Abstract
Evidence from animal self-administration and human genetics studies suggests that the serotonin1B (5-HT1B) receptor may be involved in modulating responses to cocaine or alcohol. We hypothesize that polymorphisms, including single-nucleotide polymorphisms (SNPs), in the human 5-HT1B receptor gene, may be associated with individual differences in vulnerability to cocaine or alcohol abuse or dependence. A total of 210 subjects were studied, including individuals with a primary diagnosis (DSM-IV criteria) of cocaine abuse or dependence, alcohol abuse or dependence, and controls with no history of previous or current illicit drug or alcohol abuse or dependence. Genomic DNA samples were isolated from each individual. For 157 of the subjects, polymerase chain reaction (PCR) was used to amplify the entire coding region of the 5-HT1B receptor gene as well as parts of the 5’ and 3’ untranslated regions. PCR products were sequenced in forward and reverse directions on an automated sequencer. Amplified DNA from an additional 53 subjects was sequenced in the 5’ untranslated region to gain additional data on the frequency of one identified SNP. Seven polymorphisms were identified: one novel SNP in the 5’ untranslated region (UTR) of the gene (A-161T); one SNP not reported in any published scientific communication (but found to be recorded in GenBank) in the 3’ UTR (A1180G); two novel dinucleotide deletions at positions −184/−183 −182/ −181; and three previously identified SNPs (T-261G, C129T, G861C). Data were stratified by ethnicity and pooled Relative Risk was calculated for combined alcohol abuse and dependence cases and controls, and also for combined cocaine abuse and dependence cases and controls. No significant differences between cases and controls were found.
Keywords: 5-HT1B receptor gene, single-nucleotide polymorphism, cocaine, alcohol, genetics of addiction
INTRODUCTION
Cocaine and alcohol abuse and dependence are major medical and social problems. The National Institute on Drug Abuse reports that in 1997, 1.5 million Americans age 12 and older were chronic cocaine users [National Institutes of Health, 1999]. In the United States, the economic cost of alcoholism alone was estimated to total 276 billion dollars in 1995 [National Institutes of Health, 1998]. Clinical observation suggests that self-exposed individuals vary in their vulnerability to cocaine and alcohol addiction. While the basis for these variations is not well understood, there is substantial evidence that genetic factors play a contributory role [Schuckit et al., 1985; Grove et al., 1990; Pickens et al., 1991; Lin et al., 1996; Tsuang et al., 1996, 1998].
The reinforcing or rewarding effects of cocaine and alcohol are mediated by several neurochemical systems, including the serotonin (5-HT) system. The serotonin1B (5-HT1B) receptor, one of the 20 (or more) subtypes of 5-HT receptors, has been specifically implicated in contributing to the reinforcing properties of both cocaine and alcohol [Roth et al., 2000]. Evidence for a regulatory role of 5-HT1B receptors in cocaine reinforcement is provided by experiments with genetically modified (knockout) mice lacking the 5-HT1B receptor gene. Compared to wild-type controls, mice homozygous for the 5-HT1B receptor gene deletion require a shorter latency period to meet cocaine self-administration criteria [Rocha et al., 1997]. Compared to wild-type control animals, the homozygous mutant mice display increased locomotor responses to cocaine and will perform more work to receive a cocaine infusion in self-administration studies [Rocha et al., 1998]. The behavior and biochemical state of these homozygous knockout mice resemble that of wild-type mice that have been sensitized to cocaine by repeated exposure to the drug [Rocha et al., 1998]. In addition, the effect of cocaine on c-fos induction, used as a marker of cocaine-induced neural activity, is reduced in the homozygous mutant mice, suggesting that 5-HT1B receptors also mediate cellular responses elicited by cocaine [Lucas et al., 1997].
Pharmacological experiments in animals using combinations of both nonselective and selective serotonergic agonists also support a role of the 5-HT1B receptor in modulating dopaminergic regulation of cocaine reinforcement [Guan and McBride, 1989; Parsons et al., 1996]. Also, mixed 5-HT1B/1A agonists, but not specific 5-HT1A agonists, can enhance the discriminative cue produced by cocaine in rats and partially substitute for the discriminative stimulus properties of cocaine [Callahan and Cunningham, 1997]. 5-HT1B receptor agonists have been demonstrated to reduce the self-administration of cocaine by rats in a dose-dependent manner, an effect similar to that produced by increasing the unit dose of cocaine [Parsons et al., 1998].
Evidence for a role of the 5-HT1B receptor in modulating alcohol intake is provided by genetic and animal self-administration studies. A quantitative trait locus affecting alcohol drinking phenotypes in mice has been provisionally linked to the chromosomal region where the 5-HT1B receptor gene is located [Crabbe et al., 1994; Phillips et al., 1994; Rodriguez et al., 1995]. Moreover, knockout mice lacking the 5-HT1B receptor have been reported to consume twice as much alcohol as wild-type mice in a two-bottle choice paradigm [Crabbe et al., 1996]. The knockout mutation in the 5-HT1B gene in these studies was maintained on the 129 Sv-ter strain. The homozygous 5-HT1B knockout mice are also less sensitive to alcohol-induced ataxia and appear to develop tolerance to alcohol more slowly [Crabbe et al., 1996].
The human 5-HT1B receptor gene is an intronless gene that has been cloned and sequenced [Demchyshyn et al., 1992; Hamblin et al., 1992; Jin et al., 1992; Levy et al., 1992; Weinshank et al., 1992; Lappalainen et al., 1995]. The coding region consists of 1,173 base pairs (bp). In previous scientific communications, seven naturally occurring polymorphisms have been reported within the 5-HT1B receptor gene, including six single-nucleotide polymorphisms (SNPs) and one dinucleotide deletion [Nöthen et al., 1994; Lappalainen et al., 1995; Ohara et al., 1996; Huang et al., 1999]. Several genetic studies have examined potential associations between identified SNPs in the human 5-HT1B receptor gene and specific human phenotypes, including alcoholism, antisocial alcoholism, various responses to antimigraine therapy, suicide, depression, panic disorder, and pathological aggression [Ohara et al., 1996; Lappalainen et al., 1998; MaassenVanDenBrink et al., 1998; Huang et al., 1999]. In one of these studies, Finnish antisocial alcoholics were found to have a higher frequency of one specific SNP (a G-to-C substitution at nucleotide position 861) than Finnish control subjects [Lappalainen et al., 1998].
