Abstract
Context
Gastroesophageal reflux (GER) has not previously been widely regarded as a hereditary disease. A few reports have suggested, however, that a genetic component may contribute to the incidence of GER, especially in its severe or chronic forms.
Objective
To identify a genetic locus that cosegregates with a severe pediatric GER phenotype in families with multiple affected members.
Design
A genome-wide scan of families affected by severe pediatric GER using polymorphic microsatellite markers spaced at an average of 8 centimorgans (cM), followed by haplotyping and by pairwise and multipoint linkage analyses.
Setting
General US community, with research performed in a university tertiary care hospital.
Subjects
Affected and unaffected family members from 5 families having multiple individuals affected by severe pediatric GER, identified through a patient support group.
Main Outcome Measures
Determination of inheritance patterns and linkage of a genetic locus with the severe pediatric GER phenotype by logarithm-of-odds (lod) score analysis, considering a lod score of 3 or greater as evidence of linkage.
Results
In these families, severe pediatric GER followed an autosomal dominant hereditary pattern with high penetrance. A gene for severe pediatric GER was mapped to a 13-cM region on chromosome 13q between microsatellite markers D13S171 and D13S263. A maximum multifamily 2-point lod score of 5.58 and a maximum multifamily multipoint lod score of 7.15 were obtained for marker D13S1253 at map position 35 cM when presumptively affected persons were modeled as unknown (a maximum multipoint score of 4.88 was obtained when presumptively affected persons were modeled as unaffected).
Conclusion
These data suggest that a gene for severe pediatric GER maps to chromosome 13q14.
GASTROESOPHAGEAL REFLUX (GER) afflicts persons of all age groups and is characterized by the retrograde movement of stomach contents into the esophagus.1 It is highly prevalent, and population studies indicate that as many as 20% of adults report frequent heartburn.2 In its most severe form, GER results in extensive tissue damage caused by acid reflux. This chronic mucosal insult leads to esophagitis, which can in turn lead to metaplastic and neoplastic changes.3 Severe pediatric GER can even result in death through aspiration of regurgitated stomach contents in infants and small children4 and has been implicated in sudden infant death syndrome.5 In adolescents and adults, and even infrequently in children, chronic GER is associated with the risk of developing Barrett metaplasia, a premalignant lesion of the esophageal mucosa. In turn, Barrett metaplasia is correlated with the development of adenocarcinoma of the esophagus, which is now the fifth most prevalent neoplasia in the Western world.3
The spectrum of GER includes clinical phenotypes from mild to severe.6 When GER is chronic and results in tissue damage, it is often referred to as gastroesophageal reflux disease.7 Gastroesophageal reflux is most common among infants and young children and middle-aged and older adults.3,8 Severe pediatric GER is the most common esophageal disorder of children9 and is probably the most common inherited disease of the alimentary canal.10
Nonsyndromic severe pediatric GER usually presents early in infancy, and estimates suggest that severe GER affects up to 2% or more of all infants.11 Many affected children outgrow most or all of the symptoms between ages 1 and 5 years. However, some severely affected persons remain affected into adulthood.10 Severe pediatric GER can result in a failure to thrive due to malnutrition caused by reflux of food, which can be exacerbated by avoidance of food.10 The mucosal insult resulting from acid reflux has been linked to an increase in risk and severity of asthma and other ailments of the upper aerodigestive system in children.10–14 Chief among severe pediatric GER’s sequelae is an increased incidence of acute and chronic infections in the upper respiratory tract,4,15–17 which then predispose children to conditions such as voice disorders, apnea, recurrent croup, stridor, and subglottic stenosis.4,18,19 In a controlled trial, treatment of GER resulted in the amelioration of extraesophageal symptoms12; these data and other findings strongly implicate GER in the pathogenesis of many pediatric respiratory diseases.13,14
Epidemiological studies have suggested that both pediatric and adult onset GER have major genetic components, and Barrett metaplasia has been considered to be an autosomal dominant disorder with incomplete penetrance.2,20–23 Examination of several families with multiple affected members, as part of the current study, suggested an autosomal dominant mode of inheritance of GER with nearly complete penetrance if diagnoses were made during childhood. To date, no responsible genes have been mapped or identified for nonsyndromic GER in either children or adults. Although there is a recent report24 of a rare complex syndrome mapping to chromosome 10 that includes GER as 1 of its symptoms, this disorder is distinct from nonsyndromic GER. Because of nonsyndromic GER’s relatively high prevalence in the population, gene mapping studies may be complicated by affected alleles entering a family from more than 1 source.
In this study, we approached the problem of severe pediatric GER from a genetic point of view. The study subjects were ascertained from multiplex families that were identified through a collaboration with a patient support group, PAGER (Pediatric/Adolescent Gastroesophageal Reflux Association, http://www.reflux.org/).
METHODS
Enrollment of Patients and Specimen Collection
Institutional review board approval was obtained from Allegheny General Hospital, Pittsburgh, Pa, to conduct a family-based gene mapping project for nonsyndromic severe pediatric GER. All initial family contacts were made via a recruitment letter and questionnaire developed by Center for Genomic Sciences staff in cooperation with PAGER This questionnaire was distributed by PAGER via a mailing to more than 1000 member families; those interested in participating in a genetic study were asked to indicate the number of affected persons in their families. About 80 of the initial surveys were returned. Of these, 20 identified families of sufficient size to suggest a mode of inheritance; most of the rest of the returned questionnaires identified families with concordant sib pairs. Personnel from the Center for Genomic Sciences then contacted those families indicating a willingness to participate in this study and having multiple affected individuals. Several family picnics were sponsored in the Midwest and California to obtain informed consent, accurate medical histories, and blood or buccal swab specimens for DNA extraction from potentially informative individuals. Informed consent (or assent, in the case of minors above the age of understanding) was obtained for all study subjects prior to specimen acquisition. Through this process, we obtained over 120 specimens, most of which belonged to persons in the 5 families presented herein. None of the families that were genotypically analyzed were omitted from the analysis presented herein, nor were any potentially informative family members who had agreed to participate omitted. We did, however, omit from the genotypic and linkage analyses the spouses and unaffected children of unaffected persons as these individuals are not informative. The pedigrees presented in this study indicate all participating family members and deceased individuals for whom we could infer marker data and haplotypes, but do not include family members who declined participation or who were not available for contact. Written permission to publish pedigree information was obtained from all living persons represented in the pedigrees herein; parents signed for minors, and spouses signed individually. Permission was not required for deceased persons.
