pre-processing. DTI scans were also used to perform a whole brain deterministic tractography (Yeh et al., 2013), from which we built a connectivity matrix indicating the number of bundles connecting regions of interest (ROIs) based on the Harvard-Oxford Atlas (http://www.cma.mgh.harvard.edu/fsl_atlas.html). The same processing pipeline was applied to all subject groups.
Raw images obtained from rsfMRI scans underwent realignment, slice timing correction, coregistration with the T1 weighted images, and normalization to the MNI space using SPM12. The resulting images were processed with the CONN toolbox (www.nitrc.org/projects/conn; Whitfield-Gabrieli and Nieto-Castanon, 2012) to obtain a correlation matrix, also based on the Harvard-Oxford Atlas, thus representing bivariate functional connectivity among the ROIs.
2.3. Subject division
The RRMS patients were divided according to their EDSS, considering as non-disabled those patients with an EDSS of 1.5 or lower (Kurtzke, 1983). We defined the following group of pairs for which we built different classifiers:
-
1.
All RRMS patients vs. healthy subjects
-
2.
RRMS patients with EDSS >1.5 vs. healthy subjects
-
3.
RRMS patients with EDSS ≤1.5 vs. EDSS >1.5
2.4. Classification
We developed four separate classifiers for each group pair, based on the following input types:
-
1.
FA voxel-wise value
-
2.
DTI connectivity matrix
-
3.
rsfMRI correlation matrix
-
4.
Normalized combination of DTI connectivity and rsfMRI correlation matrices
We constructed 12 final SVM linear classifiers or models (Vapnik and Lerner, 1963), one for each combination of input type (previously listed in this Section) and subject group pair (listed in the previous Section). The construction of each SVM model involved the following steps, based on Ebadi et al., 2017:
-
1.We ran a feature selection procedure to reduce input dimensionality. This is usually recommended when the number of features is higher than the number of samples, as is our case. It is based on the assumption that the data contains many redundant or irrelevant features (Ebadi et al., 2017). A Fisher score (FS) (He et al., 2006) was assigned to each feature, computed as follows:
where for each group i ∈ {1, 2}, ni is the number of subjects, μi the mean, σi2 the variance of the specific feature for group, and μT is the overall mean of the feature. Only the features with an FS above a threshold were selected for the classification. Many threshold values were explored by randomly dividing the dataset into training and testing sets and running a one-dimensional grid search over the training set using cross-validation methods. The FS threshold that gave the best results for each classifier was selected. -
2.
We built 100 instances of the SVM linear model because of the imbalance in the number of subjects in each group. In each iteration, a random undersampling over the largest group was made. Each SVM model was built using the Statistics and Machine Learning Toolbox available in Matlab 2017b (The MathWorks Inc., Natick, Massachusets). In order to verify the robustness of the classifiers we used cross-validation. For this task, the data were divided in sets for training and testing the classifier using two cross-validation methods, namely k-folding (with k = 10) and leave-one-out cross-validation (LOOCV). The results were analyzed using confusion matrix indicators to assess the classifier's performance, including accuracy, precision, sensitivity and specificity.
-
3.
These confusion matrix indicators were then calculated to evaluate the classification performance and a final SVM model was constructed by averaging out the 100 instances. The beta-values, or weights, in this final model were used to identify the most relevant brain regions and connections for the classification.
3. Results
3.1. Subjects
Three patients were excluded from the sample because their scans had artifacts. Four subjects from the control group were also excluded from the sample. Three of the subjects were removed because their scans revealed brain pathology, and the fourth was removed because of the presence of artifacts in their scans. The study thus considered two groups: a RRMS patient group of 104 subjects and a control group of 46 healthy subjects (Table 2).
Table 2.