The hypothesis underlying this study is that polymorphisms, including SNPs, in the human 5-HT1B receptor gene may be associated with individual vulnerability to cocaine or alcohol abuse or dependence, either by directly altering the function or expression of the receptor, or by being in linkage disequilibrium with other known or unknown polymorphisms that affect gene expression or function of gene products. This study population includes individuals with a primary diagnosis of cocaine abuse or dependence, or a primary diagnosis of alcohol abuse or dependence. Control individuals had no alcohol or drug abuse or dependence history. Abuse and dependence were defined by DSMIV criteria, with additional criteria defined below. The study aims were to identify known and novel polymorphisms in the 5-HT1B receptor gene; to determine genotype and allele frequencies of identified polymorphisms within the sample populations; to evaluate any differences in frequencies of polymorphisms between ethnic groups or genders; and to evaluate any differences in frequencies of polymorphisms between cocaine abuse or dependence groups and controls and between alcohol abuse or dependence groups and controls.
MATERIALS AND METHODS
Study Subjects
Study subjects were unrelated individuals with a primary diagnosis (DSM-IV criteria) of cocaine abuse or dependence or alcohol abuse or dependence and control individuals with no history of prior or current illicit drug or alcohol abuse or dependence. All subjects included in this study were identified from a larger pool of subjects recruited to an ongoing study of the genetic contribution to addictions conducted at the Laboratory of the Biology of Addictive Diseases at Rockefeller University in New York City. Subjects are recruited to this study through the posting of notices and newspaper advertisements or by referral as potential research volunteers by physicians or staff at Rockefeller University or associated clinical resources.
A total of 210 subjects were included in this study (Table I). All subjects included in this study entered the ongoing Rockefeller University study between August 1995 and June 2000. The total study population (n = 210) consists of 157 subjects who were initially studied, as well as 53 additional subjects that were identified in order to provide greater statistical power in the analysis of one specific SNP. The phenotype of all study subjects remained unknown to all laboratory researchers until the final data analysis.
TABLE I.
Demography of Total Subjects Included in Study (n = 210)
| Cocaine abuse | Cocaine dependence | Alcohol abuse | Alcohol dependence | Controls | Totals | |
|---|---|---|---|---|---|---|
| Caucasian American | 7 | 11 | 14 | 5 | 44 | 81 |
| African American | 14 | 38 | 4 | 2 | 35 | 93 |
| Hispanic American | 2 | 7 | 1 | 3 | 13 | 26 |
| Caucasian American/African American | 2 | 2 | ||||
| Caucasian American/Hispanic American | 1 | 1 | ||||
| African American/Hispanic American | 4 | 1 | 1 | 6 | ||
| African American/Native American | 1 | 1 | ||||
| Totals | 23 | 63 | 20 | 10 | 94 | 210 |
Every subject entering the study was required to demonstrate competence to understand the study procedures and to understand and sign the institutional review board–approved informed consent. All subjects were rigorously characterized by a research nurse, clinical psychologist, or psychiatrist with respect to medical history, psychological and psychiatric pro-files, chemical abuse or dependence history, and ethnic background. Subjects answered several detailed questionnaires and structured interviews, including the Addiction Severity Index (ASI) [McLellan et al., 1980]. This instrument was modified to establish DSM-IV criteria for past or current diagnoses of abuse or dependence on specific drugs or alcohol. Diagnoses were made for all subjects by the same physician by review of the modified ASI. Affected subjects were categorized into groups based on predominant drug preference, as determined by self-report of preference as well as by an assessment of which drug caused the greatest amount of interference in the subject’s life. Subjects with a clinical diagnosis of schizophrenia or any other psychotic mental illness were excluded from the study. Urine samples were obtained from all subjects, including control subjects, and toxicological analyses performed for several drugs of abuse and alcohol. Blood specimens were obtained by venipuncture for molecular biological analysis, including DNA extraction and sequencing.
All subjects in the cocaine abuse or cocaine dependence group met DSM-IV criteria for either cocaine abuse or dependence. In addition, to provide greater stringency for either of these diagnoses than DSM-IV criteria, additional inclusion criteria were as follows: subject demonstrates a regular pattern of current or previous cocaine use, including either two or more weekly binges or two or more uses per day for a period of at least 6 months; subject identifies cocaine as his or her current or previous drug of choice; subject has sustained major life alterations due to cocaine use, including problems with relationships, employment, or legal matters; and subject may or may not abuse or be addicted to other drugs, either currently or previously, but does not identify any of these other drugs as a drug of primary focus. Many of the cocaine abuse or dependent individuals also abused or were dependent on alcohol, reflecting a common pattern of coabuse or codependence (Table II). A history of opiate abuse or dependence was used as an exclusion criterion for this group.
TABLE II.
| Cocaine abuse or dependence subjects | Alcohol abuse or dependence subjects | |||
|---|---|---|---|---|
| Cocaine abuse (n = 23) | Cocaine dependence (n = 63) | Alcohol abuse (n = 20) | Alcohol dependence (n=10) | |
| Alcohol abuse | 12 | 24 | ||
| Alcohol dependence | 2 | 26 | ||
| Cocaine abuse | 0 | 0 | ||
| Cocaine dependence | 0 | 0 | ||
| Opiate abuse | 0 | 0 | 0 | 0 |
Alcohol, cocaine, or opiate only; DSM-IV criteria.
