Abstract
A gene therapeutic platform needs to be both efficient and safe. The criterion of safety is particularly important for diseases like hepatocellular carcinoma (HCC), which develop in a background of an already compromised liver. Gene vectors can be constructed either by targeting HCC or by detargeting liver and/or other major organs. miRNA-based negative detargeting has gained considerable attention in recent times due to its effectiveness and the ease with which it can be adapted into current gene delivery vectors. In this study, we provide a proof-of-concept using miRNA199a as a negative targeting agent. We introduced vectors harboring reporters with miRNA199a binding sites in cells expressing high endogenous levels of miRNA199a and compared the reporter expression in HCC cells with low endogenous miRNA199a. We observed that the expression of reporters with miRNA199a binding sites is significantly inhibited in miRNA199a-positive cells, whereas minimal effect was observed in miRNA199a-negative HCC cells. In addition, we created a post-transcriptionally regulated suicide gene therapeutic system based on cytosine deaminase (CD)/5-fluorocytosine (5-FC) exploiting miRNA199a binding sites and observed significantly lower cell death for miRNA199a-positive cells. Furthermore, we observed a decrease in the levels of miRNA199 in 3D tumorspheres of miRNA199a-positive Hepa1-6 cells and a reduction in the inhibition of reporter expression after transfection in these 3D models when compared with 2D Hepa1-6 cells. In summary, we provide evidence of miRNA199a-based post-transcriptional detargeting with relevance to HCC gene therapy.
Keywords: gene therapy, post-transcriptional targeting, heptocellular carcinoma, miRNA, miRNA199a
Introduction
Hepatocellular carcinoma (HCC) accounts for the majority of liver cancers, one of the cancer types with high incidence-to-mortality rates.1 The limitation of available therapies, especially at advanced stages, is clearly highlighted by their palliative nature and rising trends in disease-associated mortality and morbidity rates.1, 2 Moreover, the presence of underlying liver dysfunction in most patients limits the application of conventional therapies like radio and chemotherapy.3 Sorafenib, with only a modest survival benefit, is the only US Food and Drug Administration (FDA)-approved drug for late-stage HCC.4 Given the etiology of HCC, targeted therapies that can limit the treatment to HCC while sparing the liver could have a significant therapeutic benefit.5 In this regard, gene therapy, which provides several strategies that can be exploited to target a certain cell type, may be particularly attractive.5, 6
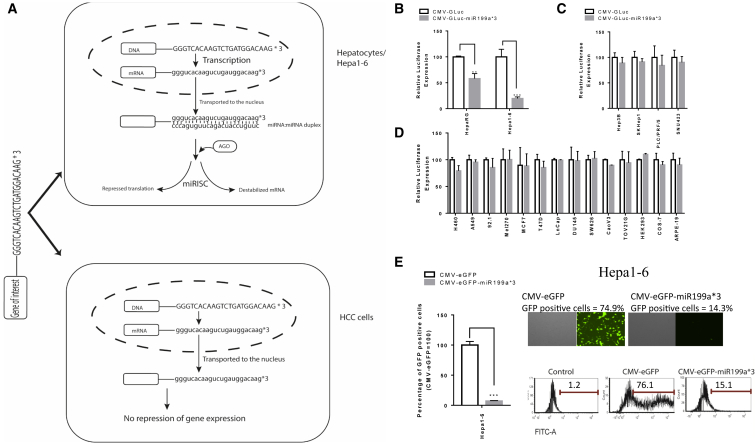
Gene therapy offers different approaches for cancer-targeted gene delivery; these include (but are not limited to) modification of gene delivery vehicle (vector) and modification of the therapeutic payload (controlled expression of the transgene).7 The former mostly comprises the use of capsid-modified viral vectors with an altered tropism, displaying a preference for cancer cells6. Limiting the expression of the therapeutic gene in cancer cells by transcriptional targeting, i.e., exploiting tumor-specific promoters, is another strategy with proven efficacy.8, 9 However, this method is limited by the number of available promoters with a strong HCC specificity.6, 10 Post-transcriptional regulation of gene expression by utilizing cell-specific endogenous microRNAs (miRNAs) is an emerging approach for targeted gene expression. Since the first evidence of effective application of this approach in antigen-presenting cells,11 a number of studies have successfully used binding sites of tissue or disease-specific miRNAs for regulating transgene expression.12, 13, 14 In this negative targeting method, binding sites of miRNA expressed at high levels in target cells are incorporated at the UTRs of transgene, and, as a result, transgene expression gets inhibited in those cells (Figure 2A).15
Figure 2.
Reporter Gene Expression under the Control of miRNA199a Binding Sites
(A) Principle of miRNA-mediated detargeting: inclusion of binding sites of miRNA expressed at high levels in a cell can limit the expression of transgene in that particular cell, while minimal effects are expected in cells that do not express the miRNA. (B) To study the effect of the presence of miRNA199a binding sites at the 3′ UTR of a transgene, miRNA199a-positive cells HepaRG and Hepa1-6 were transfected with CMV-GLuc and CMV-GLuc-miR199a*3; GLuc expression with CMV-GLuc-miR199a*3 was reported as a percentage of CMV-GLuc. Similarly, miRNA199a-negative (C) HCC cell lines and (D) non-HCC cell lines were transfected with CMV-GLuc and CMV-GLuc-miR199a*3, and the amount of secreted GLuc was quantified. (E) Further confirmation of miRNA199a-mediated gene regulation was performed by transfection of Hepa1-6 cells with CMV-EGFP and CMV-EGFP-miR199a*3; 72 hr post-transfection, the percentage of GFP-positive cells was quantified with flow cytometry, and the percentage of GFP-positive cells after transfection with CMV-EGFP-miR199a*3 was normalized with CMV-EGFP. A two-tailed t test was used for statistical analysis to compare the percent expression of CMV-GLuc-miR199a*3 or CMV-EGFP-miR199a*3 with that of CMV-GLuc or CMV-EGFP, respectively, for both HepaRG and Hepa1-6 using GraphPad Prism 7.0 (data are reported as mean ± SD, n > 3; **p < 0.005; ***p < 0.001).