Characteristics of each subject group.
| Group | Sex | Number | Age (mean ± STD) | Age (min - max) | EDSS (mean ± STD) | EDSS (min - max) |
|---|---|---|---|---|---|---|
| RRMS patients with EDSS ≤1.5 | Women | 46 | 35 ± 10 | 17–57 | 0.5 ± 0.6 | 0.0–1.5 |
| Men | 25 | 34 ± 7 | 21–50 | 0.5 ± 0.5 | 0.0–1.0 | |
| Total | 71 | 35 ± 9 | 17–57 | 0.5 ± 0.5 | 0.0–1.5 | |
| RRMS patients with EDSS >1.5 | Women | 22 | 44 ± 10 | 22–63 | 2.6 ± 0.9 | 2.0–5.5 |
| Men | 11 | 33 ± 4 | 27–37 | 2.3 ± 0.6 | 2.0–3.0 | |
| Total | 33 | 40 ± 10 | 22–63 | 2.5 ± 0.8 | 2.0–5.5 | |
| All RRMS patients | Women | 68 | 3.8 ± 11 | 17–63 | 1.2 ± 1.2 | 0.0–5.5 |
| Men | 36 | 34 ± 6 | 21–50 | 1.0 ± 1.0 | 0.0–3.5 | |
| Total | 104 | 37 ± 10 | 17–63 | 1.1 ± 1.1 | 0.0–5.5 | |
| Healthy subjects | Women | 24 | 38 ± 12 | 23–63 | – | – |
| Men | 22 | 38 ± 10 | 24–60 | – | – | |
| Total | 46 | 38 ± 11 | 23–63 | – | – |
STD: standard deviation; EDSS: expanded disability status scale; RRMS: relapsing-remitting multiple sclerosis.
3.2. Training results
Table 3 shows the mean accuracies found for the Fisher Score thresholds with best results, averaging out the results in each of the 100 instances, for each of the 12 SVM models. The results show that the classifiers were indeed able to distinguish between patients and healthy subjects, reaching accuracy levels as high as 89% ± 2%. In contrast, the classifiers' performance was noticeably weaker when comparing patients with different levels of EDSS, reaching levels of 63% ± 5%.
Table 3.
Mean accuracies found in each of the SVM classification of the 12 combinations of input features and group pairs. The results are presented as the mean percentage accuracy ± standard deviation of the 100 iterations with different subjects on the largest group, found using leave-one-out and k-folding cross-validations.
| Input type | FA | DTI connectivity | rsfMRI connectivity | Combined (DTI + rsfMRI) | ||||
|---|---|---|---|---|---|---|---|---|
| Cross-validation method | LOOCV | k-folding | LOOCV | k-folding | LOOCV | k-folding | LOOCV | k-folding |
| All RRMS vs. HS | 78.0 ± 2.7 | 78.3 ± 3.1 | 71.8 ± 4.1 | 72.5 ± 4.3 | 86.1 ± 3.0 | 85.7 ± 3.0 | 87.7 ± 3.2 | 87.8 ± 3.4 |
| EDSS >1.5 vs. HS | 83.6 ± 1.9 | 84.3 ± 2.4 | 78.8 ± 3.0 | 79.5 ± 3.8 | 88.4 ± 2.5 | 87.7 ± 3.3 | 88.9 ± 2.4 | 88.6 ± 3.2 |
| EDSS≤1.5 vs. EDSS>1.5 | 62.1 ± 4.1 | 63.0 ± 4.5 | 50.2 ± 5.3 | 47.8 ± 5.5 | 53.1 ± 11.3 | 50.8 ± 6.8 | 50.7 ± 12.2 | 51.0 ± 8.6 |
FA: Fractional anisotropy; DTI: Diffusion Tensor Imaging; rsfMRI: resting state functional Magnetic Resonance Imaging; LOOCV: leave-one-out cross-validation; RRMS: relapsing-remitting multiple sclerosis; HS: healthy subjects; EDSS: expanded disability status scale.
Taking into account two group pairs, all RRMS patients vs. healthy subjects and RRMS patients with EDSS >1.5 vs. healthy subjects, the lowest accuracies were found using DTI connectivity as input features. These group pairs were better distinguished with FA and even better with rsfMRI correlation input types. The best accuracies were found when combining DTI connectivity and rsfMRI correlation as input features.
The third group pair, comparing RRMS patients with EDSS ≤1.5 vs. patients with EDSS >1.5, obtained their best accuracies when using FA as input feature, reaching only values close to 62%. The results using other input features reached accuracies close to 50%, indicating that there was no clear distinction between the two groups.
There were no significant differences between the mean accuracies found using any cross-validation method. This, together with their low standard deviations, suggests that the models are indeed representative, not biased nor a result of overfitting.
The rest of the analysis focuses on those four SVM models with the best performances, namely, the ones built with rsfMRI correlations and the ones that combined rsfMRI and DTI input features, considering the group pairs of all RRMS patients vs. healthy subjects and, patients with EDSS >1.5 vs. healthy subjects.