All subjects included in the alcohol abuse or alcohol dependence group met DSM-IV criteria for either alcohol abuse or dependence. To provide greater stringency in the diagnosis of these disorders than described by DSM-IV criteria, the following additional inclusion criteria were used: subject demonstrates a regular pattern of current or previous alcohol use, either three or more weekly binges of four or more drinks, or daily use of three or more drinks for a period of at least 6 months; subject identifies alcohol as his or her current or previous drug of choice; subject has sustained major life alterations due to alcohol use, including problems with relationships, employment, or legal matters; and subject may or may not abuse or be addicted to other drugs, either currently or previously, but does not identify any of these other drugs as a drug of primary focus (Table II). A history of opiate abuse or dependence was used as an exclusion criterion.
All subjects included in the control group have no history of any previous or current illicit drug or alcohol abuse or dependence. Individuals who met DSM-IV criteria for any substance abuse or dependence, or who had any ongoing or prior period of drug or alcohol abuse, were excluded from this category. Additional exclusion criteria were as follows: subjects undergoing clinical management of chronic pain; subjects who do not meet DSM-IV criteria for abuse or dependence of alcohol or any other drug, including cannabis, but who have had more than two significant periods of use of alcohol or any illicit drug (excluding cannabis) during their lifetime. Individuals who had abused cannabis for no more than 12 days during the 30-day period prior to the assessment, or whose use during the 30-day period prior to the assessment did not exceed the equivalent of two cannabis cigarettes per day, were not excluded. Users of nicotine or caffeine were not excluded from the control group.
Subjects included in this study were classified as either African American, Caucasian American, or Hispanic American based on detailed information provided by the subjects regarding family origin and ethnic background. Given the ethnic composition of the study subject recruitment area, the ethnic composition of study subjects is not necessarily in proportion to national averages. Several subjects in this study reported having parents who each came from a different ethnic group. These individuals will be referred to as individuals of mixed ethnicity and are listed in Table I. All subjects for whom genotyping of each specific SNP had been successfully performed were used to calculate the overall allele frequency of each identified polymorphism and to assess any variation of allele frequencies between genders. Subjects of mixed ethnicity were not included in any analyses of the identified polymorphisms for which data were stratified by ethnicity due to the small numbers of these subjects.
Polymerase Chain Reaction (PCR) Amplification and DNA Sequencing
From genomic DNA samples isolated from each of 157 study subjects, PCR was used to amplify the entire coding region of the 5-HT1B receptor gene as well as 595 bp of the 5’ untranslated region (UTR) and 74 bp of the 3’ UTR. The GenBank sequence (accession number M75128) of the human 5-HT1B receptor gene was used to design an oligonucleotide primer pair to amplify the gene. These primers were designated H5HT1BR-595 F (5’-CAGCGCTGCTCCTAGACTTC-3’) and H5HT1BR-1247R (5’-TTCGACCTACCTGTGGAACC-3’; Table III). A PCR protocol, commonly referred to as a stepdown protocol, was performed in a 100 μl volume that contained 300–400 ng of genomic DNA, ×10 PCR buffer, 2.5 mM magnesium chloride, 0.2 mM dNTPs, 0.5 μM of each primer, and 5 U AmpliTaq Gold polymerase (Applied Biosystems, PE Biosystems, Foster City, CA). PCR was performed on a Perkin Elmer DNA Thermal Cycler model 480. An initial denaturation step at 95°C for 4 min was followed by six sets of three cycles of denaturation, annealing, and extension. In each cycle, denaturing was performed at 95°C for 30 sec and extension was performed at 72°C for 90 sec. Each set of three cycles had a progressively lower annealing temperature of 66°C, 63°C, 60°C, 57°C, and 54°C, respectively, for 30 sec (stepdown). Thirty cycles were then performed with an annealing temperature of 51°C. A final extension step was performed at 72°C for 6 min. The 1,842 bp PCR products were visualized by running 5 μl from each sample on 1% agarose gels. Prior to sequencing, the remaining PCR products were purified to remove reaction buffer and unincorporated primers using Quiagen Quiaquick kits by a Quiagen Biorobot 9600 (Quiagen, Valencia, CA).
TABLE III.
Oligonucleotide Primers Used for PCR Amplification and/or for Sequencing of the 5-HT1B Receptor Gene*
| Primer designation | Type | Sequence | Length |
|---|---|---|---|
| H5HT1BR −595F | PCR and Sequencing | 5′-CAG CGC TGC TCC TAG ACT TC-3′ | 20 |
| H5HT1BR −174F | Sequencing | 5′-GGC TGC CGC ACC CAT GAC CT-3′ | 20 |
| H5HT1BR 178F | Sequencing | 5′-TCA CCG ACC TGC TTG TGT CC-3′ | 20 |
| H5HT1BR 864F | Sequencing | 5′-CCA AGT CAA AGT GCG AGT CT-3′ | 20 |
| H5HT1BR −40R | Sequencing | 5′-ATG GAG CGG ACG AAG GAG A-3′ | 19 |
| H5HT1BR 493R | Sequencing | 5′-TCT TGG GAG TCC TTT TAG C-3′ | 19 |
| H5HT1BR 784R | Sequencing | 5′-CGG GGG AGT CGG TTA TCA C-3′ | 19 |
| H5HT1BR 1247R | PCR and sequencing | 5′-TTC GAC CTA CCT GTG GAA CC-3′ | 20 |
The designation of primers corresponds to the position within the gene using the numbering scheme that designates the first nucleotide of the start codon as position +1 (F = forward; R = reverse).