Extensive history taking was performed for all study subjects and included questions relating to GER signs and symptoms and associated pulmonary and otolaryngological sequelae. Evidence of regurgitation was assessed by questions on recurrent vomiting without fever, recurrent spitting up, recurrent nasal exudates not associated with respiratory illness, failure to thrive due to caloric restriction, recurrent or persistent hiccups, persistent or recurrent halitosis, and frequent rumination. The GER-associated sequelae were assessed by questions on abdominal and thoracic pain, choking spells, food avoidance, colic, extreme irritability or inconsolable crying, sleep difficulties, breathing problems including apnea and apparent life-threatening event episodes, stridor, recurrent cough and pharyngitis, asthma, and recurrent otitis media. Responses concerning the frequency, magnitude, and duration of symptoms were elicited for all signs suggestive of GER. Occasional symptoms associated with GER or persistent GER symptoms that resolved prior to age 1 year were not considered evidence of severe pediatric GER.
A definitive diagnosis of severe pediatric GER was accepted if made by a gastroenterologist or otolaryngologist through history and physical examination, and/or radiography or pH probe; or by endoscopic examination and biopsy during infancy or childhood.17,25 Definitive severe pediatric GER diagnoses, based on histories, were made for adults who had overwhelming GER signs and symptoms as this study. Presumptive severe pediatric GER diagnoses were established for postadolescent family members whose childhood medical histories and anecdotal family information were suggestive of severe GER. Individuals with a presumptive diagnosis were modeled both as unknowns and unaffecteds when linkage analyses were performed; both sets of data are presented. Children and adults with no symptoms consistent with severe pediatric GER elicited after extensive history taking were modeled as unaffecteds.
All blood was collected via peripheral venipuncture into acid citrate dextrose vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ). Buccal swab specimens were collected by scraping the inside of the cheek 10 times with a sterile cytobrush (Allegiance, Menlo Park, Ill); 10 to 12 brushes were collected per study subject over a period of 2 days. DNA was extracted from blood and buccal swab specimens with Puregene DNA extraction kits (Gentra Systems, Minneapolis, Minn) according to the manufacturer’s recommendations. All DNA was quantitated by UV absorption spectrophotometry at 260 and 280 nm, and adjusted to a working concentration of 50 ng/μL for use in the genotyping reactions.
Genotyping of the GER Families
A microsatellite marker–based genome-wide scan for a GER gene was performed for 34 members of families 2 through 4, essentially as we have described for other heritable traits26,27 with the following modifications. A linkage mapping set (ABI Prism, version 2; PE Biosystems, Foster City, Calif) of about 400 microsatellite markers was used to provide coverage of the genome at an average resolution of 8 centimorgans (cM) and a multiplex amplification and gel analysis format was used to reduce the time and labor involved. The multiplex polymerase chain reaction (PCR) protocol was developed such that all of the markers from an ABI Prism linkage mapping set panel (12–20 markers per panel) could be amplified in 3 reactions, grouped by fluorescent dye color. Thus, all of the primers with a blue tag were included in 1 reaction and so on for the yellow- and green-labeled primers. This system required more DNA and enzyme input per reaction, but the overall use of reagents was less and the time savings were substantial. Amplification conditions for all 3 multiplex reactions were as follows: 50 mmol/L of potassium chloride, 10 mmol/L of Tris hydrochloride (pH 8.3), 2.5 mmol/L of magnesium chloride, 400 μmol/L of each dNTP (dATP, dCTP, dGTP, and dUTP; Pharmacia, Piscataway, NJ), 50 nmol/L of each forward and reverse primer, 0.12 units of AmpliTaq Gold (PE Biosystems), and 50 ng of study subject DNA. The following thermal cycling conditions were used for all markers and were programmed into a suite of 6 PE Biosystems 9600 thermal cyclers to ensure uniformity: 1 cycle at 95°C for 12 minutes to activate the AmpliTaq Gold; 10 cycles at 94°C for 15 seconds, 55°C for 15 seconds, and 72°C for 30 seconds; 20 cycles at 89°C for 15 seconds, 55°C for 15 seconds, and 72°C for 30 seconds; 1 cycle at 72°C for 10 minutes; and 1 cycle at 4°C (holding). After amplification, the 3 dye-based multiplex reactions were combined according to the following formula (3 μL of FAM, 3 μL of HEX, and 6 μL of NED); denatured at 99°C for 2 minutes in a thermal cycler with formamide (0.5 μL of pooled amplified DNA, 1 μL of deionized formamide, 0.2 μL of ROX-labeled size standards from 70–400 base pairs [MapMarkers; Bio-Ventures, Murfreesboro, Tenn], and 0.3 μL of tracking dye solution); quenched on ice; and loaded on a 0.2-mm thick 5% Long Ranger gel (FMC Bioproducts, Rockland, Me). This permitted an entire panel of markers to be run on a single gel, thus permitting completion of the genome scan for 34 individuals with just 28 gels in less than 3 weeks. All gels were run on a pair of ABI 377 Prism automated fragment analyzers using the Genescan software package version 2.1 (PE Biosystems) for data collection.
Fine structure mapping of candidate regions identified by analyzing the original genome-scan data was accomplished by saturating the region with all additional informative microsatellite markers from the Genethon and National Center for Biotechnology Information high-resolution maps (www.genlink.wustl.edu/chrmaps and http://www.ncbi.nlm.nih.gov) that fell within 10 cM of the marker giving the highest logarithm-of-odds (lod) score on the genome scan. (The lod score statistically measures the probability of linkage between a genetic marker of known chromosomal location and a phenotypic trait. A lod score of 3 indicates odds of 1000:1 that true linkage, rather than an artifact of random chance, has been found.) All primer pairs for these confirmational analyses were custom synthesized with FAM, HEX, or TET fluorescent tags on the forward primer (Research Genetics, Huntsville, Ala). The PCR conditions were established for each of these primers by first adjusting magnesium chloride concentration and then annealing temperature, if necessary. All amplifications were performed individually and then combined for gel analyses, with care taken not to have overlapping fragment sizes with the same dyes on a single gel.