Table 4 shows the accuracy, precision, sensitivity and specificity obtained from those four models, considering a Fisher Score threshold of 0.020 for the group pair composed by all RRMS patients vs. healthy subjects and 0.017 for RRMS patients with EDSS >1.5 vs. healthy subjects. Importantly, these results show that there was no important difference between the obtained Type I and Type II errors, further supporting the reliability of the developed classifiers.
Table 4.
Confusion matrix indicators (%) for SVM models with rsfMRI and combined (rsfMRI + DTI connectivity) input features that classify RRMS patients and healthy subjects. The results are presented as the average indicator ± standard deviation percentage for 100 iterations with different subjects on the largest group, with both validation methods (LOOCV and k-folding). Fisher threshold is 0.020 for all RRMS vs. healthy subjects and 0.017 for RRMS with EDSS >1.5 vs. healthy subjects.
| Group pair |
All RRMS vs. healthy subjects |
EDSS >1.5 vs. healthy subjects |
||||||
|---|---|---|---|---|---|---|---|---|
| Input type |
rsfMRI |
Combined (rsfMRI + DTI) |
rsfMRI |
Combined (rsfMRI + DTI) |
||||
| Cross-validation method | LOOCV | k-folding | LOOCV | k-folding | LOOCV | k-folding | LOOCV | k-folding |
| Accuracy | 85.9 ± 3.0 | 85.7 ± 4.0 | 87.7 ± 3.2 | 87.8 ± 3.4 | 88.3 ± 2.6 | 87.8 ± 3.2 | 88.9 ± 2.4 | 88.6 ± 3.2 |
| Precision | 85.3 ± 3.2 | 87.1 ± 3.6 | 87.5 ± 3.6 | 89.7 ± 3.8 | 88.4 ± 3.5 | 90.0 ± 3.7 | 89.6 ± 3.3 | 91.6 ± 3.4 |
| Sensitivity | 86.8 ± 3.9 | 86.9 ± 5.3 | 88.1 ± 3.9 | 88.0 ± 4.5 | 88.3 ± 2.6 | 87.5 ± 3.9 | 88.0 ± 2.7 | 87.5 ± 4.0 |
| Specificity | 85.0 ± 3.5 | 84.5 ± 4.5 | 87.4 ± 4.0 | 87.6 ± 4.6 | 88.3 ± 3.9 | 88.1 ± 4.4 | 89.7 ± 3.6 | 89.8 ± 4.2 |
DTI: Diffusion Tensor Imaging; rsfMRI: resting state functional Magnetic Resonance Imaging; LOOCV: leave-one-out cross-validation; RRMS: relapsing-remitting multiple sclerosis; HS: healthy subjects; EDSS: expanded disability status scale.
The feature selection for the combined data for the group pair composed by all the RRMS patients vs. healthy subjects chose 5403 rsfMRI correlations and 594 DTI connectivity values out of 13,530 features in each case. Similarly, for the group pair composed by the RRMS patients with EDSS >1.5 vs. healthy subjects, the feature selection chose 5342 rsfMRI correlations and 724 DTI connectivity values. Hence, in both cases, the classifier is majorly constructed by parameters associated to rsfMRI.
3.3. Weight values of SVM models
Fig. 1 displays the 30 most important features for SVM models obtained with combined input type comparing patients with healthy subjects. It shows a comparison of their absolute weight values normalized to the highest weight for visualization. It also indicates whether the connectivity between the pair of regions increases or decreases in patients in relation to healthy subjects. In both cases, the features are mostly rsfMRI correlation measures, in accordance with the number of selected variables. Taking into consideration the proportion of areas with increased and decreased connectivity in all patients and in patients with EDSS >1.5, it is apparent that when considering all patients there are more brain regions with increased connectivity. The most important features obtained when using only rsfMRI as input were the same as the ones assigned by rsfMRI with the combined rsfMRI and DTI, but with small variations in their order of importance.
Fig. 1.
30 most important connections indicated by resulting SVM models. A) All RRMS vs. healthy subjects. B) RRMS patients with EDSS >1.5 vs. healthy subjects. The bar graph represents the weight of each of the connection pairs in the final SVM models, normalized to the highest weight for visualization. Dark green bars represent rsfMRI correlation features and light green bars represent DTI connectivity features. At the end of each bar there is an arrow indicating whether the connectivity between the areas were higher or lower in patients in relation to healthy subjects.