The sequences of each of the 159 column-purified PCR products were directly determined by the cycle-sequencing method (ABI PRISM Big Dye Terminators Cycle Sequencing Kit) using a PE Biosystems ABI PRISM 377 automated sequencer (Applied Biosystems). Four forward and four reverse oligonucleotide sequencing primers were used to obtain overlapping sequence information of the entire amplified product in both directions (Table III). The eight electrophero-grams were aligned and analyzed for the presence of known as well as novel polymorphisms. Each electro-pherogram was analyzed on separate occasions by two independent researchers trained in molecular biology. The researchers evaluating the electropherograms were blind with respect to phenotypic characterization of study subjects until the final data analysis.
An interesting result with respect to one of the SNPs was found in cocaine abuse or dependence cases compared to controls. Fifty-three additional subjects were therefore added to the study to provide additional statistical power in the analysis of this SNP. The genotype of these individuals at the position of this SNP was determined by similar methods to those described, except that only one forward and one reverse primer were used in sequencing.
Statistical Analysis
The overall frequencies for each polymorphism identified were determined. For the four most common polymorphisms, Chi-square tests were carried out to test the null hypothesis of no difference in allele frequencies between ethnic groups and genders. Since the numbers within each dependence or abuse group were small, and since additional criteria had been used to provide greater stringency than DSM-IV criteria, analyses were performed with data pooled within each substance group (i.e., individuals with cocaine abuse or dependence were pooled, and individuals with alcohol abuse or dependence were pooled). With data stratified by ethnicity, pooled Relative Risk (RR) and Mantel-Haenszel Chi-square tests were used to evaluate differences in Relative Risk between combined cocaine abuse and dependence cases and control subjects, and also between combined alcohol abuse and dependence cases and control subjects [Mantel and Haenszel, 1959]. Similar analyses were performed to compare cocaine-dependent individuals with controls. A third analysis of pooled Relative Risk between cases and controls was performed with cases defined as all substance abuse and dependence groups combined. Within each ethnic group, a Chi-square test was performed to determine whether each SNP was in Hardy-Weinberg equilibrium. The EH program was used to determine whether the most common SNPs were in linkage disequilibrium. [Xie and Ott, 1993; Terwilliger and Ott, 1994].
RESULTS
Polymorphic Sites
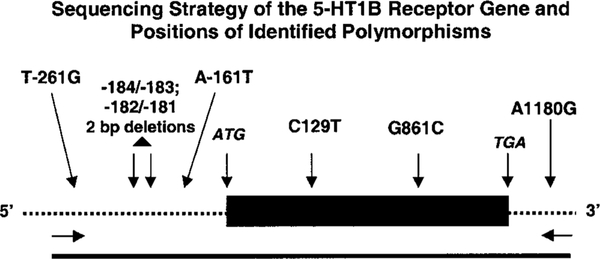
By direct sequencing of the entire coding region and portions of 5’ and 3’ noncoding sequences of the 5-HT1B gene from 157 study subjects, we identified seven polymorphic sites within the 5-HT1B receptor gene (Table IV, Fig. 1). Nucleotide positions are defined in relation to the first nucleotide of the start codon, which is designated position + 1. The term most common designates the prototypical 5-HT1B receptor gene sequence that is reported in GenBank (accession numbers M89478, M75128, M81590, D10995, A75693, AL049595).
TABLE IV.
Overall Frequencies of Identified Polymorphisms in the 5-HT1B Receptor Gene
| SNPa | Nucleotide positionb | Location in gene | Previously reported allele frequency | This study overall allele frequency (n = 157)c |
|---|---|---|---|---|
| T-261G | −261 | 5′ untranslated region | 49% (n = 46) [Nothen et al., 1994] | 32.7% |
| −184/− 183 dinucleotide deletion | −182, −181 | 5′ untranslated region | Not previously reported | 0.6% |
| −182/− 181 dinucleotide deletion | −182, −181 | 5′ untranslated region | Not previously reportedd | 2.9% |
| A-161T | −161 | 5′ untranslated region | Not previously reportedd | 18.3% |
| C129T | 129 | N-terminal domain (synonymous) | 17% (n= 178) [Huang et al., 1999] | 26.8% |
| G861C | 861 | Intracellular loop III (synonymous) | 23% (n = 640)e; 62% (n = 418)e [Lappalainen et al., 1995]; 26.8% 17% (n = 178) [Huang et al., 1999] | 26.8% |
| A1180G | 1180 | 3′ untranslated region | Not previously reportedd | 5.8% |
SNPs are named according to their position within the gene with the most commonly occurring nucleotide placed before the nucleotide position and the variant allelic nucleotide following the position.
Nucleotide position is defined according to the numbering scheme that designates the first nucleotide of the start codon as position +1.
For T-261G and A1180G, n = 156 due to ambiguous sequencing data from one subject for each allele, respectively; for A-161T, n=210 due to the genotyping of an additional 53 subjects at this position.
Following completion of the work for this study and submission of this article, these polymorphisms were found to be reposited in the National Center for Biotechnology Information SNP database, with an initial release date of 29 January, 2001. These polymorphisms have not previously been specifically reported in a scientific communication.
Two separate ethnic populations were surveyed in this report.
Fig. 1.
Amplification and sequencing strategy for the serotonin1B (5-HT1B) receptor gene. Solid rectangle represents the coding region of 5-HT1B receptor gene. Dashed lines represent 5’ and 3’ untranslated regions. Solid line represents the PCR amplified DNA used in this study. Downward arrows indicate positions of polymorphisms or start or stop codons. Horizontal arrows indicate positions of PCR primers used in amplification and sequencing.
We identified four polymorphisms in the 5-HT1B receptor gene previously unreported in scientific communications (Table IV, Fig. 1). Two novel dinucleotide deletions were observed at nucleotide positions –184/ –183 and –182/– 181. One novel SNP (A-161T) was identified in the 5’ UTR of the gene at nucleotide position – 161. One additional SNP (A1180G) that has not been reported in any published scientific communication but was found to be recorded in GenBank was identified in the 3’ UTR region of the 5-HT1B receptor gene at nucleotide position 1180. Four GenBank entries (M75128, M81590, M89478, and AL049595) report a guanine nucleotide at position 1180, while two entries (D10995, A75693) list an adenosine nucleotide at this position. Since we observed an adenosine nucleotide at position 1180 in 94.3% of the samples, we will refer to the SNP as A1180G, where the most common allele has an adenosine and the variant form has a guanine at this position.