In this proof-of-concept study, we demonstrate miRNA199a-based detargeting after gene delivery. First, we probed the expression of miRNA199a in cryopreserved human hepatocytes, HepaRG, and a panel of HCC and non-HCC cell lines. After observing a significant downregulation of miRNA199a in HCC and non-HCC tumor cell lines (except for Hepa1-6, which expressed miRNA199a at levels comparable to those of primary hepatocytes and HepaRG cells), we constructed expression vectors with miRNA199a binding sites at the 3′-UTR of reporters EGFP and Gaussia luciferase (GLuc) and transfected both miRNA199a-positive and -negative cells with these plasmids. A significant inhibition of the expression of reporter was observed in cells with high endogenous miRNA199a levels, while negligible effects were seen in others. These findings were further validated by targeted gene-directed enzyme prodrug therapy (GDEPT) using the cytosine deaminase/5-fluorocytosine (CD/5-FC) system. Next, we demonstrate that the inhibition of miRNA199 in miRNA199-positive cells can rescue the expression of transgenes with miR199a binding sites, while its overexpression with miRNA mimic can inhibit the same in miRNA199a-negative HCC cells.
Furthermore, the possibility of delivering these constructs with an adeno-associated virus (AAV)-based delivery system was explored, and the results obtained with transfection experiments were corroborated. Additionally, we discovered that the levels of miRNA199a are downregulated in CD133+ CD44+ Oct4+-expressing tumorspheres of Hepa1-6, illustrating the potential to use miRNA199a to target these 3D models of HCC. Finally, we demonstrate that tumorspheres of Hepa1-6 can be efficiently targeted with miRNA199a-binding-site-containing expression vectors. In conclusion, this study provides evidence that negative targeting after gene delivery can be achieved in hepatocytes by exploiting miRNA199a and suggests that this principle of negative targeting with miRNA199a could be exploited for other cell types expressing high endogenous miRNA199a levels.
Results
miRNA199a Is Downregulated in HCC and Other Cancer Cell Lines
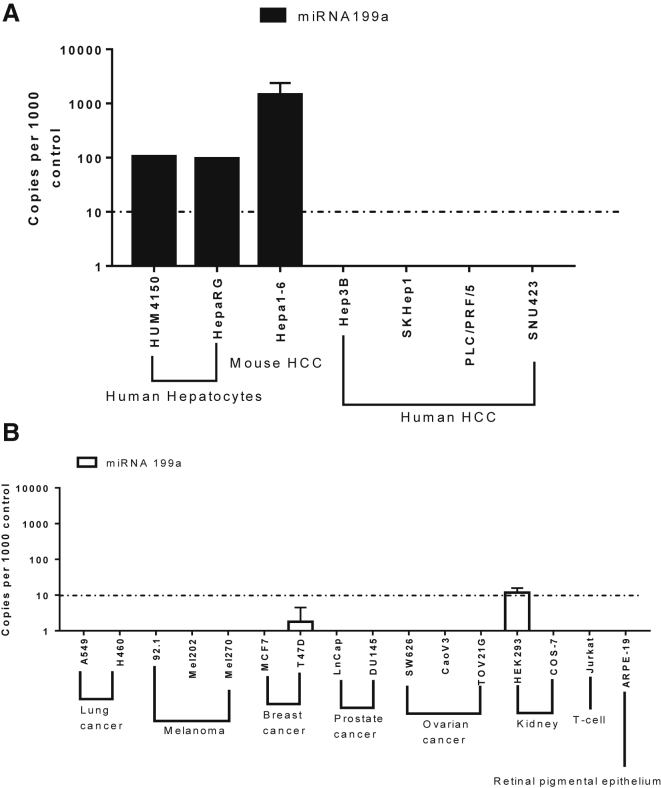
To investigate the expression pattern of miRNA199a, we performed real-time qPCR on cDNA obtained from total RNA extracted from primary hepatocytes, HepaRG, and a panel of HCC and non-HCC cell lines. After quantifying the amounts of miRNA199a with qPCR, high levels of miRNA199a were observed in cryopreserved human primary hepatocytes and HepaRG cells (92 copies per 1,000 copies of control) (Figure 1A). Similarly, the HCC cell line Hepa1-6 was found to have higher copies of miRNA199a (498 copies per 1,000 copies of RNU control) (Figure 1A). In HCC cell lines Hep3B, PLC/PRF/5, SKHep1, and SNU423, no copies of miRNA199a were detected using this method. Similarly, little or no expression of miRNA199a was observed in the non-HCC cancer cell lines used in this study (Figure 1B). Together, these results indicated that miRNA199a is downregulated in HCC as well as non-HCC tumor cell lines and established Hepa1-6 as a model for studying different aspects of miRNA199a.
Figure 1.
Expression Pattern of miRNA199a
The expression levels of miRNA199a in primary hepatocytes HepaRG and a panel of HCC (A) and non-HCC cell lines (B) as assayed by real-time qPCR. The copies of miRNA199a were normalized against those of the RNU6 control and represented as the number of copies per 1,000 control (n > 3 for all cell lines except HUM4150).
Incorporation of miRNA199a Binding Sites at the 3′ UTR of a Transgene Allows Negative Targeting of Cells with High Endogenous Expression of miRNA199a
To explore the effects of inclusion of miRNA199a binding sites at the 3′ UTR of a transgene, we constructed expression plasmids with GLuc with 3 miRNA199a binding sites at the 3′ UTR (cytomegalovirus [CMV]-GLuc-miR199a*3). These plasmids were then transfected into miRNA199a-positive cell lines HepaRG and Hepa1-6, as well as miRNA199a-negative HCC and non-HCC cell lines. The percentage of secreted luciferase after transfection with CMV-GLuc-miR199a*3 was observed to be significantly lower when compared to that after transfection with CMV-GLuc for both HepaRG (p < 0.005), and Hepa1-6 (p < 0.001) (Figure 2B). Interestingly, the decrease in luciferase expression after transfection with CMV-GLuc-miR199a*3 in HepaRG cells was observed to be 2.94 times (p < 0.001) higher than that observed for Hepa1-6, which correlated with the copies of miRNA199a present in those cells. As expected no significant decrease in GLuc expression was observed in HCC cell lines Hep3B, SKHep1, PLC/PRF/5, and SNU423 (Figure 2C), as well as non-HCC tumor cell lines (Figure 2D) after transfection with CMV-GLuc-miR199a*3. To further confirm this observed miRNA199a-mediated post-transcriptional control of gene expression, we used a second reporter EGFP. Hepa1-6 cells were transfected with either CMV-EGFP or CMV-EGFP-miR199a*3, and GFP-positive cells were quantified by flow cytometry. Like the GLuc reporter, a significantly reduced GFP expression was observed after transfection with CMV-EGFP-miR199a*3 when compared to CMV-EGFP (p < 0.001) (Figure 2E). Together, these results provided evidence that the incorporation of miRNA199a binding sites at the 3′ UTR of a gene can inhibit its expression in cells with high endogenous levels of miRNA199a.
