Abstract
Recently, there has been increased interest in using Single Nucleotide Polymorphisms (SNPs) as a method for detecting genes for complex traits. SNPs are diallelic markers that have the potential to be inexpensively produced using chip technology. It has been suggested that SNPs will be beneficial in study designs that utilize trio data (father, mother, child). In our previous work, we calculated the probability of detecting Mendelian errors at a SNP locus for a trio randomly selected from a population in Hardy-Weinberg equilibrium. The highest error-detection rate was 30%. Here we investigate the error-detection rate when additional sibs are genotyped. We define an error to be a change from a 1 allele to a 2 allele, or vice versa. Typing one additional sib increases the detection rate on average by 10 – 13%. Typing two additional sibs increases the detection rate on average by 14-19%. The increase in the detection rate is dependent on the allele frequencies. Equal allele frequencies produce the lowest detection rates, independent of true error rates and number of offspring genotyped. Typing additional siblings not only improves error-detection rates, but can also provide additional linkage information. In order to increase linkage information and error-detection rates, at least two additional siblings should be ascertained when available.
1. Introduction
Recently, there has been an increased interest in the use of Single Nucleotide Polymorphisms (SNPs) loci as a possible method for detecting genes of modest effect, i.e., for complex traits1. SNPs occur in large numbers across the human genome and usually consist of two alleles2. Statistical methods proposed for detection of genes for complex traits using SNPs include haplotype-relative risk tests3, and transmission-disequilibrium tests4 (TDT). In each of these tests, the sampling frame is a family trio, consisting of a father, a mother, and an affected child genotyped at a locus. In the case of TDT when testing for linkage in the presence of association, additional affected siblings may also be used5.
One important methodologic issue concerning markers in general and SNPs in particular is that of errors. Buetow6 found that, in constructing high resolution human linkage maps (3 cM or less) with highly polymorphic markers, typing errors, even at the rate of 1.5%, led to a reduction of power to discriminate orders, a dramatic inflation of map length, and significant support for incorrect over correct orders. Shields et al.7 also determined that the introduction of typing errors when constructing human linkage maps led to support for incorrect orders and map length inflation. Terwilliger, Weeks and Ott8 showed in simulation studies for a multi-allelic system that misreading an allele results in overestimation of recombination fraction and a decrease in expected lod score. Using simulated data for a multi-allelic marker, Gordon et al.9 showed that errors in pedigree data can significantly reduce evidence for linkage in the presence of association when using a transmission disequilibrium test. Several authors10,11,12,13 considered the issue of errors in pedigree data, and devised methods for detecting such errors.
Regarding SNPs, Gordon et al.14 investigated the error-detection rate in trios by detecting errors through deviations from Mendel’s laws. The authors defined an error, as we do in this article, to be anything that causes a 1 allele to change to a 2 allele, or vice versa. Such errors may result from non-paternity, sample swaps in the lab, or genotyping errors. The highest detection rate for trios was slightly less than 31%. Given such a low detection rate, we seek to quantify the improvement in error-detection rates by genotyping additional siblings.
2. Materials and Methods
2.1. Detection Rate for Sib Pairs (Genotype Quartets)
For a diallelic marker with allele numbers 1 and 2, there are (up to symmetry) three possible genotypes: 1/1, 1/2, and 2/2. A genotype quartet is defined to be a 4-tuple of genotypes (Parent1 Allele 1/Parent1 Allele 2, Parent2 Allele 1/Parent2 Allele 2, Child1 Allele 1/Child1 Allele 2, Child2 Allele 1/Child2 Allele 2) in which the set of alleles is consistent with Mendel’s laws. In this 4-tuple, no distinction is made between Parent1 and Parent2, or Child1 and Child2. For example, the genotype quartets (1/2, 1/1, 1/1, 1/2) and (1/1, 1/2, 1/2, 1/1) are considered to be equivalent. No distinction is made between genotypes 2/1 and 1/2. Thus, for example, the quartet (2/1, 2/1, 2/1, 2/1) is equivalent to (1/2, 1/2, 1/2, 1/2), and so forth. For consistency, the genotypes 1/2 and 2/1 shall always be referred to as 1/2. A general quartet is defined to be any 4-tuple of genotypes in which each person has the genotypes 1/1, 1/2, or 2/2, without the restriction that the quartet displays Mendelian consistency. Here and elsewhere, the term consistency (respectively, inconsistency) implies Mendelian consistency (respectively, Mendelian inconsistency). Note that the set of genotype quartets is a subset of the set of general quartets. Also, the conjugate of a genotype quartet M (denoted M) is defined as the genotype quartet that results when each value of 1 in M is replace by a 2, and each value of 2 in M is replaced by a 1. For example, the conjugate of the quartet (1/1, 1/1, 1/1, 1/1) is (2/2, 2/2, 2/2, 2/2), the conjugate of (1/2, 1/1, 1/1, 1/1) is (1/2, 2/2, 2/2, 2/2), and so forth. The list of all genotype quartets and their conjugates may be found in Table 1. An error in a genotype quartet is defined to be one or more changes in genotypes of the quartet. For example, if the original quartet is (2/2, 2/2, 2/2, 2/2) then one error is introduced into the quartet if one and only one of the 2 alleles is replaced by a 1 allele. It is assumed that errors are introduced randomly and independently into genotype quartets at a constant rate, denoted by α.