Allele Calling, Haplotyping, and Linkage Analyses
We wrote a suite of computer programs in Visual Basic, version 6.0 (Microsoft, Redmond, Wash) for Macintosh, to assist in allele calling, binning of alleles, inheritance checking, data transformations necessary to compile the genotypic data in a form suitable for the Linkage program (see below), and lod score data extraction from the Linkage output files. These programs will be more fully described in a future article (L.W.K., F.Z.H., R.A.P., G.D.E., unpublished data, 1999). Our automated allele calling program was more than 95% accurate and compared favorably with commercial software packages; however, all calls were checked manually for accuracy and all allele calls within the region of linkage were rechecked following the initial data analyses. Phasing and haplotype construction were performed by hand for all pedigree members (when possible) for all chromosomes, using a maximum parsimony approach. These data were then visually inspected to identify candidate regions for further analyses. Two-point (pairwise) linkage analyses were carried out on all autosomal markers using the Mlink routine of the Linkage 5.1 software package.28,29 Multipoint linkage analysis was carried out for the region on chromosome 13 between D13S175 and D13S156 using Linkmap of the Fastlink30 package. For the multipoint linkage analysis, map distances between markers were obtained from the Marshfield map (http://www.marshmed.org/genetics).31 Severe pediatric GER was modeled as an autosomal dominant disorder with incomplete penetrance, with an allele frequency of 0.02 in the population. Since the true allele frequencies are unknown, equal frequencies were used in the analysis. The phenocopy rate was set at 0.001. The penetrance function was set at 70% because of the difficulty in establishing a diagnosis of severe pediatric GER in postadolescents; individuals with a presumptive diagnosis were modeled both as unknowns and unaffecteds, thereby producing 2 parallel sets of lod scores. Regions exhibiting lod scores greater than 1.5 in analysis of the genome scan were chosen for fine structure analysis using additional microsatellite markers, additional family members (families 3 and 4), and additional families (1 and 5). A lod score of 3.0 or more was required for evidence of linkage.
RESULTS
Delineation of GER Pedigrees
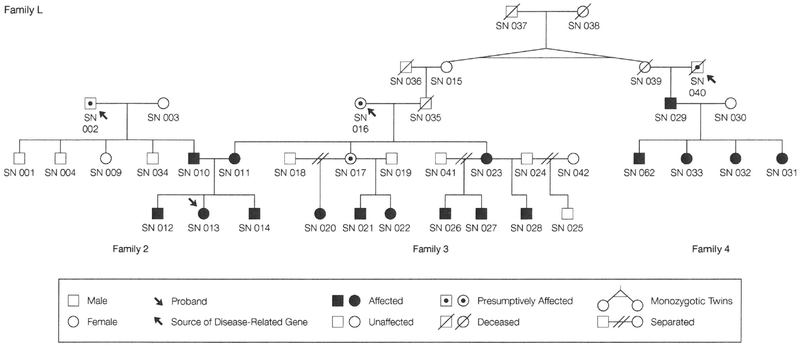
Five families with a high proportion of members affected with severe pediatric GER were chosen for genotypic and linkage analyses after evaluating the pedigrees of more than 20 prospective GER families identified through PAGER. Methods used for making definitive diagnoses of severe pediatric GER for persons within these kindreds are detailed in Table 1. Severely affected persons requiring surgical intervention to control GER are noted as having had fundoplication. Postadolescent persons considered to be presumptively affected, ie, having a history consistent with severe pediatric GER but not having a definitive childhood diagnosis, were modeled in 2 ways for linkage analyses. In the first analysis, they were listed as unknowns and in the second analysis, as unaffecteds. All persons modeled only as unaffecteds (with the exceptions noted below in family 5) had extensive histories taken and reported no childhood symptoms consistent with severe pediatric GER. To ensure as accurate diagnoses as possible for postadolescent study members, parents and older siblings, whenever available, were independently queried on the subject’s medical history relating to GER. Three of the 5 families (2,3, and 4) were related by marriage and were collectively designated as family L (L for large) (Figure 1). Families 2 and 3 were always analyzed together as a single family because study numbers (SNs) 010 and 011, who are married to each other, are both affected and all 3 of their children are severely affected. This latter point suggested that some of the children might inherit a severe pediatric GER allele from both sides, seen for SN 012 (see “Haplotyping” subsection). Families 3 and 4 are linked through SNs 015 and 039, who are unaffected identical twin sisters who married SNs 036 and 040, respectively. Detailed discussions with SN 015 revealed that neither she nor her sister, their parents, nor any of her children had ever been affected with GER as children or adults. These phenotypic findings essentially rule out the possibility of a GER gene coming from the twin sisters (SNs 015 and 039) or their ancestors; however, we performed linkage analyses of families 2, 3, and 4 together (family L) (Table 2 and 3), as well as analyzing family 4 separately and then combining these results with results from families 2 and 3 (Table 3). Family 1 consists of 3 persons who are definitely affected and several others with presumptive diagnoses. Family 5 consists of 17 persons of whom 6 are definitively affected; we have no phenotypic data for SNs 109 through 113, who were all modeled as unaffected.
Table 1.
Methods Used for Diagnosis of Severe Pediatric Gastroesophageal Reflux for Study Subjects
| Diagnosis | Diagnostic Methods | Subject Study Numbers |
|---|---|---|
| Affected (n = 26) | Endoscopy and biopsy | 010, 012, 021, 022, 023, 029, 033,* 056, and 060 (n = 9) |
| Barium swallow | 026 and 027 (n = 2) | |
| pH probe | 117 (n= 1) | |
| Barium swallow and pH probe | 011, 028, 031,* 062, 103,* 105, and 106 (n = 7) | |
| Endoscopy, barium swallow, and pH probe | 013,* and 032* (n = 2) | |
| History† | 014, 020, 048, 101, and 104 (n = 5) | |
| Presumptively affected (n = 10) | History | 002, 016, 017,‡ 040, 043, 045, 054, 058, 108, and 114 |
Subject underwent fundoplicatioπ to surgically prevent gastroesophageal reflux (GER).
Definitive severe pediatric GER diagnoses, based on histories, were made for children (study numbers [SNs] 014, 020, and 104) who exhibited multiple signs and symptoms consistent with GER; had siblings who displayed the same signs and symptoms and who had been diagnosed using objective methods of analysis; and resolved their GER symptoms when treated with antireflux medications, providing a posteriori evidence of severe pediatric GER. Definitive severe pediatric GER diagnoses, based on histories, were made for adults (SNs 048 and 101) who had overwhelming GER signs and symptoms as children that persisted unabated throughout adolescence and extended to the time of this study. Both of these persons also had children diagnosed with severe pediatric GER; however, SN 048’s son did not participate in this study and is not shown on the pedigree.
Individual was diagnosed with esophagitis by endoscopy as a young adult.
Figure 1.
Pedigree of Family L (Families 2, 3, and 4) Showing Multiple Members Affected With Severe Pediatric Gastroesophageal Reflux
Table 2.