Overexpression of miRNA199a in HCC Cells Inhibits Expression of Transgene with Its Binding Sites at the 3′ UTR, while Its Inhibition Rescues the Expression of the Same in Cells with High Endogenous Expression Levels
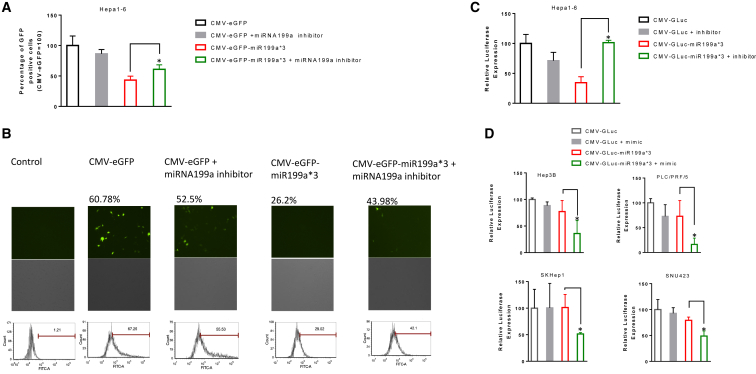
Next, we altered miRNA199a levels in Hepa1-6 and HCC cells using inhibitors and mimics to define miRNA199a’s role in the expression of reporters with binding sites incorporated in the 3′ UTR. We co-transfected miRNA199a inhibitor with either CMV-EGFP-199a*3 or CMV-GLuc-miR199a*3 and compared expression to CMV-GLuc or CMV-EGFP controls. A significant increase (p < 0.05) in the percentage of GFP-positive cells was observed after co-transfection of Hepa1-6 with CMV-EGFP-miR199a*3 and miRNA199a inhibitor when compared to transfection with CMV-EGFP-miR199a*3 (both reported as relative to CMV-EGFP) (Figures 3A and 3B). Similarly, miRNA199a inhibition by co-transfection of Hepa1-6 with CMV-GLuc-miR199a*3 and miRNA199a inhibitor resulted in a 2.9-fold increase (p < 0.05) of GLuc expression when compared to GLuc expression after transfection with CMV-GLuc-miR199a*3 alone (both normalized to CMV-GLuc control) (Figure 3C). In contrast, co-transfection of CMV-GLuc-miR199a*3 and miRNA199a mimic in miRNA199a-negative HCC cell lines Hep3B, PLC/PRF/5, SKHep1, and SNU423 resulted in a significant decrease (Figure 3D) (p < 0.05) in GLuc expression when compared to CMV-GLuc-miR199a alone (all normalized to CMV-GLuc control). Taken together, these results further strengthen the observation of miRNA199a-mediated post-transcriptional regulation of expression of reporter gene containing its binding sites at the 3′ UTR and provide evidence that its inhibition can rescue the expression of these reporters.
Figure 3.
Overexpression and Inhibition of miRNA199a and Its Effect on the Expression of Reporters Containing miRNA199a Binding Sites at the 3′ UTR
To investigate the effects of inhibiting miRNA199a on the expression of reporter genes with binding site at the 3′ UTR, Hepa1-6 cells were co-transfected with either CMV-EGFP or CMV-EGFP-miR199a*3 and miRNA199a inhibitor (A) and either CMV-GLuc or CMV-GLuc-miR199a*3 and miRNA199a inhibitor (C). The percentage of GFP-positive cells for each group was quantified with flow cytometry and expressed as the percentage of GFP-positive cells after transfection with CMV-EGFP. Similarly, the amount of secreted luciferase was quantified and expressed as a percentage of quantity of secreted luciferase after transfection with CMV-GLuc for each treatment group. (B) Representative fluorescent microscopic and flow-cytometric images for each treatment group. (D) The effects of overexpressing miRNA199a on reporter expression in HCC cell lines by co-transfecting either CMV-GLuc or CMV-GLuc-miR199a*3 and miRNA199a mimic were studied. The secreted luciferase was quantified and represented as the percentage of secreted luciferase after transfection with CMV-GLuc for each treatment group. A two-tailed t test was performed to compare the difference between indicated groups using GraphPad 7.0 (data are reported as mean ± SD, n > 3; *p < 0.05).
Post-transcriptionally Controlled GDEPT for HCC
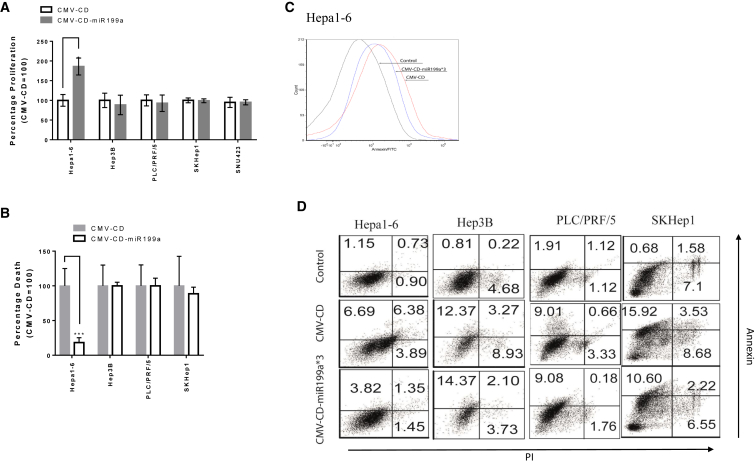
Next, we constructed a post-transcriptionally regulated GDEPT platform based on suicide gene CD and prodrug 5-FC for gene therapy. Either CMV-CD or CMV-CD-miR199a*3 was transfected into miRNA199a-positive Hepa1-6 as well as miRNA199a-negative HCC cell lines Hep3B, PLC/PRF/5, SKHep1, and SNU423. We studied both cell proliferation and cell death (Figure 4) after transfection with CMV-CD and with CMV-CD-miR199a*3. A 1.8-fold higher proliferation rate was observed for Hepa1-6 after transfection with CMV-CD-miR199a*3 when compared to transfection with CMV-CD (p < 0.05) and incubation with prodrug 5-FC, whereas no significant difference was observed in HCC cell lines Hep3B, PLC/PRF/7, SKHep1, and SNU423 (Figure 4A). Similarly, total cell death (quantified by propidium iodide [PI]/annexin staining) was significantly lower (p < 0.001) in Hepa1-6 after transfection with CMV-CD-miR199a*3 and incubation with 5-FC when compared to CMV-CD-positive control (Figures 4B and 4C), while no significant effects were observed in HCC cell lines Hep3B, PLC/PRF/7, and SKHep1 (Figures 4B and 4D). These results demonstrate that a liver detargeted GDEPT system that simultaneously targets HCC can be constructed using miRNA199a-mediated post-transcriptional gene regulation.