Table 1.
List of all genotype quartets and their conjugates
| Genotype Quartet M | Conjugate Genotype Quartet M |
|---|---|
| (1/1, 1/1, 1/1, 1/1) | (2/2, 2/2, 2/2, 2/2) |
| (1/1, 1/2, 1/1, 1/1) | (2/2, 1/2, 2/2, 2/2) |
| (1/1, 1/2, 1/1, 1/2) | (2/2, 1/2, 2/2, 1/2) |
| (1/1, 1/2, 1/2, 1/2) | (2/2, 1/2, 1/2, 1/2) |
| (1/1, 2/2, 1/2, 1/2) | (2/2, 1/1, 1/2, 1/2) |
| (1/2, 1/2, 1/1, 1/1) | (1/2, 1/2, 2/2, 2/2) |
| (1/2, 1/2, 1/1, 1/2) | (1/2, 1/2, 2/2, 1/2) |
| (1/2, 1/2, 1/1, 2/2) | (1/2, 1/2, 2/2, 1/1) |
| (1/2, 1/2, 1/2, 1/2) | (1/2, 1/2, 1/2, 1/2) |
| (1/2, 1/2, 1/2, 2/2) | (1/2, 1/2, 1/2, 1/1) |
| (1/2, 1/2, 2/2, 2/2) | (1/2, 1/2, 1/1, 1/1) |
| (1/2, 2/2, 1/2, 1/2) | (1/2, 1/1, 1/2, 1/2) |
| (1/2, 2/2, 1/2, 2/2) | (1/2, 1/1, 1/2, 1/1) |
| (1/2, 2/2, 2/2, 2/2) | (1/2, 1/1, 1/1, 1/1) |
| (2/2, 2/2, 2/2, 2/2) | (1/1, 1/1, 1/1, 1/1) |
For each of the fifteen quartets in Table 1, anywhere from 0 to 8 errors may be introduced. In the case of either 0 or 8 errors, the resulting general quartet will always display consistency. We calculate the probability that all errors go undetected for a collection of genotype quartets in which at least one error has been introduced, and denote this quantity by β. It follows that the detection rate is 1 - β. Using basic probability theory,
| (1) |
The term i = 0 is not included in the sum (1) because only genotype quartets in which errors have been introduced are considered. If B(α; i) is defined by
then because error introduction is assumed to be random and independent for each allele in a genotype quartet, and because only those genotype quartets that contain at least one error are considered, the quantity Pr(i errors in quartet) in formula (1) is given by
| (1a) |
Note that the expression B(α;i) is the probability density function, evaluated at i, 1≤ i ≤ 8, for a binomial distribution with constant success rate α in each of 8 independent events. Using basic probability theory, the quantity Pr(undetected errors | i errors in quartet) in formula (1) is calculated as
| (2) |
where S is the set of all fifteen genotype quartets in the first column of Table 1, N0 is the event that all errors in a quartet go undetected, Pr(N0 |M,i) is the probability that i errors introduced into a genotype quartet M go undetected, and Pr(M) is the population frequency of the genotype quartet M. Pr (M) is calculated using the allele frequencies p for allele 1 and q (= 1 – p) for allele 2, assuming that the alleles are in Hardy-Weinberg equilibrium. For each genotype quartet M in Table 1, Pr(M) is calculated in Table 2. For each M and each i, the probability Pr(N0 | M,i) is equal to the proportion of resulting quartets that, after i errors have been introduced, show consistency. For example, if M = (1/1, 1/1, 1/1, 1/1) and i = 1, Pr(N0 | M,i) = 4/8, or 1/2; of the eight general quartets that result from a change in one of the alleles, a change in any of the four parental alleles will display consistency and a change in any of the children’s alleles will display inconsistency.