Two-Point Lod Scores for the Severe Pediatric GER Locus and Microsatellite Markers on Chromosome 13q for Family L (Families 2, 3, and 4)*
| Microsatellite Marker | Map Distance, cM | Recombination Fraction, θ | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 5 | 10 | 20 | 30 | 40 | ||
| D13S175 | 6 | −1.79 (−2.08) | −1.39 (−1.70) | −0.68 (−0.99) | −0.31 (−0.59) | 0.00 (−0.23) | 0.09 (−0.08) | 0.08 (−0.02) |
| D13S217 | 17 | −2.14 (−2.31) | −1.61 (−1.76) | −0.75 (−0.93) | −0.27 (−0.49) | 0.09 (−0.11) | 0.14 (0.00) | 0.08 (0.02) |
| D13S171 | 25 | −0.36 (−0.33) | −0.24 (−0.29) | 0.01 (−0.17) | 0.13 (−0.07) | 0.18(0.02) | 0.15 (0.05) | 0.09 (0.04) |
| D13S218 | 33 | 2.03 (1.64) | 2.00 (1.61) | 1.85 (1.47) | 1.65 (1.30) | 1.24 (0.94) | 0.81 (0.57) | 0.37 (0.22) |
| D13S1288 | 34 | 4.05 (3.58) | 3.97(3.51) | 3.68 (3.23) | 3.30 (2.87) | 2.49(2.11) | 1.63 (1.31) | 0.75 (0.51) |
| D13S1253 | 35 | 3.26 (2.79) | 3.20 (2.73) | 2.95 (2.50) | 2.63 (2.21) | 1.97 (1.61) | 1.28 (0.98) | 0.59 (0,36) |
| D13S263 | 38 | 4.05 (3.58) | 3.97(3.51) | 3.68 (3.23) | 3.30 (2.87) | 2.49 (2.11) | 1.63 (1.31) | 0.75 (0.51) |
| D13S1297 | 39 | 2.29 (1.86) | 2.25 (1.81) | 2.06 (1.63) | 1.82 (1.41) | 1.31 (0.94) | 0.80 (0.50) | 0.33 (0.16) |
| D13S1272 | 42 | 2.06(1.62) | 2.03 (1.59) | 1.88 (1.46) | 1.68 (1.28) | 1.25 (0.91) | 0.79 (0.53) | 0.35 (0.19) |
| D13S153 | 46 | 1.34 (0.90) | 1.29 (0.86) | 1.19(0.79) | 1.16(0.79) | 0.99 (0.70) | 0.68 (0.46) | 0.32 (0.18) |
| D13S155 | 46 | −0.26 (−0.46) | −0.12 (−0.39) | 0.16 (−0.19) | 0.38 (0.02) | 0.54 (0.21) | 0.45(0.19) | 0.24 (0.09) |
| D13S1296 | 53 | −2.13 (−2.21) | −1.76 (−1.93) | −0.87 (−1.17) | −0.31 (−0.63) | 0.16 (−0.12) | 0.28 (0.06) | 0.20 (0.07) |
| D13S156 | 56 | −2.29 (−2.40) | −1.95 (−1.98) | −1.34 (−1.25) | −0.97 (−0.83) | −0.54 (−0.38) | −0.25 (−0.14) | −0.07 (−0.03) |
| D13S170 | 65 | −2.07 (−2.06) | −1.78 (−1.75) | −1.27 (−1.17) | −0.94 (−0.83) | −0.47 (−0.41) | −0.15 (−0.15) | 0.00 (−0.02) |
| D13S265 | 71 | −2.23 (−2.39) | −1.86 (−1.98) | −1.31 (−1.30) | −0.97 (−0.88) | −0.53 (−0.39) | −0.24 (−0.14) | −0.07 (−0.03) |
| D13S159 | 74 | −2.12 (−2.41) | −1.85 (−2.15) | −1.36 (−1.63) | −0.99 (−1.23) | −0.47 (−0.65) | −0.16 (−0.29) | −0.02 (−0.08) |
| D13S158 | 82 | −0.86 (−1.00) | −0.83 (−0.91) | −0.73 (−0.65) | −0.62 (−0.42) | −0.42 (−0.17) | −0.24 (−0.06) | −0.09 (−0.01) |
| D13S173 | 96 | −2.03 (−2.19) | −1.76 (−1.96) | −1.18 (−1.43) | −0.65 (−0.92) | −0.04 (−0.30) | 0.18 (−0.03) | 0.17 (0.04) |
| D13S1265 | 102 | −1.29 (−1.35) | −1.25 (−1.32) | −1.01 (−1.12) | −0.61 (−0.78) | −0.05 (−0.26) | 0.16 (−0.03) | 0.16 (0.03) |
| D13S285 | 113 | −3.67 (−3.71) | −3.13 (−3.19) | −2.12 (−2.23) | −1.41 (−1.54) | −0.57 (−0.70) | −0.16 (−0.26) | 0.00 (−0.06) |
Lod indicates logarithm of odds; GER, gastroesophageal reflux; and cM, centimorgan. Data are lod scores unless otherwise indicated. For each value of θ for each microsatellite marker, the first lod score shown resulted from running the Linkage program with all presumptively affected individuals (ie, those whose informal medical history indicated severe pediatric GER but who were not definitively diagnosed by a physician) modeled as “unknown” phenotype, a conservative assumption. The second lod score in each cell, in parentheses, resulted from rerunning Linkage with a worst-case assumption, in which all presumptively affected individuals were modeled as “unaffected” phenotype. Lod scores of 3.0 or greater are shown in boldface. Distances in cM are rounded to the nearest whole number.
Table 3.