Figure 4.
Targeted GDEPT Utilizing miRNA199a Binding Site
(A) Cells were seeded in a 96-well plate and transfected with CMV-CD and CMV-CD-miR199a*3; after incubation with 5-FC, an MTS assay was performed to quantify cell proliferation. Percent proliferation with CMV-CD-199a*3 was normalized with CMV-CD for each cell line. (B) In order to quantify total cell death after GDEPT, cells were transfected in 24-well plates with CMV-CD and CMV-CD-miR199a*3 and incubated with media containing 5-FC. Percentage of apoptotic cells was then calculated by annexin/propidium iodide (PI) staining. Percent death after GDEPT with CMV-CD-199a*3 was normalized against that with CMV-CD. (C) Representative flow-cytometric analysis of Hepa1-6 showing the difference of apoptotic cells after GDEPT with CMV-CD and with CMV-CD-miR199a*3 is indicated. (D) Representative flow-cytometric images are shown of miRNA199a-positive Hepa1-6 and miRNA199a-negative Hep3B, PLC/PRF/5, and SKHep1, indicating the percentage of annexin- and PI-positive cells after GDEPT with CMV-CD and CMV-CD-miR199a*3. Experiments were repeated thrice in triplicates, and the difference between CMV-CD and CMV-CD-miR199a groups was checked for statistical significance using the two-tailed t test in GraphPad Prism 7.0 (data are reported as mean ± SD, *p < 0.05; ***p < 0.005).
AAV-Mediated Delivery of Transgenes with miR199a Binding Site for Detargeting Cells Containing High Endogenous miRNA199a Levels
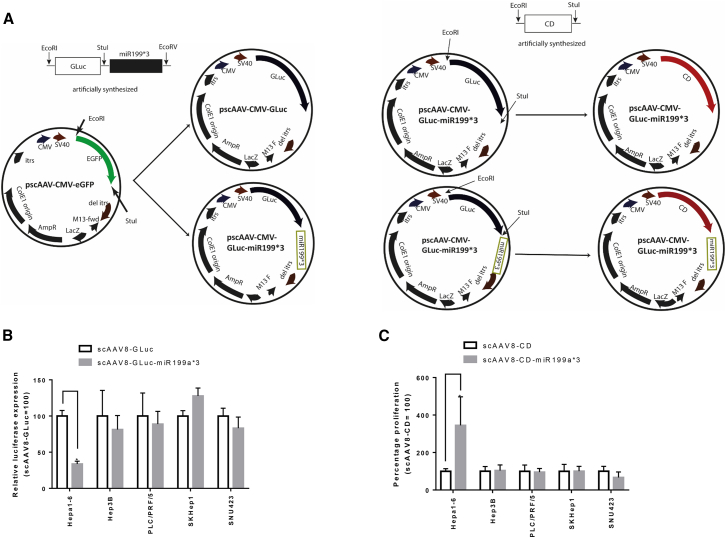
To examine the possibility of delivering miRNA199a-based post-transcriptionally regulated reporter and therapeutic genes using adeno-associated virus (AAV) vectors, we constructed self-complementary AAV serotype 8 with or without miR199a binding sites at the 3′ UTR of the transgenes (scAAV8-GLuc, scAAV8-GLuc-miR199a*3, scAAV8-CD, and scAAV8-CD-miR199a*3). Transduction of the miRNA199a-positive Hepa1-6 cell line with scAAV8-GLuc-miR199a*3 resulted in significantly reduced GLuc expression (p < 0.05) when compared to scAAV8-GLuc, while no significant difference in GLuc expression was observed in miRNA199a-negative HCC cell lines Hep3B, PLC/PRF/5, SKHep1, and SNU423 (Figure 5B). Similarly, after transduction with scAAV8-CD-miR199a*3 and subsequent incubation with prodrug 5-FC, a 3.4-fold higher proliferation rate was observed in Hepa1-6 cells (p < 0.05) when compared to scAAV8-CD, whereas no significant difference in cell proliferation was observed for both groups in HCC cell lines Hep3B, PLC/PRF/5, SKHep1, and SNU423 (Figure 5C). These results demonstrate the compatibility of our miRNA199a-based post-transcriptionally targeted gene delivery and therapy system with the AAV-vector-based delivery system, the most widely used delivery vector for therapeutic purposes.
Figure 5.
AAV-Vector-System-Mediated Targeted Gene Therapy Based on Post-transcriptional Action of miRNA199a
To investigate the possibility of vector-mediated delivery of a transgene harboring miR199a binding site, AAV8 harboring GLuc as a reporter and cytosine deaminase (CD) as a therapeutic gene was constructed with or without miR199a binding sites at the 3′ UTR of the transgenes, and cells were transduced at an MOI of 100,000 vgs per cell. (A) Construction of plasmid harboring GLuc is indicated, as well as CD flanked by self-complementary inverted terminal repeats of the AAV. (B) Reporter expression after delivery with AAV: both miRNA199a-positive and -negative cells were transduced with scAAV8-GLuc and scAAV8-GLuc-miR199a*3, and the amount of secreted GLuc was quantified. Relative expression of GLuc after transduction with scAAV8-GLuc-miR199a*3 was reported as a percentage of that after transduction with scAAV8-GLuc. (C) Targeted GDEPT after AAV-mediated suicide gene therapy: cells were transduced with scAAV8-CD and scAAV8-CD-miR199a*3 and incubated with the prodrug 5-FC. The percent proliferation was then calculated for scAAV8-CD-miR199a*3 and represented as a percentage of that for scAAV8-CD for each cell type. The significant difference between groups was tested by two-tailed t test using GraphPad Prism v7.0 (data are reported as mean ± SD, n > 3; *p < 0.05).