Three SNPs that have been previously reported were identified (T-261G, C129T, G861C; Table IV, Fig. 1). Each of these SNPs was found to occur at similar overall frequencies to those previously published. As noted in prior studies, the G129T and G861C SNPs were in absolute linkage disequilibrium with each other [Huang et al., 1999]. The previously reported G-511T, G276A, and T371G SNPs, as well as the dinucleotide deletion at positions – 179/– 178, were not found in this study population [Nöthen et al., 1994; Ohara et al., 1996].
Overall frequencies for the identified polymorphisms in the study population are presented in Table IV. For most of the polymorphisms, the study population consists of 157 subjects. For the A-161T SNP, the study population consists of 210 subjects since the genotypes of an additional 53 subjects were determined at position –161. In two separate samples, electropherograms at either position – 261 or 1180 could not be clearly interpreted due to poor quality of the sequencing electropherogram at that position; the total number of subjects included in the analysis of the T-261G and A1180G SNPs is therefore 156. For each identified polymorphism except the two rare dinucleotide deletions at positions – 184/–183 and – 182/–181, individuals heterozygous and homozygous for each substitution were identified. Genotype distributions for the four most common SNPs are given in Table V.
TABLE V.
Genotype Frequencies of Common SNPs
| T-261G(n=156) | A-161T(n=210) | C129T and G861Ca (n=157) | A1180G(n=156) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TG | GG | AA | AT | TT | CC/GG | CT/GC | TT/CC | AA | AG | GG | |
| Number of subjects (%) | 68 (43.6%) | 74 (47.4%) (9.0%) | 14(9.0%) | 139 (66.2%) | 65 (30.9%) | 6(2.9%) | 79 (50.3%) | 72 (45.9%) | 6 (3.8%) | 140 (89.7%) | 14 (9.0%) | 2 (1.3%) |
The C129T and C861C SNPs have been previously reported to be in complete linkage disequilibrium [Huang et al., 1999]. This finding was also observed in the study group reported here.
Finally, in this study three cytosine residues were found preceding the guanine nucleotide at position –183 in every sample analyzed. This sequence is in agreement with GenBank entries M89478 and AL049595. However, GenBank entry M75128 records only two cytosine residues preceding the guanine at position – 183. This GenBank entry may represent a sequencing error or, alternatively, a rare polymorphism.
Each ethnic group was evaluated for deviation from Hardy-Weinberg equilibrium for the four most common SNPs (T-261G, A-161T, and combined allele frequencies for the C129T and G861C SNPs, which were found in absolute linkage disequilibrium). In no case could the null hypothesis of Hardy-Weinberg equilibrium be rejected. In addition, it was tested whether these SNPs were in linkage disequilibrium with each other. Within each of the three ethnic groups, there was significant evidence for linkage disequilibrium among all of these common SNPs. This finding is not surprising given their close physical proximity.
Allelic Variation Between Genders and Ethnicities
No statistically significant differences in allele frequencies for any of the identified SNPs were found between genders. Allele frequencies of all four SNPs (T-261G, A-161T, and the combined C129T/G861C SNPs) were found to differ significantly between African Americans, Hispanic Americans, and Caucasian Americans (Table VI). The possible significance of the variation in allele frequencies of the remaining three polymorphisms (–184/–183 dinucleotide deletion –182/– 181 dinucleotide deletion, A1180G) was not assessed due to their low frequencies in the populations studied. We also found no evidence for differences in haplotypes between cases and controls. Because the number of individuals homozygous for the variant alleles was relatively small, allele frequencies rather than genotype frequencies were used to test for significant differences.
TABLE VI.
Allele Distribution and Analysis of Common SNPs Grouped by Ethnicity*
| T-261G(n=147) | A-161T(n = 200) | C129T and G861T (n= 148)a | ||||
|---|---|---|---|---|---|---|
| T | G | A | T | C/G | T | |
| Caucasian American | 68 (56.7%) | 52 (43.3%) | 115 (71.0%) | 47 (29.0%) | 79 (64.8%) | 43 (35.2%) |
| African American | 100 (78.1%) | 28 (21.9%) | 174 (93.5%) | 12 (6.5%) | 105 (82.0%) | 23 (18.0%) |
| Hispanic American | 30 (65.2%) | 16 (34.82%) | 37 (71.1%) | 15 (28.9%) | 31 (67.4%) | 15 (32.6%) |
| Chi-square=13.08; | P=0.0014 | Chi-square=33.47; | P<0.0001 | Chi-square=10.13; | P=0.0063 | |
Individuals of mixed ethnicity are not included in analysis.
The C129T and G861C SNPs have been previously reported to be in complete linkage disequilibrium [Huang et al., 1999]. This finding was also observed in the study group reported here.
Allelic Variation Between Cocaine or Alcohol Abuse or Dependence Groups and Controls
Given the significant differences in allele frequencies between ethnic groups and the unequal distribution of ethnicities within each specific combined drug abuse and dependence group, the combined cocaine abuse and dependence group, the combined alcohol abuse and dependence group, and the control group were stratified by ethnicity to reduce the potential for problems that might be introduced into the analyses by population admixture. Because of small subject numbers and with the increased stringency of categorization beyond DSM-IV criteria for abuse and for dependence, the pooled cocaine abuse and dependence group was compared to controls, and the pooled alcohol abuse and dependence group was compared to controls. Using data stratified by ethnicity, pooled Relative Risk and Mantel-Haenszel Chi-square analysis was performed for each of the four most frequent polymorphisms. No statistically significant differences in pooled Relative Risk were found for any of the SNPs analyzed (Table VII). Additional comparisons were also made for pooled Relative Risk between cocaine-dependent cases (without abuse group) and controls; no significant differences were observed in this separate analysis. A similar analysis of alcohol-dependent cases compared to controls was not performed because of the small number of alcohol-dependent subjects. A third analysis was also performed with all substance and abuse groups (all cocaine and alcohol cases combined) compared to controls. No significant differences were found in allele frequencies between controls and combined cases. The low overall frequencies of the –184/–183 and –182/–181 dinucleotide deletions and the A1180G SNP did not allow statistical analysis to be performed on these polymorphisms.