Multifamily 2-Point Lod Scores for the Severe Pediatric GER Locus on Chromosome 13q at θ = 0*
| Microsatellite Marker | Map Distance, cM | Family | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 2&3 | 4 | 2&3 + 4 | 2&3&4 | 1 | 5 | 2&3 + 4 + 1+5 | 2&3&4 + 1+5 | ||
| D13S175 | 6 | −0.69 | −0.97 | −1.66 | −1.79 (−2.08) | 0.00 (0.00) | 0.00 (0.00) | −1.66 | −1.79 (−2.08) |
| D13S217 | 17 | −1.70 | −1.18 | −2.88 | −2.14 (−2.31) | 0.00 (0.00) | 0.00 (0.00) | −2.88 | −2.14 (−2.31) |
| D13S171 | 25 | −0.64 | 0.00 | −0.64 | −0.36 (−0.33) | −0.65 (−1.19) | 0.17(0.08) | −1.12 | −0.84 (−1.44) |
| D13S218 | 33 | 2.10 | 0.00 | 2.10 | 2.03 (1.64) | 1.03 (−0.63) | 0.46 (0.27) | 3.59 | 3.52 (1.28) |
| D13S1288 | 34 | 2.96 | 1.13 | 4.09 | 4.05 (3.58) | 0.17 (−1.14) | 1.06 (0.87) | 5.31 | 5.27 (3.31) |
| D13S1253 | 35 | 2.18 | 1.13 | 3.31 | 3.26 (2.79) | 0.46 (−0.41) | 1.81 (1.61) | 5.58 | 5.53 (3.99) |
| D13S263 | 38 | 2.96 | 1.13 | 4.09 | 4.05 (3.58) | 1.03 (−0.62) | −0.67 (−0.87) | 4.45 | 4.40 (2.09) |
| D13S1297 | 39 | 1.22 | 1.13 | 2.35 | 2.29 (1.86) | 0.73 (−0.70) | −0.73 (−0.60) | 2.36 | 2.30 (0.56) |
| D13S1272 | 42 | 2.14 | −0.01 | 2.13 | 2.06 (1.62) | 1.03 (−0.63) | −2.23 (−2.42) | 0.93 | 0.86 (−1.43) |
| D13S153 | 46 | 2.41 | −0.93 | 1.48 | 1.34 (0.90) | 0.44 (−0.77) | 0.00 (0.00) | 1.92 | 1.78 (0.13) |
| D13S155 | 46 | 0.46 | −0.93 | −0.48 | −0.26 (−0.46) | 1.03 (−0.30) | 0.00 (0.00) | 0.55 | 0.77 (−0.76) |
| D13S1296 | 53 | −1.51 | −1.08 | −2.59 | −2.13 (−2.21) | 0.00 (0.00) | −2.40 (−2.33) | −4.99 | −4.53 (−4.54) |
| D13S156 | 56 | −2.09 | 0.00 | −2.09 | −2.29 (−2.40) | 0.00 (0.00) | 0.00 (0.00) | −2.09 | −2.29 (−2.40) |
| D13S170 | 65 | −2.15 | −0.41 | −2.56 | −2.07 (−2.06) | 0.00 (0.00) | 0.00 (0.00) | −2.56 | −2.07 (−2.06) |
| D13S265 | 71 | −0.87 | −1.18 | −2.05 | −2.23 (−2.39) | 0.00 (0.00) | 0.00 (0.00) | −2.05 | −2.23 (−2.39) |
| D13S159 | 74 | −1.00 | −0.98 | −1.98 | −2.12 (−2.42) | 0.00 (0.00) | 0.00 (0.00) | −1.98 | −2.12 (−2.42) |
| D13S158 | 82 | −0.98 | 0.00 | −0.98 | −0.86 (−1.00) | 0.00 (0.00) | 0.00 (0.00) | −0.98 | −0.86 (−1.00) |
| D13S173 | 96 | −1.99 | −0.99 | −2.98 | −2.03 (−2.19) | 0.00 (0.00) | 0.00 (0.00) | −2.98 | −2.03 (−2.19) |
| D13S1265 | 102 | −1.21 | −1.01 | −2.21 | −1.29 (−1.35) | 0.00 (0.00) | 0.00 (0.00) | −2.21 | −1.29 (−1.35) |
| D13S285 | 113 | −2.54 | −1.01 | −3.54 | −3.67 (−3.71) | 0.00 (0.00) | 0.00 (0.00) | −3.54 | −3.67 (−3.71) |
Lod indicates logarithm of odds; θ, recombination fraction; cM, centimorgan; &, that the numbered families shown were analyzed together using the Linkage program with all available common ancestors included; and +, that the lod scores were arithmetically combined for the families shown after being run individually. Data are lod scores unless otherwise Indicated. In columns 6, 7, 8, and 10, the first lod score shown for each microsatellite marker resulted from running the program with all presumptively affected individuals modeled as “unknown” phenotype, and the second lod score (in parentheses) resulted from rerunning it with these individuals modeled as “unaffected.” Lod scores of 3.0 or greater are shown In boldface.
Genome Scan
Pairwise linkage analyses of all genotypic data from the genome scan resulted in a single marker, D13S263, on chromosome 13 giving strong evidence of linkage with a lod score greater than 3.0. This marker was flanked on both sides by markers that also yielded positive lod scores. About 80% of the genome was eliminated from further consideration by using a lod score o less than −2.0 as an exclusion criterion. An additional 10% of the markers produced lod scores of less than 1.0 Except for the region on chromosome 13, only 16 markers gave positive lod scores, of which only 3 were greater than 1.0. All of these were less than 1.5. Two of these markers, D3S1304 and D11S4175, were flanked by markers with lod scores of less than −6.0 and −6, and −3 and −4, respectively. The third marker in this group, D19S226, had a lod score of 1.15 and was flanked telomerically by a marker with a lod score of less than −2.0 and centromerically by a marker, D19S221, with a lod score of 0.62. Thus, only the region on chromosome 13 met the predefined criteria (lod score >1.5) for further evaluation.
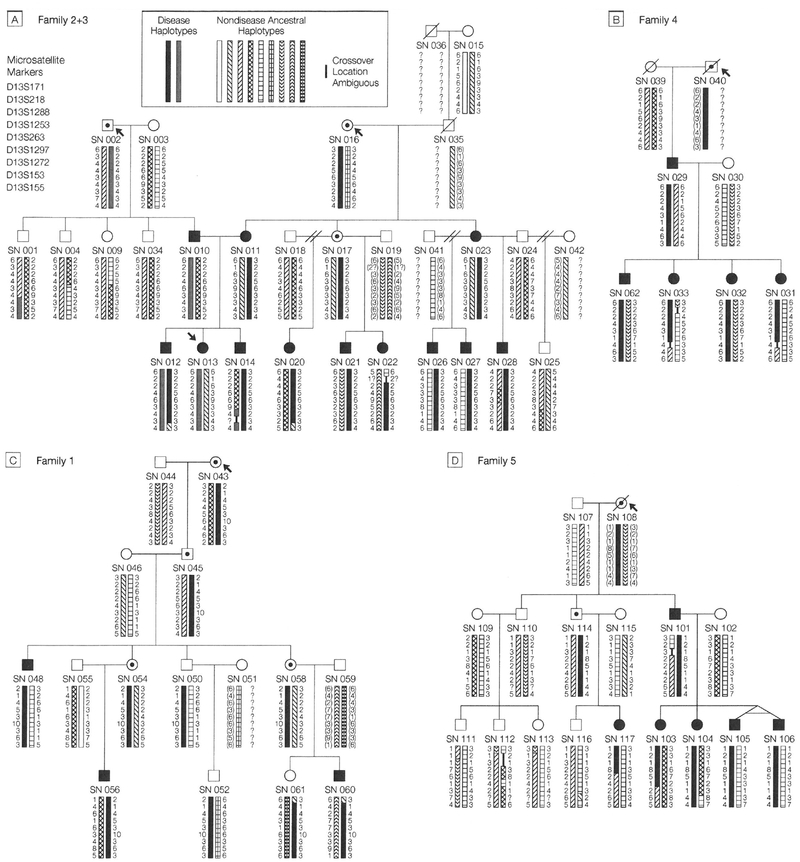
Haplotyping
Figure 2A through D displays the complete pedigrees of all study families and haplotypic data for the GER critical region on chromosome 13. In examining the haplotypic data for families 2 and 3, it can be seen that SN 010 (diagnosed with Barrett metaplasia early in the third decade of life) is the only 1 of 5 children to have severe pediatric GER, and he is the only one to have received chromo-some 13a (a indicates affected) from his father (who was probably affected based on history). The 4 siblings all received the father’s chromosome 13u (u indicates unaffected). SN 010 passed his chromosome 13a on to 2 of his children (SNs 012 and 013). His wife, SN 011, who is also affected (and who has 2 affected siblings, each with 3 affected children) passed on her 13a chromosome to 2 children (SNs 012 and 014). Thus, SN 012 would appear to have received 2 GER alleles. Both of SN 011’s affected sisters (SN 017 has a presumptive diagnosis of severe pediatric GER, being affected as a young adult and having childhood symptoms consistent with GER) have the same maternal chromosome, which they have each passed on to 3 children, all 6 of whom are affected (SN 020s diagnosis of GER was made without invasive procedures by the same physician who diagnosed her siblings).