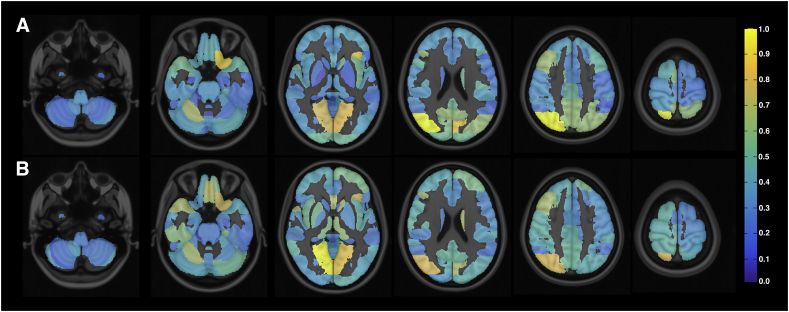
Fig. 2 presents the normalized sum of weights of different brain areas for the two analyzed cases considering the DTI connectivity and rsfMRI correlation features, thus representing the importance of each area for the classification. The most important areas when comparing all the RRMS patients with healthy subjects were (in descending order) the right superior division of the occipital cortex, left frontal orbital cortex, left cuneal cortex, and bilateral lingual gyrus. The most important areas when comparing only the RRMS patients with EDSS >1.5 with healthy subjects were practically the same, but in a different order: the bilateral lingual gyrus, right superior division of the occipital cortex, left frontal orbital cortex, and medial frontal cortex.
Fig. 2.
Discriminating brain regions according to absolute values of feature weights of resulting SVM models. A) Relative weights of SVM classifier between all RRMS patients vs. healthy subjects. B) Relative weights of SVM classifier between RRMS patients with EDSS >1.5 vs. healthy subjects. The results were computed by adding all the weights associated with each area and then normalizing the values. The most discriminating areas are associated with higher weight values.
4. Discussion
Advances in Radiological Sciences allow an accurate diagnosis of multiple sclerosis. The McDonald Criteria is widely used and has been proven to reach high levels of sensitivity (Thompson et al., 2017). Highly accurate biomarkers have already been reported in literature. For instance, accuracies as high as 96% have been achieved using microRNA expression profiles (Keller et al., 2009). Lesser accuracies (92%) have also been reported in literature using MRI, specifically with T1 and T2 weighted images (Eshaghi et al., 2016). However, considering the existing clinico-radiological paradox, different approaches for studying MS are needed to fully understand the diease, and to accomplish more effective diagnosis and treatment.
We have shown that rsfMRI and DTI based machine learning techniques are able to distinguish between RRMS patients and healthy subjects reliably and with high accuracy. This classifying technique also showed to be representative, avoiding bias and overfitting, as represented by the similar results obtained using different cross-validation methods. Training accuracies using rsfMRI in this study were slightly better than those found in a previous study that used another machine learning approach applied to graphs to classify patients and healthy subjects (Richiardi et al., 2012). The difference in connectivity, with increased and decreased rsfMRI activity is also in accordance with previous reports (Filippi and Rocca, 2011).
Taking only into account the models that distinguished between RRMS patients and healthy subjects, different results were obtained according to the input features assigned to develop the models. DTI connectivity input data by itself reached accuracies between 70% and 80% when classifying patients from healthy subjects. These results might be explained by the fiber tracts' diversion due to the presence of MS-related lesions. On the other hand, rsfMRI input data lead to better classifiers, reaching accuracies between 85% and 90%. The difference in these results could be due to the adaptive mechanism of the functional system when facing disease related damage. The patients included in this study had low levels of disability, which could be associated to low levels of structural damage, thus explaining the poor results found using only structural connectivity. However, functional plasticity mechanisms could be inefficiently increased (Rocca et al., 2017), resulting in great difference between patients and healthy subjects.
The combined input, involving DTI connectivity values and rsfMRI correlations, gave the highest accuracy, which was slightly higher than those obtained using rsfMRI as input features. This suggests that DTI and rsfMRI data can be complemented and used to analyze the brain as a whole, taking into account both, structural and functional information. Although this has been done before in MS (Sbardella et al., 2017; Rocca et al., 2010), those approaches are based on a specific network, and were not analyzed from a machine learning perspective using whole brain data.