TABLE VII.
Allele Distribution and Analysis of Common SNPs Stratified by Ethnicity and Grouped by Combined Drug Abuse and Dependence Categories*
| Cocaine | Alcohol | Controls | ||||
|---|---|---|---|---|---|---|
| T-261G(n=147) | T | G | T | G | T | G |
| Caucasian American | 16 (57.1%) | 12 (42.9%) | 20 (55.6%) | 16 (44.4%) | 32 (57.1%) | 24 (42.9%) |
| African American | 56 (84.8%) | 10 (15.2%) | 10 (83.3%) | 2 (16.7%) | 34 (68.0%) | 16 (32.0%) |
| Hispanic American | 9 (75.00%) | 3 (25.00%) | 4 (50.0%) | 4 (50.0%) | 17 (65.4%) | 9 (34.6%) |
| Cocaine | Alcohol | Controls | ||||
| A-161T(n = 200) | A | T | A | T | A | T |
| Caucasian American | 30 (83.3%) | 6 (16.7%) | 27 (71.1%) | 11 (28.9%) | 58 (65.9%) | 30 (34.1%) |
| African American | 96 (92.3%) | 8 (7.7%) | 12 (100.0%) | 0 (0.0%) | 66 (94.3%) | 4 (5.7%) |
| Hispanic American | 15 (83.3%) | 3 (16.7%) | 4 (50.0%) | 4 (50.0%) | 18 (69.2%) | 8 (30.8%) |
| Cocaine | Alcohol | Controls | ||||
| C129T/G861C(n=148) | C | T | C | T | C | T |
| Caucasian American | 19 (67.9%) | 9 (32.1%) | 23 (60.5%) | 15 (39.5%) | 37 (66.1%) | 19 (33.9%) |
| African American | 53 (80.3%) | 13 (19.7%) | 8 (66.7%) | 4 (33.3%) | 44 (88.0%) | 6 (12.0%) |
| Hispanic American | 9 (75.0%) | 3 (25.0%) | 5 (62.5%) | 3 (37.5%) | 17 (65.4%) | 9 (34.6%) |
Individuals of mixed ethnicity are not included in calculations. Pooled Relative Risk (RR) and Mantel-Haenszel Chi-square test on combined cocaine abuse and dependence and control subjects stratified by ethnicity are as follows. For T-261G, RR (cocaine vs. controls) =1.63, Chi-square =2.26, P =0.133. For A-161T, RR (cocaine vs. controls) = 1.72, Chi-square = 2.13, P = 0.145. For C129T/G861C, RR (cocaine vs. controls) =0.90, chi-square =0.032, P = 0.857. Pooled relative risk and Mantel-Haenszel chi-square test on combined alcohol abuse and dependence and control subjects stratified by ethnicity are as follows. For T-261G, RR (alcohol vs. controls) =1.02, Chi-square =0.01, P =0.919. For A-161T, RR (alcohol vs. controls) =1.11, Chi-square =0.01, P =0.922. For C129T/G861C, RR (alcohol vs. controls =0.66, Chi-square =1.12, P =0.291.
DISCUSSION
In this study, we sequenced the entire coding region of the 5-HT1B receptor gene, as well as 595 bp of the 5’ UTR and 74 bp of the 3’ UTR, from 157 subjects that included individuals with a primary diagnosis of cocaine abuse or dependence or alcohol abuse or dependence and control subjects with no current or prior history of illicit drug or alcohol abuse or dependence. Part of the 5’ UTR was also sequenced in an additional 53 study subjects in order to gain more information about the frequency of the A-161T SNP.
We identified seven polymorphisms in the 5-HT1B receptor gene: one novel SNP of high allelic frequency in the 5’ UTR of the gene (A-161T); a second SNP of high allelic frequency not reported in any published scientific communication (but recorded as a sequence discrepancy in GenBank) in the 3’ UTR (A1180G); two novel dinucleotide deletions at positions –184/–183 and –182/–181, one of moderate frequency and one of rare frequency; and three previously reported high-allelic-frequency SNPs (T-261G, C129T, G861C). The seven polymorphisms that were identified are either in nontranslated regions or are synonymous changes that do not alter the predicted amino acid sequence of the receptor. In addition, an extra cytosine residue preceding position –183 that is not recorded in one of three GenBank entries was observed in both chromosomes of every subject analyzed.
Provocatively, three different dinucleotide deletions at positions –179/–178, –182/–181, and –184/–183 have been identified between the findings in this study or in previous studies (Nöthen et al. [1994] and National Center for Biotechnology Information SNP database). A hypervariable region of the gene may therefore exist at this position. We found three cytosine residues preceding the guanine at position –183, confirming the published sequence of two GenBank entries (M89478, AL049595) but disagreeing with a third GenBank entry (M75128) that reports only two cytosine residues at this position. Given the several (albeit rarely occurring) polymorphisms in region surrounding position – 183, it is possible that GenBank entry M75128 might represent the polymorphic sequence of a rare single-nucleotide deletion rather than a sequencing error.
Four SNPs identified in this study were found to vary significantly between ethnic groups. This is not unexpected since allele frequencies are known to vary between ethnic groups and significant differences in allele frequencies between ethnic groups have been reported for SNPs in a study of individuals recruited from the same geographic area [Bond et al., 1998].