Figure 2.
Pedigrees and Haplotype Data for All Study Families Showing Multiple Members Affected With Severe Pediatric Gastroesophageal Reflux
See key to Figure 1 for explanation of symbols. The bars in Panels A through D represent haplotypes including markers D13S171, D13S218, D13S1288, D13S1253, D13S263, D13S1297, D13S1272, D13S153, and D13S155 (from centromere to telomere, respectively). Numbers beside the bars indicate allele sizes according to a simple ordered numbering of allele values for each marker (thus, a size of 7 indicates that at least 7 alleles exist for a marker). The symbol”?” indicates allele was not determined or inferable. Parenthetical allele sizes indicate an inferred haplotype. A bar of 1 pattern indicates no recombinations from the previous generations assuming maximum parsimony; wherever parental homozygosity makes it impossible to determine phase a question mark is placed beside the binned allele values. Within each family, individuals with the same bar type share a common haplotype. SN indicates study number. Study numbers 015 and 039 were identical twins (see Figure 1); because SN 039 was deceased, we used data from SN 015 in haplotype construction. Allele comparison between SN 015 and nephew SN 029 showed 399/400 markers as identical, confirming identical twin status.
In family 1, the disease penetrance appears to be lower than in families 2 through 5. The GER chromosome appears to come from SN 043. Three descendents have definitively diagnosed GER and SNs 054 and 058 have histories consistent with GER. SN 048 also has an affected child who was unavailable for study participation. In family 4, all 4 children are affected, and all inherited the same chromosome from their affected father; the same pattern is observed for SN 101 in family 5 and his 4 children. In addition, SN 101’s niece is also affected, and her father has symptoms consistent with GER.
Linkage Analyses
Generation of 2-Point Lod Scores for Chromosome 13 in Family L. The initial 2-point lod scores for markers D13S218, D13S263, and D13S153, generated by the Linkage program, supported our visual inspection of the haplotypic data for family L that had suggested linkage with the GER phenotype (Tables 2 and 3). In addition to these markers, families 2 through 4 were also analyzed with markers D13S1288, D13S1253, D13S1297, and D13S1272, which provided confirmational linkage data.
Rather than only modeling the 3 branches of family L together, we performed additional linkage analyses to confirm our results because it appeared that disease alleles were entering family L from multiple sources. Families 2 and 3 were always analyzed together with both SN 010 and SN 011 modeled as affected and with the GER disease allele frequency set at 0.02, the phenocopy rate set at 0.001, and the penetrance set at 0.7. Given this bilineal family configuration, the Mlink function of Linkage 5.1 assigned to SN 010 a probability of 0.19801 that he is homozygous wild type when his father was coded as unaffected, and 0.09968 when coded as unknown. This mode of analysis results in the generation of very conservative lod scores; however, we still achieved lod scores of 2.96 for both of the markers D13S1288and D13S263 at θ =0 (Table 3, column 3) (θ indicates recombination fraction; θ=0, no recombination). These settings also produced lod scores between 1.22 and 2.41 for 5 other contiguous markers on either side of D13S263, including a score of 2.41 for D13S153. We believe that this marker has been misplaced on the Genethon map (http://wvvw.genlink.wustl.edu/chrmaps).
We next analyzed family 4 and obtained identical maximal lod scores at θ = 0 of 1.13 for markers D13S1288, D13S1253, D13S263, and D13S1297 (Table 3). These values are near the theoretical maximum for such a small family. Our linkage analyses of family L were completed by combining families 2 through 4 (all of family L) prior to linkage analysis. In this analysis, all available common ancestors of families 3 and 4 were included. Using this configuration, maximal lod scores at θ = 0 of 4.05 were obtained at D13S1288 and D13S263 for family L when we modeled presumptively affected individuals as unknowns (Tables 2 and 3). When all of the presumptively affected persons were modeled as unaffecteds, the lod scores were reduced by approximately 0.5 for family L but still indicated linkage at markers D13S1288 and D13S263 (Table 2, parenthetical values). The combined lod scores were similar whether the various branches of family L were run individually and then combined (Table 3, column 5) or combined prior to linkage (Table 3, column 6).
Recombinations in family 4 reduced the size of the critical region, defined by families 2 and 3, by excluding the region telomeric to D13S1297; thus, D13S153 and D13S155 were excluded and the telomeric limit can be stated as being between 39 cM and D13S153. Data from this family and families 2 and 3 suggest that D13S153 is, in fact, centromeric to D13S155 rather than telomeric as noted on the Genethon map; otherwise, it would be necessary to invoke double crossovers within a 2-cM region for SNs 031 and 033 in family 4 and explain how D13S155 can have a low lod score (0.46) for families 2&3 while D13S153 has a high lod score (2.41). Furthermore, the Whitehead Institute map (http://www-genome.wi.mit.edu/) agrees with our suggested placement of these 2 markers.
Multifamily 2-Point Linkage Analyses.
We chose a subset of chromosomal markers within and adjacent to the presumptive GER locus, as identified by analysis of family L data, consisting of D13S171, D13S218, D13S1288, D13S1253, D13S263, D13S1297, D13S1272, D13S153, and D13S155 to analyze family 1. For this small family, there were no recombinants within this interval and maximal pairwise lod scores of 1.03 at θ = 0 were recorded at each of 4 markers (D13S218, D13S263, D13S1272, and D13S155) when the presumptively affected individuals were modeled as unknowns (Table 3). The relatively larger proportion of presumptively affected persons in this family may account for the low lod scores not supporting linkage when these persons are modeled as unaffecteds. The same markers used to examine family 1, with D13S1296 substituted for D13S155, were used to analyze family 5. We obtained lod scores of 1.81 and 1.06 at markers DOS 1253 and D13S1288 at θ = 0 when the presumptively affected persons in family 5 were modeled as unknowns and lod scores of 1.61 and 0.87 at these markers when these persons were modeled as unaffecteds.