Our technique did not distinguish with high accuracy between RRMS patients with EDSS above and below 1.5. This could be due to the little difference between disability levels between both groups, especially considering that the patients with EDSS above 1.5 have a mean EDSS of 2.5, which is very low. However, since we have greater accuracy to distinguish healthy volunteers from patients, this issue revealed that the imaging modalities as DTI and rsfMRI in our classifiers were not able to pick those patients with low disability. In other words, using these imaging techniques for the proposed classifiers, we were not able to identify the low levels of disability associated to the disease. Another imaging modality, such as Quantitative Susceptibility Mapping, which is highly sensitive to myelin changes (Wisnieff et al., 2015), or other clinical indicator, needs to be further investigated as input feature for the classifier for this case.
The resulting models were able to identify areas that may be affected by the disease. The highlighted areas give an objective perspective, thus providing another view of the problem which needs further research with clinical data. The brain regions found to affect MS patients in comparison to healthy controls in this study have also been associated with MS in previous studies. For instance, the right superior occipital cortex, left cuneus, lingual gyrus and, middle frontal cortex have been reported to have significant atrophies in MS patients (Gobbi et al., 2014). The left orbitofrontal cortex has been reported to have an increase, while the lingual gyrus a decrease in their functional activity in MS patients (Sweet et al., 2004). Increases in functional connectivity have also been found between the left lingual gyrus, middle occipital gyrus, and cuneus in RRMS in MS patients when compared to healthy subjects (Faivre et al., 2012).
This study has some limitations. We only considered the data from DTI and rsfMRI for the combined classifier, even though SVM models with FA voxel-wise values as inputs also performed reliably. The amount and properties of FA data is considerably different from the DTI and rsfMRI connectivity matrices, therefore it is harder to include in the group of combined inputs. Even though including all features might be useful, this might also incorporate redundancy and noise related compontets, which may require more training samples for optimal classification. This issue would be very interesting to explore in a future study. Furthermore, the patient cohort included patients with very low levels of disability, which limited our threshold options for classification. It would perhaps be more effective for classification and also informative of brain regins affecting the disease to classify patients with EDSS above and below 3, which is the threshold for moderate disability.
5. Conclusion
In conclusion, in the current study we developed reliable linear classifiers that reached accuracies as high as 89% ± 2%, and indicated functional and structural connections and specific brain areas that are relevant for characterizing RRMS patients. We have presented an evidence-based perspective on MS imaging analysis which will contribute towards a better understanding of this complex disease.
Acknowledgments
This work was supported by Comisión Nacional de Investigación Científica y Tecnológica de Chile (Conicyt) through Fondecyt (project no 1171320, 1171313 and 1181057), CONICYT-PIA Anillo ACT172121, ACT 1416 and from Millennium Science Iniciative of the Ministry of Economy, Development and Tourism, grant Nucleos for Cardiovascular Magnetic Resonance.
References
- Barkhof F., Filippi M. Multiple sclerosis: MRI—the perfect surrogate marker for multiple sclerosis? Nat. Rev. Neurol. 2009;5(4):182–183. doi: 10.1038/nrneurol.2009.31. [DOI] [PubMed] [Google Scholar]
- Bendfeldt K., Klöppel S., Nichols T.E., Smieskova R., Kuster P., Traud S.…Borgwardt S.J. Multivariate pattern classification of gray matter pathology in multiple sclerosis. NeuroImage. 2012;60(1):400–408. doi: 10.1016/j.neuroimage.2011.12.070. [DOI] [PubMed] [Google Scholar]