Table 2.
Conditional probability that a quartet M displays consistency if i errors are introduced, 1 ≤i ≤4
| Quartet = M | Pr(M) | Pr(N0|M,i) | |||
|---|---|---|---|---|---|
| i=1 | i=2 | i=3 | i=4 | ||
| (1/1, 1/1, 1/1, 1/1) | P4 | 1/2 | 5/7 | 4/7 | 16/35 |
| (1/1, 1/2, 1/1, 1/1) | P3q | 7/8 | 15/28 | 15/28 | 4/7 |
| (1/1, 1/2, 1/1, 1/2) | 2p3q | 5/8 | 17/28 | 31/56 | 19/35 |
| (1/1, 1/2, 1/2, 1/2) | p3q | 5/8 | 17/28 | 4/7 | 18/35 |
| (1/1, 2/2, 1/2, 1/2) | 2p2q2 | 1/2 | 1/2 | 4/7 | 22/35 |
| (1/2, 1/2, 1/1, 1/1) | 1/4 p2q2 | 3/4 | 17/28 | 15/28 | 19/35 |
| (1/2, 1/2, 1/1, 1/2) | p2q2 | 3/4 | 9/14 | 31/56 | 17/35 |
| (1/2, 1/2, 1/1, 2/2) | 1/2 p2q2 | 1/2 | 9/14 | 4/7 | 4/7 |
| (1/2, 1/2, 1/2, 1/2) | p2q2 | 1 | 9/14 | 1/2 | 18/35 |
| (1/2, 1/2, 1/2, 2/2) | p2q2 | 3/4 | 9/14 | 31/56 | 17/35 |
| (1/2, 1/2, 2/2, 2/2) | 1/4 p2q2 | 3/4 | 17/28 | 15/28 | 19/35 |
| (1/2, 2/2, 1/2, 1/2) | pq3 | 5/8 | 17/28 | 4/7 | 18/35 |
| (1/2, 2/2, 1/2, 2/2) | 2pq3 | 5/8 | 17/28 | 31/56 | 19/35 |
| (1/2, 2/2, 2/2, 2/2) | pq3 | 7/8 | 15/28 | 15/28 | 4/7 |
| (2/2, 2/2, 2/2, 2/2) | q4 | 1/2 | 5/7 | 4/7 | 16/35 |
We now state some lemmas whose application simplifies the calculation of Pr(N0 |M,i) for any of the fifteen genotype quartets M in Table 1 and any i, 0 ≤ i ≤ 8. These lemmas are extensions of ones proved previously14. The lemmas in that reference apply to genotype trios, and the lemmas below follow from the observation that two genotype trios may be uniquely formed (corresponding to the two children) from any genotype quartet. For example, the quartet (1/2, 1/2, 1/1, 2/2) maps uniquely to the set of genotype trios {(1/2, 1/2, 1/1), (1/2, 1/2, 2/2)}.
Lemma 1. For any genotype quartet M and for any i, 0 ≤ i ≤ 8,
Pr(N0 |M,i) = Pr(N0 |M,i).
Lemma 2. For any genotype quartet M and for any i, 0 ≤ i ≤ 8,
Pr(N0 |M,i) = Pr(N0 |M,8 - i).