CD133+ CD44+ Oct4+-Enriched 3D Tumorspheres Can Be Targeted with Vectors Harboring Transgenes with miRNA199a Binding Sites at the 3′ UTR
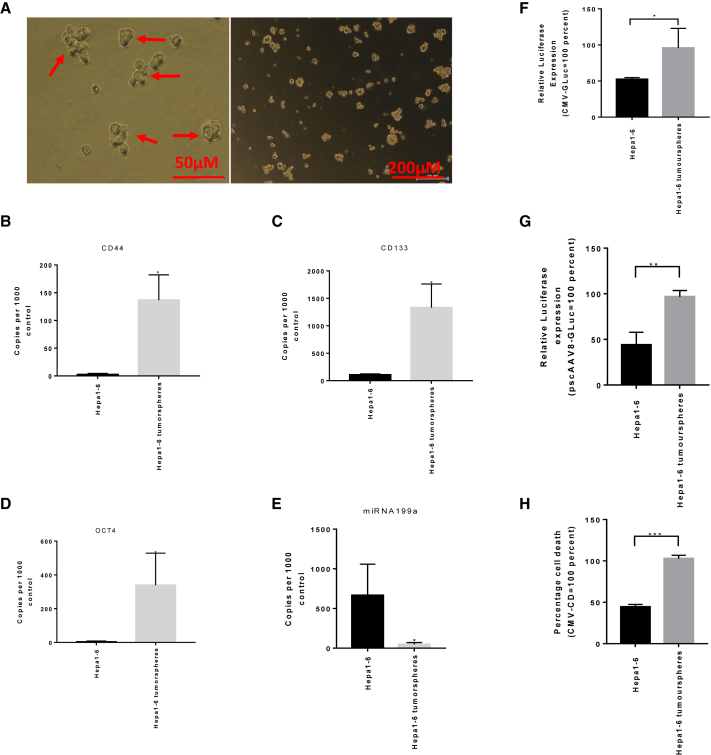
In order to assess the efficacy of miRNA199a targeting in a more complex tumor-like environment, we utilized Hepa1-6 cells grown under 3D tumorsphere culture conditions (Figure 6A). These heterogeneous 3D tumor-like cultures have also been shown to enrich for cancer stem cells.16 Given the reported tumor-suppressing roles of miRNA199a and its downregulation in cancer stem cells for other cancer types, we first investigated whether maintaining Hepa1-6 cells as 3D tumorspheres could enrich for classic cancer stem cell markers. We quantified the levels of expression of HCC stemness markers CD44 (Figure 6B), CD133 (Figure 6C), and Oct4 (Figure 6D) in these tumorspheres and observed a significant upregulation of these genes when compared to 2D-grown Hepa1-6 cells (p < 0.05). The increase in stemness markers correlated to a downregulation in levels of miRNA199a (Figure 6E).
Figure 6.
Expression of miRNA199a in Tumorspheres of Hepa1-6 and Its Implication for Targeted Gene Delivery
(A) Photomicrographs of tumorspheres of Hepa1-6 after 5 days in culture in serum-free, stem-cell-enriching media (scale bars, 50 μM on the left and 200 μm on the right). Real-time qPCR was performed on cDNA of the Hepa1-6 and Hepa1-6 tumorspheres to compare the markers of stemness, (B) CD44, (C) CD133, and (D) Oct4, as well as (E) miRNA199a. Comparison of miRNA199a levels in Hepa1-6 and Hepa1-6 tumorspheres: (F) Hepa1-6 and Hepa1-6 tumorspheres were transfected with CMV-GLuc-miR199a*3, and secreted GLuc was reported as percentage of CMV-GLuc. (G) Hepa1-6 and Hepa1-6 tumorspheres were transduced with pscAAV8-GLuc and pscAAV8-GLuc-miR199a*3, and relative GLuc expression was reported. (H) Similarly, Hepa1-6 and Hepa1-6 tumorspheres were transfected with CMV-CD and CMV-CD-miR199a*3, and percent proliferation was calculated following incubation with 5-FC. Percent proliferation for CMV-CDmiR199a*3 was normalized with CMV-CD. A two-tailed t test was performed to analyze the difference between groups (data are reported as mean ± SD, n > 3; *p < 0.05; **p < 0.01; ***p < 0.001).
Next, we examined whether Hepa1-6 cell tumor-like spheroids could be efficiently targeted by the miR199a target sequence (TS) vectors. To do this, we transfected 3D tumorspheres of Hepa1-6 and 2D Hepa1-6 with CMV-GLuc-miR199a*3 and examined the reported expression. We observed a significant increase in the reporter expression in 3D tumorspheres compared to the miRNA199a-expressing 2D cultures (p < 0.05) (Figure 6F). Similarly, a 2.19-fold increase in GLuc expression (p < 0.005) was observed after transduction of 3D tumorspheres of Hepa1-6 with scAAV8-GLuc-miR199a*3 when compared to 2D culture (Figure 6G). Using GDEPT with the CD/5-FC system also resulted in significantly higher cell death (p < 0.001) in 3D tumorspheres of Hepa1-6 cells when compared to the 2D culture (Figure 6H). These results suggest that miRNA199a-targeting strategies have utility in targeting more complex tumor-like spheroids with decreased miRNA199 expression.
Discussion
miRNAs are small, untranslated, endogenous RNA molecules that efficiently regulate the expression of a gene by binding to a specific sequence in its mRNA (binding sites). In addition to having a cell-specific expression pattern and being involved in several important biological processes ranging from development to apoptosis, several miRNAs have been reported to be dysregulated in several diseases, including cancer. Inclusion of binding sites of miRNAs that are downregulated in cancer while being expressed at high levels in normal cells is an attractive approach for limiting transgene expression in cancer cells.