Maximal combined multifamily 2-point lod scores of 3.59,5.31,5.58, and 4.45 were obtained for the 4 contiguous markers D13S218, D13S1288, D13S1253, and D13S263 at θ = 0 when the lod scores for the individual families (2 and 3 combined, 4,1, and 5) were arithmetically summed (Table 3, column 9). Similar values (when presumptively affecteds were modeled as unknowns) were obtained when family L was run together and then combined with families 1 and 5 (column 10).
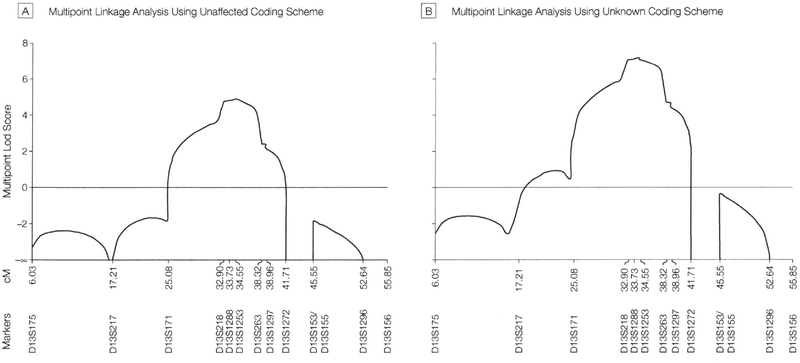
Multipoint Linkage Analyses.
Maximum multifamily multipoint lod scores of 4.88 and 7.15 were obtained for marker D13S1253 at 34.55 cM, for the unaffected and unknown coding schemes, respectively. For both coding schemes the 1-unit support interval ranged from 32.02 cM to 38.35 cM (interval, 6.33 cM) and the 3.0-unit support interval from 25.98 cM to 39.86 cM (interval, 13.88 cM). A graph of the results of the multipoint analysis is displayed in Figure 3A for the unaffected coding scheme and Figure 3B for the unknown coding scheme.
Figure 3.
Multipoint Lod Scores
Graphical representation of the multipoint data generated by Linkmap of the Fastlink software program showing the logarithm-of-odds (lod) score on the y axis and the microsatellite marker positions in cM on the x axis. Panel A displays data obtained when all individuals who were presumptively (but not definitively) affected were modeled as unaffected. Panel B displays data obtained when presumptively affected individuals were modeled as unknown. These computer-generated curves do not show the x axis drawn exactly to scale.
Establishment of a GER Gene Locus
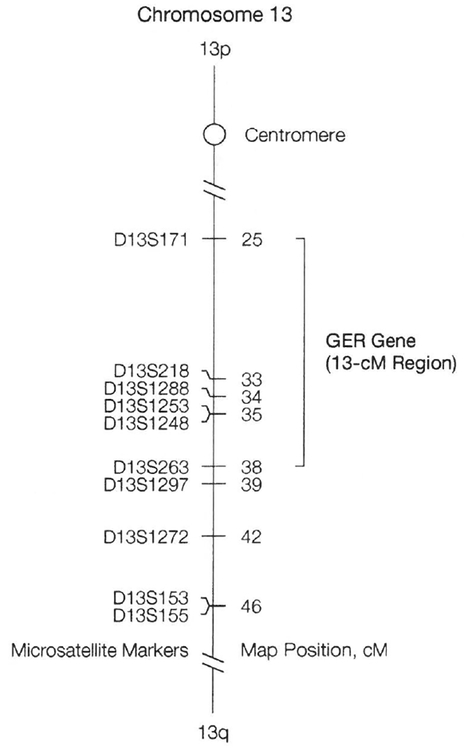
The centromeric limit of the GER interval is currently defined by an obligate recombination between D13S1253 and D13S171 (located at map positions 35 and 25, respectively) in SN 022, an affected child in family 3. It is not possible currently to define the centromeric limit any closer to D13S1253 because the marker D13S218 (located between D13S1253 and D13S171) is not informative for many study subjects. The telomeric limit for the GER gene within family L is defined by a recombination between D13S1297 and D13S153 (D13S1272 is uninformative in this family) in 2 affected individuals, SNs 031 and 033. However, haplotypic analyses of SN 117 in family 5 revealed an obligate recombination between marker D13S1248 (located at map position 35.3 and used only for analyzing this recombination) and D13S263 (located at map position 38.3) thereby setting the telomeric limit for the GER gene at D13S263. Thus, we have mapped a gene for severe pediatric GER to a region no larger than 13 cM on chromosome 13 (Figure 4).
Figure 4.
Location of the Severe Pediatric GER Gene on Chromosome 13
Line diagram of chromosome 13 showing the position of the severe pediatric gastroesophageal reflux (GER) gene and the distance of the microsatellite markers in cM from the p telomere that were used in mapping and refining the locus, p Indicates the short arm of the chromosome; q, the long arm of the chromosome.
COMMENT
Pediatric GER is a major disease of childhood that affects not only the gastrointestinal system, but also profoundly affects the upper and lower respiratory tracts.14 In one study, 63% of pediatric patients with chronic pulmonary disease without overt symptoms of GER were identified by 24-hour pH probe analysis and scintiscanning as having GER. Treatment with anti-GER regimens resulted in disappearance of the respiratory problems in 82% of patients.12 In another study, 28% of infants hospitalized with congenital airway abnormalities were diagnosed with GER.32 These studies highlight 2 important points, first that GER often goes un-diagnosed, even when causing symptoms, and second, that it is responsible for considerable respiratory morbidity.
Current understanding of pediatric GER is limited to the gross phenotypic effects described above. Since the biochemical and molecular biological mechanisms of GER are unknown, the disease is treated empirically. To design more effective, rational therapies, an understanding of the physiological processes that underlie severe pediatric GER is needed. This need drove our search to begin to find the genetic basis for severe pediatric GER.
Using PAGER’s substantial family database, we are now recruiting numerous other small GER families to determine the applicability of the 13q14 mapping to GER patients generally, both pediatric and adult, and to identify potential genetic heterogeneity. However, the finding that all 5 families map to the same region argues that there may be limited heterogeneity among families with severely affected children, and recent additional analyses of 3 sib-pair families has provided data consistent with linkage to 13ql4 in 2 of the 3 families (G.D.E., unpublished data, 1999). It is possible that milder forms of GER are associated with a gene, or genes, that map to a different location, and that some cases of pediatric GER occur sporadically, but family studies of our probands with severe pediatric GER suggest that most such cases are hereditary. Efforts to narrow the locus and identify the gene itself are ongoing.
Acknowledgment:
We wish to thank Miles Ehrlich and Stephanie Petiprin, MS, fortheir help in delineating the pedigrees and obtaining specimens and for providing editorial assistance. We also wish to thank Sandy Johnson, BS, and Jay Hayes, BS, for technical assistance.