Note that any quartet M falls uniquely into one of two categories: either M = M or not. There are exactly three quartets M such that M = M : (1/2, 1/2, 1/2, 1/2), (1/1, 2/2, 1/2, 1/2) and (1/2, 1/2, 1/1, 2/2). For the remaining twelve, we divide them into six genotype quartets and their conjugates, compute Pr(N0 |M,i) for the six quartets, and use Lemma 1 to compute this probability for the remaining six. With this information, the value Pr(undetected errors|i errors in quartet) in formula (2) is calculated. Using Table 2, it follows:
| (2.2a) |
Applying Lemma 2, we observe that
| (2.2b) |
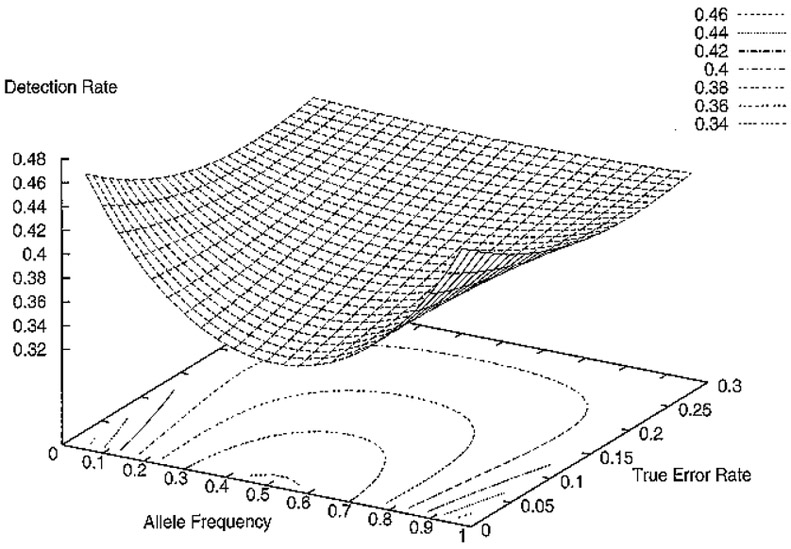
Equations (2.2a) and (2.2b) are substituted into formula (2), which are then substituted into formula (1) along with the term Σ Pr(N0 |M,8) Pr(M) = 1, to determine β. In Table 4, error-detection rates for various values of α and p are calculated, and in Figure 1, error-detection rates for a range of values of α and p are plotted. It follows immediately from equations (2.2a) and (2.2b) that the detection rate is symmetric about the line p = 0.5. This symmetry is seen in Figure 1 as well. Therefore, for a fixed value of α, the detection rate will be the same for frequencies p and 1 – p.
Table 4.
Detection Rate 1 - β for Trios, Quartets and Quintets with various values of true error rate α and various allele frequencies p at diallelic locus.
| Frequency p of one Quartets allele = .1 | Frequency p of Quartets one allele = .5 | |||||
|---|---|---|---|---|---|---|
| True error rate | Trios* | Quintets | Quintets | Trios* | Quintets | Quintets |
| .0010 | .3032 | .4352 | .5064 | .2501 | .3361 | .3706 |
| .0050 | .3025 | .4337 | .5045 | .2506 | .3370 | .3718 |
| .0100 | .3017 | .4319 | .5022 | .2512 | .3380 | .3732 |
| .0200 | .3001 | .4284 | .4978 | .2525 | .3400 | .3762 |
| .0500 | .2960 | .4193 | .4856 | .2562 | .3467 | .3859 |
| .1000 | .2909 | .4081 | .4705 | .2622 | .3586 | .4039 |
| .2000 | .2862 | .4017 | .4631 | .2732 | .3841 | .4441 |
| .3000 | .2865 | .4117 | .4825 | .2820 | .4084 | .4830 |
Figure 1.
Error-Detection Rate for quartets as function of allele frequency and true error rate
2.2. Detection Rate for Genotype Quintets
In the case where three offspring are available for study, definitions from the previous section are extended in the natural way to speak of genotype quintets, general quintets, conjugates and errors. The list of all genotype quintets may be found in Table 3. To compute β and the detection rate 1 - β for genotype quintets, we need only change the index in formulas (1) and (1a) so that the sum goes to 10 (corresponding to the 10 alleles in a genotype quintet). Also, in formula B(α; i) and in Lemmas 1 and 2, the value 10 is substituted for 8 everywhere. As above, extensions of Lemmas 1 and 2 are proved using the observation that any genotype quintet maps uniquely to a set of three genotype trios.
Table 3.