In this study, we first investigated the expression levels of miRNA199a in hepatocytes and a panel of HCC and non-HCC cell lines. We showed that miRNA199a was significantly downregulated in HCC and non-HCC lines while remaining high in primary hepatocytes, HepRG cells, and Hepa1-6. The observation that the murine Hepa1-6 cell line contains miRNA199a at levels comparable to that of hepatocytes allowed us to use it as a model for normal hepatocyte expression. These results were in line with other studies showing miRNA199a downregulation in HCC but expression at high levels in the cells of the liver, including hepatocytes,17, 18 hepatic stellate cells,19 and liver sinusoid endothelial cells.20 It has been reported as one of the key miRNAs dysregulated in HCC development and progression, with a role as a diagnostic marker21, 22 as well as a therapeutic target.23, 24 miRNA199a has also been shown to be downregulated in a number of non-liver cancers such as breast cancer,25 renal cell cancer,26 osteosarcoma,27 thyroid cancer,28 and bladder cancer.29
In line with previous studies performed with liver-specific miRNA122a,30, 31 transfection of reporters with three miRNA199a binding sites at the 3′ UTR resulted in a significant inhibition of the reporter expression in miRNA199a-positive primary hepatocytes, HepaRG cells, and Hepa1-6 cells, while no significant effects were observed in miRNA199a-negative HCC and non-HCC cell lines. No effect in expression was seen when a control miRNA binding site was incorporated at the 3′ UTR (Figure S1). Next, we generated plasmids harboring CD and miRNA199a binding sites and observed significantly reduced cell death in miRNA199a-positive cell Hepa1-6 after transfection and incubation with the prodrug 5-FC, while, in miRNA199a-negative HCC cells, cell death equivalent to CMV-driven CD expression was observed. These results validated a post-transcriptionally targeted suicide gene therapeutic system for HCC. Given the complete homology between murine and human miRNA199a,32 we were able to model the effects of incorporating binding sites of miRNA199a at the 3′ UTR using Hepa1-6, which was useful especially for targeted GDEPT studies, which is often problematic in primary hepatocytes due to culture conditions. We chose to use perfectly complementary binding sites, as imperfectly complementary binding sites can cause saturation of endogenous miRNA, even at a low concentration of transcripts15, 33; similarly, utilizing perfectly complementary binding induces target RNA degradation, facilitating quick turnover.34, 35
Next, we explored the possibility of constructing a targeted AAV vector harboring our miRNA199a-based post-transcriptionally regulated gene therapeutic system. Similar to reports by Peruta et al., who utilized the liver-specific nature of AAV8 to construct miRNA122a-based post-transcriptionally liver detargeted gene delivery system, we observed that transduction of Hepa1-6 with scAAV8-GLuc-miR199a*3 leads to a significant reduction of reporter expression when compared to scAAV8-GLuc.36 This observation was further corroborated by post-transcriptionally detargeted suicide gene therapy with scAAV8-CD-miR199a*3 in Hepa1-6. Given the ability of AAV8 to transduce liver and tissues of liver origin, our system could provide an option to target disseminated tumors.
In an interesting observation, we observed reduced levels of miRNA199a in 3D Hepa1-6 tumorspheres of enriched in stemness markers, including CD44. This observation is in line with the reported tumor-suppressive role of miRNA199a in HCC and the fact that miRNA199a directly regulates the expression of stemness marker CD44 in other cancers.37, 38 While we have not directly shown the existence of cancer stem cells in this study, others have shown that tumorspheres derived from HCC are enriched in stem-like cells and exhibit high chemoresistance.39 The potential utility of miRNA199 to target a sub-population of cancer stem cells is intriguing and could be explored in future studies. Although this particular study does not answer the questions of how and what roles miRNA199a might play in HCC stem cell biology, the decreased levels of miRNA199a in Hepa1-6 tumorspheres not only allowed us to assess the effectiveness of miRNA199a targeting in a miRNA199a-negative cell population but also showed its effectiveness in terms of increased expression and increased death when used in conjunction with GDEPT in a complex 3D tumor-like environment. Similar 3D tumorsphere culture systems are widely used to recapitulate some of the tumor heterogeneity seen in vivo and to screen for novel drug candidates while reducing the need of animal models.40, 41
In conclusion, this proof-of-concept study establishes negative targeting based on post-transcriptional gene regulation by miRNA199a in the context of HCC gene therapy. This system was found to efficiently target HCC cells with downregulation of miRNA199a while, at the same time, detargeting miRNA199a-positive HepaRG and Hepa1-6. Furthermore, AAV-based delivery of this system was found to be feasible and effective. Finally, given that miRNA199a has been reported to be downregulated in multiple cancer types, this system could be exploited to detarget any cell type with high endogenous levels of miRNA199a.
Materials and Methods
Cell Culture
The Hepa1-6 cell line, which expresses high levels of miRNA199a, and HCC cell lines Hep3B, PLC/PRF/5, SKHep1, and SNU423 with miR199a downregulation were obtained from ATCC and maintained in DMEM media (Thermo Fisher Scientific, Scoresby, Australia) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Australia) and 1% penicillin-streptomycin (P/S) (GIBCO, Australia). The Australian Genome Research Facility (AGRF) cell line ID service was used to confirm the identity of the human cell lines. Breast cancer cell lines T47D and MCF-7 were maintained in standard DMEM media. Melanoma cell lines 92.1 and Mel270 (gifted by Nicholas Hayward) and ovarian cancer lines SW626, CAOV3, and TOV21G were grown in RPMI media (Thermo Fisher Scientific) supplemented with 10% FBS and 1% P/S. Prostate cancer cell lines LnCap and DU145 were maintained in standard DMEM media. All the other cell lines were maintained as per the ATCC recommendations. Cryopreserved primary human hepatocytes (HUM4150) and NoSpin HepaRG (NSHPRG) cells were obtained from Lonza (Sydney, Australia) and maintained as per the manufacturer’s protocol.
Real-time qPCR and Quantification of miRNA Levels
To quantify the endogenous expression levels of miRNA199a, total RNA was isolated with TRIzol, and cDNA was synthesized with the MystiCq microRNA cDNA Synthesis Mix (Sigma Aldrich, St. Louis, MO, USA) as per the manufacturer’s protocol. The synthesized cDNA was then used for real-time qPCR with the Bioline SYBR Lo-ROX system (Alexandria, Australia) in a ViiA7 RT-PCR machine (Thermo Fisher Scientific) in the following conditions: 95°C for 10 min followed by 40 cycles of 95°C for 5 s, 60°C for10 s, and 70°C for10 s. The MystiCq Universal PCR (MIRUP, Sigma) and 5′-CCCAGTGTTCAGACTACCTG-3′ primers were used to amplify miRNA199a, and the amount of miR199a was calculated as the number of copies per 1,000 copies of RNU6 (MIRCP00001) control using the formula 2ˆ(Ct control − Ct sample) ⋅ 1,000. A 100% homologous nature of murine and human miRNA199a allowed usage of the same primer for amplification. Similarly, markers for the level of stemness (CD44, CD133, and Oct4) were measured relative to GAPDH control (primers are listed in Table S1).