Funding/Support: This work was supported by Allegheny General Hospital, Allegheny University of the Health Sciences, NIDCD grants DC02148 and DC04591 (G.D.E.) and DC02398 (J.C.P.), and NIHG grant HG00008 (J.O.).
REFERENCES
- 1.Fonkalsrud EW, Ament ME. Gastroesophageal reflux in childhood. Curr Probl Surg. 1996;33:1–70. [PubMed] [Google Scholar]
- 2.Romero Y, Locke GR III. Is there a GERD gene? Am J Gastroenterol. 1999;94:1127–1129. [DOI] [PubMed] [Google Scholar]
- 3.Lagergren J, Bergstrom R, Lindgren A, Nyren O. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med. 1999; 340:825–831. [DOI] [PubMed] [Google Scholar]
- 4.Euler AR. Upper respiratory tract complications of gastroesophageal reflux in adult and pediatric-age patients. Dig Dis. 1998;16:111–117. [DOI] [PubMed] [Google Scholar]
- 5.McMurray JS, Holinger LD. Otolaryngic manifestations in children presenting with apparent life-threatening events. Otolaryngol Head Neck Surg. 1997; 116:575–579. [DOI] [PubMed] [Google Scholar]
- 6.Stephen TC, Younoszai MK, Massey MP, Fellows RA. Diagnosis of gastroesophageal reflux in pediatrics. J Ky Med Assoc. 1994;92:188–191. [PubMed] [Google Scholar]
- 7.Tolaymat N, Chapman DM. Gastroesophageal reflux disease in children older than two years of age. W V Med J. 1998;94:22–25. [PubMed] [Google Scholar]
- 8.Callahan CW. Increased gastroesophageal reflux in infants. Acta Paediatr. 1998;87:1219–1223. [DOI] [PubMed] [Google Scholar]
- 9.del Rosario JF, Orenstein SR. Common ped¡atricesophageal disorders. Gastroenterologist. 1998;6:104–121. [PubMed] [Google Scholar]
- 10.Faubion WA Jr, Zein NN. Gastroesophageal reflux in infants and children. Mayo Clin Proc. 1998; 73:166–173. [DOI] [PubMed] [Google Scholar]
- 11.Orenstein SR. Infantile reflux: different from adult reflux. Am J Med. 1997;103(5A):114S–119S. [DOI] [PubMed] [Google Scholar]
- 12.Malfroot A, Vandenplas Y, Verlinden M, et al. Gastroesophageal reflux and unexplained chronic respiratory disease in infants and children. Pediatr Pulmonol. 1987;3:208–213. [DOI] [PubMed] [Google Scholar]
- 13.Orenstein SR. Gastroesophageal reflux In: Wyllie R, Hyams JS, eds. Pediatric Gastrointestinal Disease. Philadelphia, Pa: WB Saunders Co; 1993:337–369. [Google Scholar]
- 14.Orenstein SR, Orenstein DM. Gastroesophageal reflux and respiratory disease in children. J Pediatr. 1988;112:847–858. [DOI] [PubMed] [Google Scholar]
- 15.Giannoni C, Sulek M, Friedman EM, Duncan NO III. Gastroesophageal reflux association with laryngomalacia. Int J Pediatr Otorhinolaryngol. 1998;43:11–20. [DOI] [PubMed] [Google Scholar]
- 16.Halstead LA. Role of gastroesophageal reflux in pediatric upper airway disorders. Otolaryngol Head Neck Surg. 1999;120:208–214. [DOI] [PubMed] [Google Scholar]
- 17.Little JP, Matthews Bl, Glock MS, et al. Extra-esophageal pediatric reflux. Ann Otol Rhinol Laryngol Suppl. 1997;169:1–16. [PubMed] [Google Scholar]
- 18.Walner DL, Stern Y, Gerber ME, et al. Gastroesophageal reflux in patients with subglottic stenosis. Arch Otolaryngol Head Neck Surg. 1998;124:551–555. [DOI] [PubMed] [Google Scholar]
- 19.Halstead LA. Gastroesophageal reflux: a critical factor in pediatric subglottic stenosis. Otolaryngol Head Neck Surg. 1999;120:683–688. [DOI] [PubMed] [Google Scholar]
- 20.Crabb DW, Berk MA, Hall TR, et al. Familial gastroesophageal reflux and development of Barrett’s esophagus. Ann Intern Med. 1985;103:52–54. [DOI] [PubMed] [Google Scholar]
- 21.Fahmy N, King JF. Barrett’s esophagus: an acquired condition with genetic predisposition. Am J Gastroenterol. 1993;88:1262–1265. [PubMed] [Google Scholar]
- 22.Romero Y, Cameron AJ, Locke GR III, et al. Familial aggregation of gastroesophageal reflux in patients with Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 1997;113:1449–1456. [DOI] [PubMed] [Google Scholar]
- 23.Trudgill NJ, Kapur KC, Riley SA. Familial clustering of reflux symptoms. Am J Gastroenterol. 1999; 94:1172–1178. [DOI] [PubMed] [Google Scholar]
- 24.Seri M, Cusano R, Forabosco P, et al. Genetic mapping to 10q23.3-q24.2, in a large Italian pedigree, of a new syndrome showing bilateral cataracts, gastroesophageal reflux, and spastic paraparesis with amyotrophy. Am J Hum Genet. 1999;64:586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcon MA. Advances in the diagnosis and treatment of gastroesophageal reflux disease. Curr Opin Pediatr. 1997;9:490–493. [DOI] [PubMed] [Google Scholar]
- 26.Preston RA, Post JC, Keats BJ, et al. A gene for Crouzon craniofacial dysostosis maps to the long arm of chromosome 10. Nat Genet. 1994;7:149–153. [DOI] [PubMed] [Google Scholar]
- 27.Whitcomb DC, Preston RA, Aston CE, et al. A gene for hereditary pancreatitis maps to chromosome 7q35. Gastroenterology. 1996; 110:1975–1980. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger JD, Ott J. Handbook of Human Genetic Linkage. Baltimore, Md: Johns Hopkins University Press; 1994. [Google Scholar]
- 29.Ott J Analysis o f Human Genetic Linkage. 3rd ed Baltimore, Md: Johns Hopkins University Press; 1999. [Google Scholar]
- 30.Cottingham RW Jr, Idury RM, Schaffer AA Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 31.Broman KW, Murray JC, Sheffield VC, et al. Comprehensive human genetic maps. Am J Hum Genet. 1998;63:861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altman KW, Wetmore RF, Marsh RR. Congenital airway abnormalities in patients requiring hospitalization. Arch Otolaryngol Head Neck Surg. 1999; 125:525–528. [DOI] [PubMed] [Google Scholar]