- Budde M.D., Xie M., Cross A.H., Song S.K. Axial diffusivity is the primary correlate of axonal injury in the experimental autoimmune encephalomyelitis spinal cord: a quantitative pixelwise analysis. J. Neurosci. 2009;29(9):2805–2813. doi: 10.1523/JNEUROSCI.4605-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droby A., Yuen K.S., Muthuraman M., Reitz S.C., Fleischer V., Klein J.…Groppa S. Changes in brain functional connectivity patterns are driven by an individual lesion in MS: a resting-state fMRI study. Brain Imaging Beha. 2016;10(4):1117–1126. doi: 10.1007/s11682-015-9476-3. [DOI] [PubMed] [Google Scholar]
- Ebadi A., Dalboni Da Rocha J.L., Nagaraju D.B., Tovar-Moll F., Bramati I., Coutinho G.…Rashidi P. Ensemble classification of Alzheimer's disease and mild cognitive impairment based on complex graph measures from diffusion tensor images. Front. Neurosci. 2017;11:56. doi: 10.3389/fnins.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A., Wottschel V., Cortese R., Calabrese M., Sahraian M.A., Thompson A.J.…Ciccarelli O. Gray matter MRI differentiates neuromyelitis optica from multiple sclerosis using random forest. Neurology. 2016;87(23):2463–2470. doi: 10.1212/WNL.0000000000003395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre A., Rico A., Zaaraoui W., Crespy L., Reuter F., Wybrecht D.…Pelletier J. Assessing brain connectivity at rest is clinically relevant in early multiple sclerosis. Mult. Scler. J. 2012;18(9):1251–1258. doi: 10.1177/1352458511435930. [DOI] [PubMed] [Google Scholar]
- Filippi M., Agosta F. Imaging biomarkers in multiple sclerosis. J. Magn. Reson. Imaging. 2010;31(4):770–788. doi: 10.1002/jmri.22102. [DOI] [PubMed] [Google Scholar]
- Filippi M., Rocca M.A. MR imaging of multiple sclerosis. Radiology. 2011;259(3):659–681. doi: 10.1148/radiol.11101362. [DOI] [PubMed] [Google Scholar]
- Filippi M., Agosta F., Spinelli E.G., Rocca M.A. Imaging resting state brain function in multiple sclerosis. J. Neurol. 2013;260(7):1709–1713. doi: 10.1007/s00415-012-6695-z. [DOI] [PubMed] [Google Scholar]
- Fu L., Matthews P.M., De Stefano N., Worsley K.J., Narayanan S., Francis G.S.…Arnold D.L. Imaging axonal damage of normal-appearing white matter in multiple sclerosis. Brain. 1998;121(1):103–113. doi: 10.1093/brain/121.1.103. [DOI] [PubMed] [Google Scholar]
- Gobbi C., Rocca M.A., Riccitelli G., Pagani E., Messina R., Preziosa P.…Filippi M. Influence of the topography of brain damage on depression and fatigue in patients with multiple sclerosis. Mult. Scler. J. 2014;20(2):192–201. doi: 10.1177/1352458513493684. [DOI] [PubMed] [Google Scholar]
- Griva I., Nash S., Sofer A. 2nd ed. Society for Industrial and Applied Mathematics; 2009. Linear and Nonlinear Optimization. [Google Scholar]
- He X., Cai D., Niyogi P. Advances in Neural Information Processing Systems. 2006. Laplacian score for feature selection; pp. 507–514. [Google Scholar]
- Keller A., Leidinger P., Lange J., Borries A., Schroers H., Scheffler M.…Meese E. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocevar G., Stamile C., Hannoun S., Cotton F., Vukusic S., Durand-Dubief F., Sappey-Marinier D. Graph theory-based brain connectivity for automatic classification of multiple sclerosis clinical courses. Front. Neurosci. 2016;10:478. doi: 10.3389/fnins.2016.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzke J.F. Rating neurologic impairment in multiple sclerosis an expanded disability status scale (EDSS) Neurology. 1983;33(11):1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Lin X., Tench C.R., Morgan P.S., Niepel G., Constantinescu C.S. Importance sampling in MS: use of diffusion tensor tractography to quantify pathology related to specific impairment. J. Neurol. Sci. 2005;237(1):13–19. doi: 10.1016/j.jns.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Loizou C.P., Kyriacou E.C., Seimenis I., Pantziaris M., Petroudi S., Karaolis M., Pattichis C.S. Brain white matter lesion classification in multiple sclerosis subjects for the prognosis of future disability. Intell. Decis. Technol. 2013;7(1):3–10. [Google Scholar]