Construction of Expression Plasmids
GLuc with three miRNA199a-5p binding sites (GGGTCACAAGTCTGATGGACAAG*3) at the 3’-UTR flanked by StuI and EcoVI was artificially synthesized (ThermoFisher Scientific). GLuc with and without miRNA binding sites was then cloned in the pscAAV-GFP (a gift from John T Gray, Addgene plasmid #32396) using enzymes EcoRI and StuI to generate pscAAV-CMV-GLuc (CMV-GLuc) or EcoRI and EcoRV to generate pscAAV-CMV-GLuc-miR199a*3 (CMV-GLuc-miR199a*3). For the construction of CD-expressing plasmids, the gene was artificially synthesized separately and cloned in the aforementioned plasmids replacing GLuc to obtain CMV-CD and CMV-CD-miR199a*3. A control miRNA binding site GGGTCACAAGTCTGATGGACAAG *3 was also incorporated at the 3′ UTR of reporters (Figure S1). A representation of plasmid construction has been included in Figure 5A.
Transfection and Gaussia Luciferase Reporter Assays
All transfection studies for investigating the reporter expression were performed with Lipofectamine 3000 (Thermo Fisher Scientific) in a 24-well plate as per the manufacturer’s protocol. Briefly, 30,000 cells were seeded in a 24-well plate, and transfection was performed with 500 ng of plasmids. 72 hr post-transfection, the amount of GLuc secreted in the media was quantified with the Pierce Gaussia Luciferase Glow Assay Kit (Thermo Fisher Scientific) as per the manufacturer’s recommendations. The chemiluminescence measurement was conducted with the Infinite 200 Pro NanoQuant (Tecan Trading, Zurich, Switzerland). In order to regulate the difference in transfection efficiencies across cell lines, chemiluminescence detected with CMV-GLuc-miR199a*3 was normalized with CMV-GLuc for individual cell type.
Inhibition and Overexpression of miR199a
For knockdown experiments in Hepa1-6 cells, 5 pmol of miRNA199a inhibitor (4464084, Life Technologies, Mulgrave, Australia) was co-transfected using Lipofectamine 3000 with either CMV-GLuc or CMV-GLuc-199a*3 and either CMV-EGFP or CMV-EGFP-miR199a*3 in a 24-well plate as per the manufacturer’s protocol. Similarly, for overexpression, miRNA199a mimic (4464066, Life Technologies) was co-transfected with either CMV-GLuc or CMV-GLuc-miR199a*3. 72 hr post-transfection, the percentage of GFP-positive cells or secreted GLuc was quantified for each group and expressed as the percentage of either CMV-EGFP or CMV-GLuc respectively.
Cell Proliferation Assay
Cell proliferation assay was performed with the CellTiter96 Aqueous One Solution Cell Proliferation Assay Kit (Promega, Madison, WI, USA). Briefly, 10,000 cells were seeded in a 96-well plate and transfected with either CMV-CD or CMV-CD-199a*3. 24 hr post-transfection, fresh media containing 10 μm 5-FC were added. After 48 hr, the cells were incubated with MTS (tetrazolium compound, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) reagent as per the manufacturer’s protocol, and absorbance was measured at 540 nm with the Infinite 200 Pro NanoQuant. Percent proliferation was calculated for CMV-CD and CMV-CD-199a*3 for each cell line, and percent proliferation with CMV-CD-199a*3 was subtracted from that with CMV-CD to account for the difference in transfection efficiencies.
Cell Death Assay with Annexin V/PI
In order to quantify the amount of cell death after the expression of the suicide gene CD with or without an miRNA199a binding site at the 3′ UTR, flow cytometry based on annexin V/PI staining (Life Technologies) was performed as suggested by the supplier. Briefly, 30,000 cells were transfected with CMV-CD or CMV-CD-199a*3 in a 24-well plate. 48 hr post-transfection, the media were replaced with fresh media containing 10 μM 5-FC. After 24 hr, the cells were washed, collected, and stained with annexin V/PI. Annexin V/PI-positive cells were quantified using the BD FACSCanto II, and data analysis was performed with FCS Express 3 (BD Biosciences; North Ryde, Australia). The percentage of apoptotic cells after transfection with CMV-CD-199a*3 was normalized with CMV-CD for each cell type.
AAV Production and Transduction
AAV serotype 8 was produced using the triple transfection method using polyethylenimine (PEI). Briefly, HEK293 cells were seeded in 15-cm plates and transfected with pHelper (Agilent Technologies, Mulgrave, Australia), AAV8 capsid, and either psc-CMV-GLuc, psc-CMV-GLuc-miR199*3, psc-CMV-CD, or psc-CMV-CD-miR199*3 in a 2:1:1 ratio. After 48–72 hr, the cells were washed with PBS and subjected to 3 freeze-thaw cycles with ethanol and dry ice followed by incubation at 37°C to release the AAV. The crude lysate was then treated with benzonase (Sigma-Aldrich) and passed through the Amicon Ultra 100-kDa filter (Sigma-Aldrich), and buffer exchange was performed three times with PBS, after which the end product was filtered through 0.22-μm filters. The number of vector genomes (vgs) was quantified with qPCR (primers have been listed in Table S1). All transduction experiments to study GLuc expression were performed in 96-well plates at an MOI of 100,000 vgs per cell, and luminescence was measured as previously described. Similarly, suicide gene therapy was performed with an MTS assay after transduction of 10,000 cells with 100,000 vgs per cell of either pscAAV-CMV-CD or pscAAV-CMV-CD-miR199a*3 as previously described.