Conditional probability that a quintet M displays consistency if i errors are introduced, 1 ≤i ≤ 5
| Quintet = M | Pr(M) | Pr(N0|M,i) | ||||
|---|---|---|---|---|---|---|
| i=1 | i=2 | i=3 | i=4 | i=5 | ||
| (1/1, 1/1, 1/1, 1/1, 1/1) | P4 | 2/5 | 28/45 | 3/5 | 46/105 | 8/21 |
| (1/1, 1/2, 1/1, 1/1, 1/1) | 1/2 p3q | 9/10 | 3/5 | 29/60 | 16/35 | 3/7 |
| (1/1, 1/2, 1/1, 1/1, 1/2) | 3/2 p3q | 7/10 | 5/9 | 59/120 | 19/42 | 29/63 |
| (1/1, 1/2, 1/1, 1/2, 1/2) | 3/2 p3q | 3/5 | 8/15 | 59/120 | 33/70 | 28/63 |
| (1/1, 1/2, 1/2, 1/2, 1/2) | 1/2 p3q | 3/5 | 8/15 | 61/120 | 16/35 | 19/42 |
| (1/1, 2/2, 1/2, 1/2, 1/2) | 2 p2q2 | 2/5 | 19/45 | 13/30 | 52/105 | 65/126 |
| (1/2, 1/2, 1/1, 1/1, 1/1) | 1/16 p2q2 | 4/5 | 2/3 | 29/60 | 3/7 | 29/63 |
| (1/2, 1/2, 1/1, 1/1, 1/2) | 3/8 p2q2 | 4/5 | 3/5 | 61/120 | 16/35 | 26/63 |
| (1/2, 1/2, 1/1, 1/1, 2/2) | 3/16 p2q2 | 3/5 | 7/15 | 29/60 | 33/70 | 10/21 |
| (1/2, 1/2, 1/1, 1/2, 1/2) | 3/4 p2q2 | 4/5 | 31/45 | 31/60 | 91/210 | 3/7 |
| (1/2, 1/2, 1/1, 1/2, 2/2) | 3/4 p2q2 | 3/5 | 29/45 | 1/2 | 7/15 | 55/126 |
| (1/2, 1/2, 1/1, 2/2, 2/2) | 3/16 p2q2 | 3/5 | 7/15 | 29/60 | 33/70 | 10/21 |
| (1/2, 1/2, 1/2, 1/2, 1/2) | 1/2 p2q2 | 1 | 31/45 | 1/2 | 46/105 | 56/126 |
| (1/2, 1/2, 1/2, 1/2, 2/2) | 3/4 p2q2 | 4/5 | 31/45 | 31/60 | 91/210 | 3/7 |
| (1/2, 1/2, 1/2, 2/2, 2/2) | 3/8 p2q2 | 4/5 | 3/5 | 61/120 | 16/35 | 26/63 |
| (1/2, 1/2, 2/2, 2/2, 2/2) | 1/16 p2q2 | 4/5 | 2/3 | 29/60 | 3/7 | 29/63 |
| (1/2, 2/2, 1/2, 1/2, 1/2) | 1/2 pq3 | 3/5 | 8/15 | 61/120 | 16/35 | 19/42 |
| (1/2, 2/2, 1/2, 1/2, 2/2) | 3/2 pq3 | 3/5 | 8/15 | 59/120 | 33/70 | 28/63 |
| (1/2, 2/2, 1/2, 2/2, 2/2) | 3/2 pq3 | 7/10 | 5/9 | 59/120 | 19/42 | 29/63 |
| (1/2, 2/2, 2/2, 2/2, 2/2) | 1/2 pq3 | 9/10 | 3/5 | 29/60 | 16/35 | 3/7 |
| (2/2, 2/2, 2/2, 2/2, 2/2) | q4 | 2/5 | 28/45 | 3/5 | 46/105 | 8/21 |
Proceeding as above and using the quantities from Table 3, we compute:
| (2.3a) |
As above, note that
| (2.3b) |
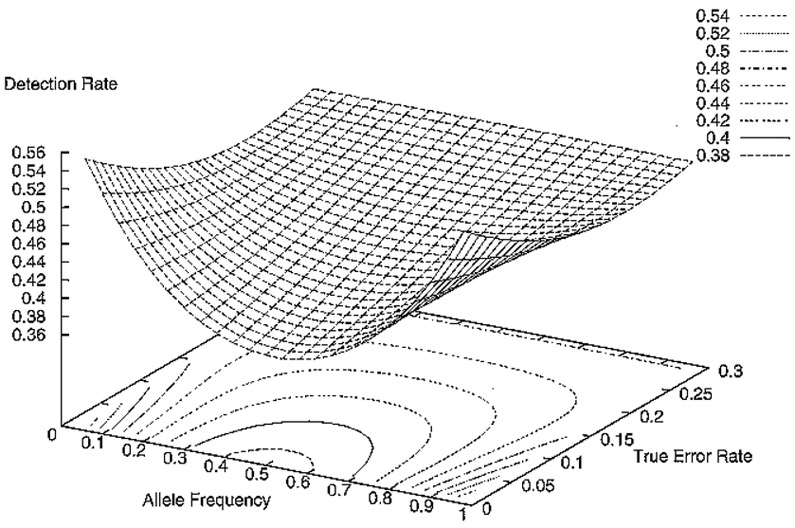
These equations are used to compute the detection rate for any randomly selected genotype quintet from a population in Hardy-Weinberg equilibrium. As in the case of genotype trios14 and genotype quartets above, the detection rate 1 - β is symmetric about the line p = 0.5. In Table 4, error-detection rates for various values of α and p are presented. In Figure 2, error-detection rates for a range of values of α and p are plotted.