- Mollison D., Sellar R., Bastin M., Mollison D., Chandran S., Wardlaw J., Connick P. The clinico-radiological paradox of cognitive function and MRI burden of white matter lesions in people with multiple sclerosis: a systematic review and meta-analysis. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuraman M., Fleischer V., Kolber P., Luessi F., Zipp F., Groppa S. Structural brain network characteristics can differentiate CIS from early RRMS. Front. Neurosci. 2016;10:14. doi: 10.3389/fnins.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman C.H., Reingold S.C., Banwell B., Clanet M., Cohen J.A., Filippi M.…Lublin F.D. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J., Gschwind M., Simioni S., Annoni J.M., Greco B., Hagmann P.…Van De Ville D. Classifying minimally disabled multiple sclerosis patients from resting state functional connectivity. NeuroImage. 2012;62(3):2021–2033. doi: 10.1016/j.neuroimage.2012.05.078. [DOI] [PubMed] [Google Scholar]
- Rocca M.A., Colombo B., Falini A., Ghezzi A., Martinelli V., Scotti G.…Filippi M. Cortical adaptation in patients with MS: a cross-sectional functional MRI study of disease phenotypes. Lancet Neurol. 2005;4(10):618–626. doi: 10.1016/S1474-4422(05)70171-X. [DOI] [PubMed] [Google Scholar]
- Rocca M.A., Absinta M., Moiola L., Ghezzi A., Colombo B., Martinelli V.…Filippi M. Functional and structural connectivity of the motor network in pediatric and adult-onset relapsing-remitting multiple sclerosis. Radiology. 2010;254(2):541–550. doi: 10.1148/radiol.09090463. [DOI] [PubMed] [Google Scholar]
- Rocca M.A., Bonnet M.C., Meani A., Valsasina P., Colombo B., Comi G., Filippi M. Differential cerebellar functional interactions during an interference task across multiple sclerosis phenotypes. Radiology. 2012;265(3):864–873. doi: 10.1148/radiol.12120216. [DOI] [PubMed] [Google Scholar]
- Rocca M.A., Valsasina P., Leavitt V.M., Rodegher M., Radaelli M., Riccitelli G.C.…Filippi M. Functional network connectivity abnormalities in multiple sclerosis: Correlations with disability and cognitive impairment. Mult. Scler. J. 2017 doi: 10.1177/1352458517699875. [DOI] [PubMed] [Google Scholar]
- Roosendaal S.D., Geurts J.J.G., Vrenken H., Hulst H.E., Cover K.S., Castelijns J.A.…Barkhof F. Regional DTI differences in multiple sclerosis patients. NeuroImage. 2009;44(4):1397–1403. doi: 10.1016/j.neuroimage.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Sbardella E., Upadhyay N., Tona F., Prosperini L., De Giglio L., Petsas N.…Pantano P. Dentate nucleus connectivity in adult patients with multiple sclerosis: functional changes at rest and correlation with clinical features. Mult. Scler. J. 2017;23(4):546–555. doi: 10.1177/1352458516657438. [DOI] [PubMed] [Google Scholar]
- Stamile C., Kocevar G., Hannoun S., Durand-Dubief F., Sappey-Marinier D. Medical Learning Meets Medical Imaging. Springer International Publishing; 2015, July. A graph based classification method for multiple sclerosis clinical forms using support vector machine; pp. 57–64. [Google Scholar]
- Sweet L.H., Rao S.M., Primeau M., Mayer A.R., Cohen R.A. Functional magnetic resonance imaging of working memory among multiple sclerosis patients. J. Neuroimaging. 2004;14(2):150–157. [PubMed] [Google Scholar]
- Thompson A.J., Banwell B.L., Barkhof F., Carroll W.M., Coetzee T., Comi G.…Fujihara K. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2017 doi: 10.1016/S1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- Vapnik V.N. Wiley; New York: 1998. Statistical Learning Theory. [Google Scholar]
- Vapnik V., Lerner A. Pattern recognition using generalized portrait method. Autom. Remote Control. 1963;24:774–780. [Google Scholar]
- Whitfield-Gabrieli S., Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012 doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- Wisnieff C., Ramanan S., Olesik J., Gauthier S., Wang Y., Pitt D. Quantitative susceptibility mapping (QSM) of white matter multiple sclerosis lesions: interpreting positive susceptibility and the presence of iron. Magn. Reson. Med. 2015;74(2):564–570. doi: 10.1002/mrm.25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh F.C., Verstynen T.D., Wang Y., Fernández-Miranda J.C., Tseng W.Y.I. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0080713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Chen D.Q., Nantes J.C., Holmes S.A., Hodaie M., Koski L. Combined structural and functional patterns discriminating upper limb motor disability in multiple sclerosis using multivariate approaches. Brain Imaging Behav. 2017;11(3):754–768. doi: 10.1007/s11682-016-9551-4. [DOI] [PubMed] [Google Scholar]