With data stratified by ethnicity, no difference in pooled Relative Risk was observed for any of the polymorphisms between any of the group comparisons made. It should be noted that the sample sizes for this study were small. For example, given the samples sizes for the cocaine cases compared to controls, in order to detect a difference in allele frequency of the T-216G SNP between the two at the level of α= 0.05 and (1 – β) = 0.08, with the observed allele frequency of 62.9% in controls, an odds ratio of 2.4 (allele frequency80.0% in cases) would be required. The observed combined allele frequency for the cases was 76.4%. For each of the common SNPs studied, given the allele frequencies of these SNPs in the study groups, larger sample sizes would be necessary to assess the significance of the observed differences in allele frequencies between cases and controls.
While this study is the first association study of SNPs in the 5-HT1B receptor gene and cocaine abuse or dependence, three separate populations of individuals with histories of alcoholism have previously been analyzed for a potential association with one SNP, G861C. In one study, no association was found between 64 individuals with history of alcoholism and 60 controls [Huang et al., 1999]. In another study, with a population sample of 418 Southwestern American Indians, the G861C SNP was not found to be associated with either antisocial alcoholism, nonantisocial alcoholism, or combined antisocial and nonantisocial alcoholics [Lappalainen et al., 1998]. However, in the same report, a significantly higher frequency of the 861C allele was observed in a sample of 183 Finnish antisocial alcoholics compared to 457 Finnish controls (P = 0.005). No association was found between non-antisocial alcoholics and controls [Lappalainen et al., 1998]. Our study, with a small number of alcohol abuse or dependence subjects, does not support an association between combined alcohol abuse and dependence and the G861C SNP.
For complex diseases such as specific substance addictions, it is likely that a combination of specific variations in multiple genes, rather than a single variation in a single gene, will confer vulnerability or protection. By identifying SNPs and other polymorphisms in specific genes and analyzing potential associations between specific alleles and various phenotypes, our study represents a necessary step toward ultimately identifying such combinations. The novel polymorphism identified in this study may also be useful in future association studies of the wide variety of other conditions in which the serotonin system has been implicated, including depression, anxiety, aggression, obsessive-compulsive disorder, attention-deficit disorder, and schizophrenia. Also, the identified SNPs may prove useful for linkage disequilibrium studies and may serve as valuable markers for scanning the genome.
ADDENDUM
Following completion of all work for this study and initial review of this article, several of the polymorphisms identified in this report were released in the SNP database maintained by the National Center for Biotechnology Information. The initial release date for these SNPs in the database was 29 January 2001.
ACKNOWLEDGMENTS
We extend our gratitude to all subjects who participated in the study. Special thanks to Ann Ho and Susan Russo of the Laboratory of the Biology of Addictive Diseases, to Jurg Ott of the Laboratory of Statistical Genetics, and to the staff at the Protein DNA Technology Center at Rockefeller University. We also thank the Howard Hughes Medical Institute and the National Institutes of Health for their support.
Grant sponsor: NIH-NIDA; Grant numbers: DA05130, DA00049, DA12848; Grant sponsor: NIH-NIMH; Grant number: MH44292; Grant sponsor: NIH-NCRR; Grant number: RR00102; Grant sponsor: Howard Hughes Medical Institute Research Fellowship for Medical Students 1999–2000.
REFERENCES
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. 1998. Single-nucleotide polymorphism in the human mu opioid receptor gene alters b-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA 95:9608–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan PM, Cunningham KA. 1997. Modulation of the discriminative stimulus properties of cocaine: comparison of the effects of fluoxetine with 5-HT1A and 5-HT1B receptor agonists. Neuropharmacology 36: 373–381. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Belknap JK, Buck KJ. 1994. Genetic animal models of alcohol and drug abuse. Science 264:1715–1723. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Feller DJ, Hen R, Wenger CD, Lessov CN, Schafer GL. 1996. Elevated alcohol consumption in null mutant mice lacking 5-HT1B serotonin receptors. Nat Genet 14:98–101. [DOI] [PubMed] [Google Scholar]
- Demchyshyn L, Sunahara RK, Miller K, Teitler M, Hoffman BJ, Kennedy JL, Seeman P, Van Tol HHM, Niznik HB. 1992. A human serotonin 1D receptor variant (5HT1Dβ) encoded by an intronless gene on chromo-some 6. Proc Natl Acad Sci USA 89:5522–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove WM, Eckert ED, Heston L, Bouchard TJ Jr, Segal N, Lykken DT. 1990. Heritability of substance abuse and antisocial behavior: a study of monozygotic twins reared apart. Biol Psychiatry 27:1293–1304. [DOI] [PubMed] [Google Scholar]
- Guan X-M, McBride WJ. 1989. Serotonin microinfusion into the ventral tegmental area increases accumbens dopamine release. Brain Res Bull 23:541–547. [DOI] [PubMed] [Google Scholar]
- Hamblin MW, Metcalf MA, McGuffin RW, Karpells S. 1992. Molecular cloning and functional characterization of a human 5-HT1B serotonin receptor: a homologue of the rat 5-HT1B receptor with 5-HT1D-like pharmacological speci®city. Biochem Biophys Res Comm 184:752–759. [DOI] [PubMed] [Google Scholar]