Figure 2.
Error-Detection Rate for quintets as function of allele frequency and true error rate
3. Results
In Table 4, error-detection rates for various error rates and allele frequencies when typing genotype trios (one child), quartets, and quintets are presented. Detection rates for trios were presented previously14. The main result is that typing additional siblings increases the detection rate by at least 9%. By computing the average detection rate for each sampling type (trio, quartet, and quintet), it may be seen that typing quartets or quintets increases the detection rate on average by 13% or 19% respectively when one allele frequency is 0.1. For equal allele frequencies, the change in sampling types increases the detection rate on average by 10% and 14% respectively.
This increase in detection rate holds for all error rates and allele frequencies. These results are displayed in Figures 1 and 2, which plot the detection rates for a range of true error rates from 1% to 30% and for allele frequencies (one allele) from 0.02 to 0.98. Note that detection rates are symmetric about the plane p = 0.5.
The second result is that, independent of the sampling type, detection rates are always the lowest when allele frequencies are equal (see Figures 1 and 2). However, the graphs indicate that the detection rates converge to a common detection rate, independent of allele frequencies, as the true error increases. This result holds for all sampling types.
At low error rates, a significant difference is observed between detection rates for extreme allele frequencies (one allele has frequency ≤ 0.05) and equal allele frequencies, regardless of the sampling type. To explain this observation, consider genotype quartets. When allele frequencies are extreme, then the majority of quartets sampled will have both parents homozygous for the same allele. Without loss of generality, let us assume that the 1 allele at a SNP locus has allele frequency greater than 0.95. Then the majority of the quartets are of the form M = (1/1, 1/1, 1/1, 1/1). When error rates are very low, the majority of quartets with errors will have only one error introduced. A way to reach this conclusion quantitatively is to evaluate formula (1a) for small values of α. When i in formula (1a) is greater than 1, the probability is approximately 0. It follows that the detection rate for extreme allele frequencies and small true error rates is approximately 1 - Pr(N0| M,1) = 1 – 1/2 = 1/2, or 0.50 (Table 2).
By comparison, for the case of equal allele frequencies, all genotype quartets M appear with probability Pr(M) >> 0 (Table 2). When α is small, the detection rate is approximately 1 - Σ Pr(N0 |M,1) Pr(M). Note that Pr(N0 |M,1) > 1/2 for all genotype quartets M, with the exception of the quartets (1/1, 1/1, 1/1, 1/1) and (2/2, 2/2, 2/2, 2/2), and for the quartet M = (1/2, 1/2, 1/2, 1/2), Pr(N0 |M,1) = 1, so that in computing the sum, the detection rate for equal allele frequencies is considerably less than 0.5. The same reasoning applies for trios and quintets.
4. Discussion
In this article, an analytic solution to SNP error-detection rates in nuclear families is provided. For quartets (two sibs) or quintets (three sibs), the detection rate is quantified as a function of the true error rate at a SNP locus in Hardy-Weinberg equilibrium and the allele frequency of one of the alleles. It is shown in our analysis that genotyping at least two additional siblings, when available, provides a considerable improvement in error-detection rates, on the order of 14%-19%. The authors therefore recommend that researchers who are analyzing SNP data consider results of these formulas (Table 4) when designing linkage and association studies.
There is an added benefit to genotyping additional siblings, even if unaffected. Such siblings can provide information for linkage studies. For example, in studying quantitative phenotypes with a recurrence risk of 75%, Risch and Zhang15 showed that extreme discordant sib-pairs may provide a significant amount of linkage information.
In addition, genotyping additional siblings, even if unaffected, is typically much less expensive than ascertaining an additional family with an affected child. While it is true that collection of additional families increases power to detect linkage in the presence of association, allowing errors to go undetected in pedigree data can reduce power to detect that linkage9, potentially negating the effects of the additionally ascertained families. Also, errors in data can make statistical tests like the TDT invalid by increasing the type I error rate16.
It is important to recognize that our formulas calculate the probability that a family is brought to the researcher’s attention because of inconsistency. Our method does not suggest the most likely location of the error. Ehm et al.11 presents a likelihood method for determining typing errors in pedigree data displaying consistency, while other authors12, 13 present methods for determining (statistically) incorrect genotypes in pedigree data displaying inconsistency. These methods are implemented in freeware programs (see Electronic Database Information).
Regarding ascertainment, we assume that the locus studied is in Hardy-Weinberg equilibrium. However, for most studies, families are ascertained on the basis of disease. While it is true that the detection rates for SNP loci in linkage disequilibrium with a disease locus will differ from the results presented in Table 4, our recommendations regarding genotyping of additional siblings are still relevant. As previously shown14, for low error rates there is a theoretical maximum error-detection rate of 33% when sampling trios, independent of linkage disequilibrium between marker and locus, ascertainment scheme, or allele frequencies at the marker locus. This detection rate can be improved only by genotyping additional siblings, if consistency is used as the sole check for errors.
Acknowledgments
The authors acknowledge grants HG00008 from the National Human Genome Research Institute and DC03594 from the National Institutes of Health. Also, the authors acknowledge the helpful comments of anonymous reviewers.
Footnotes
Electronic Database Information
The freeware program GENOCHECK11 is available via ftp from the URL ftp://softlib.cs.rice.edu/pub/GenoCheck. The freeware program PEDCHECK13 is available from the URL ftp://watson.hgen.pitt.edu/pub/pedcheck. The USERM1412 module, part of MENDEL freeware, is available via the URL http://www.sph.umich.edu/group/statgen/klange/. Links to these programs are also available at the URL http://linkage.rockefeller.edu/soft/.
References
- 1.Risch N, Merikangas K, Science 273, 1516 (1996) [DOI] [PubMed] [Google Scholar]
- 2.Wang DG et al. , Science 280, 1077 (1998) [DOI] [PubMed] [Google Scholar]
- 3.Falk CT, Rubinstein P, Ann Hum Genet 51, 227 (1987) [DOI] [PubMed] [Google Scholar]
- 4.Spielman RS, McGinnis RE, Ewens WJ, Am J Hum Genet 52(3), 506 (1993) [PMC free article] [PubMed] [Google Scholar]
- 5.Spielman RS, Ewens WJ, Am J Hum Genet 59(5), 983 (1996) [PMC free article] [PubMed] [Google Scholar]
- 6.Buetow KH, Am J Hum Genet 49, 985 (1991) [PMC free article] [PubMed] [Google Scholar]
- 7.Shields DC, Collins A, Buetow KH, Morton NE, Proc Natl Acad Sci USA 88, 6501(1991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terwilliger JD, Weeks DE, Ott J, Am J Hum Genet 47, A201 (1990) [Google Scholar]
- 9.Gordon D, Matise TC, Heath SC, Ott J, Genet Epidemiol (in press) (December 1999) [DOI] [PubMed] [Google Scholar]
- 10.Ott J, Hum Hered 43: 25 (1993) [DOI] [PubMed] [Google Scholar]
- 11.Ehm MG, Kimmel M, Cottingham RW, Am J Hum Genet 58(1), 225 (1996) [PMC free article] [PubMed] [Google Scholar]
- 12.Stringham HM, Boehnke M, Am J Hum Genet 59(4), 946 (1996) [PMC free article] [PubMed] [Google Scholar]
- 13.O’Connell JR, Weeks DE, Am J Hum Genet 63(1), 259 (1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon D, Heath SC, Ott J, Hum Hered 49(2), 65 (1999) [DOI] [PubMed] [Google Scholar]
- 15.Risch N, Zhang H, Science 268, 1584 (1995) [DOI] [PubMed] [Google Scholar]
- 16.Heath SC, Am J Hum Genet 63(4), Supplement, A292 (1998) [Google Scholar]