- Huang Y-Y, Grailhe R, Arango V, Hen R, Mann JJ. 1999. Relationship of psychopathology to the human serotonin1B genotype and receptor binding kinetics in postmortem brain tissue. Neuropsychopharmacology 21:238–246. [DOI] [PubMed] [Google Scholar]
- Jin H, Oksenberg D, Ashkenazi A, Peroutka SJ, Duncan AMV, Rozmahel R, Yang Y, Mengod G, Palacios JM, O’Dowd BF. 1992. Characterization of the human 5-Hydroxytryptamine1B receptor. J Biol Chem 267:5735–5738. [PubMed] [Google Scholar]
- Lappalainen J, Dean M, Charbonneau L, Virkkunen M, Linnoila M, Goldman D. 1995. Mapping of the serotonin 5-HT1Dβ autoreceptor gene on chromosome 6 and direct analysis for sequence variants. Am J Med Genet 60:157–161. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Long JC, Eggert M, Ozaki N, Robin RW, Brown GL, Naukkarinen H, Virkkunen M, Linnoila M, Goldman D. 1998. Linkage of antisocial alcoholism to the serotonin 5-HT1B receptor gene in 2 populations. Arch Gen Psychiatry 55:989–994. [DOI] [PubMed] [Google Scholar]
- Levy FO, Gudermann T, Perez-Reyes E, Birnbaumer M, Kaumann AJ, Birnbaumer L. 1992. Molecular cloning of a human serotonin receptor (S12) with a pharmacological profile resembling that of the 5-HT1D subtype. J Biol Chem 267:7553–7562. [PubMed] [Google Scholar]
- Lin N, Eisen SA, Scherrer JF, Goldberg J, True WR, Lyons MJ, Tsuang MT. 1996. The influence of familial and non-familial factors on the association between major depression and substance abuse/dependence in 1874 monozygotic male twin pairs. Drug Alc Depend 43:49–55. [DOI] [PubMed] [Google Scholar]
- Lucas JJ, Segu L, Hen R. 1997. 5-Hydroxytryptamine1B receptors modulate the effect of cocaine on c-fos expression: converging evidence using 5-hydroxytryptamine1B knockout mice and the 5-hydroxytryptamine1B/1D antagonist GR127935. Mol Pharmacol 51:755–763. [DOI] [PubMed] [Google Scholar]
- MaassenVanDenBrink A, Vergouwe MN, Ophoff RA, Saxena PR, Ferrari MD, Frants RR. 1998. 5-HT1B receptor polymorphism and clinical response to sumatriptan. Headache 38:288–291. [DOI] [PubMed] [Google Scholar]
- Mantel N, Haenszel W. 1959. Statistical aspects of analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748. [PubMed] [Google Scholar]
- McLellan AT, Luborsky L, Woody GE, O’Brien CP. 1980. An improved diagnostic evaluation instrument for substance abuse patients: the addiction severity index. J Nerv Ment Dis 168:26–33. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health. 1998. Economic costs of alcohol and drug abuse estimated at $246 billion in the United States. National Institutes of Health-National Institute on Drug Abuse News Release, 13 May 1998. [Google Scholar]
- National Institutes of Health. 1999. Cocaine abuse and addiction National Institute on Drug Abuse Research Report Series. Rockville, Maryland: National Clearinghouse on Alcohol and Drug Information. [Google Scholar]
- Nöthen MM, Erdmann J, Shimron-Abarbanell D, Propping P. 1994. Identification of genetic variation in the human serotonin1Dβ receptor gene. Biochem Biophys Res Comm 205:1194–1200. [DOI] [PubMed] [Google Scholar]
- Ohara K, Xie D-W, Ishigaki T, Deng Z-L, Nakamura Y, Suzuki Y, Miyasato K, Ohara K. 1996. The genes encoding the 5HT1Dα and 5HT1Dβ receptors are unchanged in patients with panic disorder. Biol Psychiatry 39:5–10. [DOI] [PubMed] [Google Scholar]
- Parsons LH, Weiss F, Koob GF. 1996. Serotonin1B receptor stimulation enhances dopamine-mediated reinforcement. Psychopharmacology 128:150–160. [DOI] [PubMed] [Google Scholar]
- Parsons LH, Weiss F, Koob GF. 1998. Serotonin1B receptor stimulation enhances cocaine reinforcement. J Neurosci 18:10078–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK. 1994. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res 18:931–941. [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. 1991. Heterogeneity in the inheritance of alcoholism. Arch Gen Psychiatry 48:19–28. [DOI] [PubMed] [Google Scholar]
- Rocha BA, Ator R, Emmett-Oglesby MW, Hen R. 1997. Intravenous cocaine self-administration in mice lacking 5-HT1B receptors. Pharmacol Biochem Behav 57:407–412. [DOI] [PubMed] [Google Scholar]
- Rocha BA, Scearce-Levie K, Lucas JJ, Hiroi N, Castanon N, Crabbe JC, Nestler EJ, Hen R. 1998. Increased vulnerability to cocaine in mice lacking the serotonin-1B receptor. Nature 393:175–178. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. 1995. Alcohol acceptance, preference, and sensitivity in mice. II. Quantitative trait loci mapping analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res 19:367–373. [DOI] [PubMed] [Google Scholar]
- Roth BL, Lopez E, Patel S, Kroeze WK. 2000. The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist 6:252–262. [Google Scholar]
- Schuckit MA, Li TK, Cloninger CR, Deitrich RA. 1985. Genetics ofAlcoholism. Alcohol Clin Exp Res 9:475–492. [DOI] [PubMed] [Google Scholar]
- Terwilliger J, Ott J. 1994. Handbook of human genetic linkage. Baltimore:Johns Hopkins University Press. [Google Scholar]
- Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, Meyer JM, Toomey R, Faraone SV, Eaves L. 1996. Genetic influences on DSM-IIIR drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet 67:473–477. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. 1998. Co-occurrence of abuse of different drugs in men: the role of drug-speci®c and shared vulnerabilities. Arch Gen Psychiatry 55:967–972. [DOI] [PubMed] [Google Scholar]
- Weinshank RL, Zgombick JM, Macchi MJ, Branchek TA, Hartig PR. 1992. Human serotonin 1D receptor is encoded by a subfamily of two distinct genes: 5-HT1Dα and 5-HT1Dβ. Proc Natl Acad Sci USA 89:3630–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Ott J. 1993. Testing linkage disequilibrium between a disease gene and marker loci. Am J Hum Genet 53:1107.8105690 [Google Scholar]