3D Culture of Hepa1-6
3D tumorspheres of Hepa1-6 were maintained in stem-cell conditioned, serum-free neurosphere assay (NSA) media containing DMEM/F12 (Thermo Fisher Scientific), 10 ng/mL recombinant human basic fibroblast growth factor (rhFGF) (Lonza), 20 ng/mL recombinant human epidermal growth factor (rhEGF) (Lonza), BSA (Sigma-Aldrich), 4 μg/mL heparin sulfate (Sigma-Aldrich), and 1% P/S (Thermo Fisher Scientific), as previously described.42 Briefly, 30,000 cells were collected, washed thrice with PBS, and seeded in ultra-low attachment plates (Corning, Corning, NY, USA). Images of tumorspheres were taken with a digital camera (Olympus DP21, Tokyo, Japan) connected to an inverted microscope (Olympus CKX41) with imaging software (CellSens, Olympus, Tokyo, Japan). Cells were either collected for RNA extraction (day 5), transfected at day 3 (with CMV-GLuc or CMV-GLuc-miR199a*3), or transduced at day 3 (with pscAAV8-GLuc) or pscAAV8-GLuc-miR199a*3).
Statistical Analysis
All experiments were repeated at least thrice, and data are represented as mean ± SD. To test whether there were significant differences in experimental results between groups, a two-tailed t test was performed with GraphPad Prism 7.0 (GraphPad Software) (*p < 0.05, **p < 0.01, and ***p < 0.001).
Author Contributions
B.D. and J.C.S. conceived and designed the experiments; B.D. and C.A.R.-S. conducted the experiments and wrote the manuscript; J.C.S. and C.J.L. edited the manuscript.
Conflicts of Interest
The authors have no conflict of interest.
Acknowledgments
This work was supported by the Gallipoli Medical Research Foundation.
Footnotes
Supplemental Information includes one figure and one table and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.08.016.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Bupathi M., Kaseb A., Meric-Bernstam F., Naing A. Hepatocellular carcinoma: Where there is unmet need. Mol. Oncol. 2015;9:1501–1509. doi: 10.1016/j.molonc.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cidon E.U. Systemic treatment of hepatocellular carcinoma: Past, present and future. World J. Hepatol. 2017;9:797–807. doi: 10.4254/wjh.v9.i18.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao J.J., Shi Z.Y., Xia J.F., Inagaki Y., Tang W. Sorafenib-based combined molecule targeting in treatment of hepatocellular carcinoma. World J. Gastroenterol. 2015;21:12059–12070. doi: 10.3748/wjg.v21.i42.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y.G., Huang P.P., Zhang R., Ma B.Y., Zhou X.M., Sun Y.F. Targeting adeno-associated virus and adenoviral gene therapy for hepatocellular carcinoma. World J. Gastroenterol. 2016;22:326–337. doi: 10.3748/wjg.v22.i1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhungel B., Jayachandran A., Layton C.J., Steel J.C. Seek and destroy: targeted adeno-associated viruses for gene delivery to hepatocellular carcinoma. Drug Deliv. 2017;24:289–299. doi: 10.1080/10717544.2016.1247926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Siriwon N., Rohrs J.A., Wang P. Generation of targeted adeno-associated virus (AAV) vectors for human gene therapy. Curr. Pharm. Des. 2015;21:3248–3256. doi: 10.2174/1381612821666150531171653. [DOI] [PubMed] [Google Scholar]
- 8.Chiba T., Iwama A., Yokosuka O. Cancer stem cells in hepatocellular carcinoma: Therapeutic implications based on stem cell biology. Hepatol. Res. 2016;46:50–57. doi: 10.1111/hepr.12548. [DOI] [PubMed] [Google Scholar]
- 9.Foka P., Pourchet A., Hernandez-Alcoceba R., Doumba P.P., Pissas G., Kouvatsis V., Dalagiorgou G., Kazazi D., Marconi P., Foschini M. Novel tumour-specific promoters for transcriptional targeting of hepatocellular carcinoma by herpes simplex virus vectors. J. Gene Med. 2010;12:956–967. doi: 10.1002/jgm.1519. [DOI] [PubMed] [Google Scholar]
- 10.Robson T., Hirst D.G. Transcriptional targeting in cancer gene therapy. J. Biomed. Biotechnol. 2003;2003:110–137. doi: 10.1155/S1110724303209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown B.D., Venneri M.A., Zingale A., Sergi Sergi L., Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 12.Brown B.D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 13.Xie J., Xie Q., Zhang H., Ameres S.L., Hung J.H., Su Q., He R., Mu X., Seher Ahmed S., Park S. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol. Ther. 2011;19:526–535. doi: 10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhungel B., Ramlogan-Steel C.A., Layton C.J., Steel J.C. miRNA122a regulation of gene therapy vectors targeting hepatocellular cancer stem cells. Oncotarget. 2018;9:23577–23588. doi: 10.18632/oncotarget.25280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhungel B., Ramlogan-Steel C.A., Steel J.C. MicroRNA-regulated gene delivery systems for research and therapeutic purposes. Molecules. 2018;23:E1500. doi: 10.3390/molecules23071500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayachandran A., Shrestha R., Dhungel B., Huang I.T., Vasconcelos M.Y.K., Morrison B.J., Ramlogan-Steel C.A., Steel J.C. Murine hepatocellular carcinoma derived stem cells reveal epithelial-to-mesenchymal plasticity. World J. Stem Cells. 2017;9:159–168. doi: 10.4252/wjsc.v9.i9.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Di C., Li W., Cai W., Tan X., Xu L., Yang L., Lou G., Yan Y. Oncomirs miRNA-221/222 and tumor suppressors miRNA-199a/195 are crucial miRNAs in liver cancer: a systematic analysis. Dig. Dis. Sci. 2016;61:2315–2327. doi: 10.1007/s10620-016-4156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia X.Q., Cheng H.Q., Qian X., Bian C.X., Shi Z.M., Zhang J.P., Jiang B.H., Feng Z.Q. Lentivirus-mediated overexpression of microRNA-199a inhibits cell proliferation of human hepatocellular carcinoma. Cell Biochem. Biophys. 2012;62:237–244. doi: 10.1007/s12013-011-9263-8. [DOI] [PubMed] [Google Scholar]
- 19.Murakami Y., Toyoda H., Tanaka M., Kuroda M., Harada Y., Matsuda F., Tajima A., Kosaka N., Ochiya T., Shimotohno K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE. 2011;6:e16081. doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo G., Bala S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Y., Yan J.L., Fang A.N., Zhou W.F., Huang L. Circulating miRNAs as novel diagnostic biomarkers in hepatocellular carcinoma detection: a meta-analysis based on 24 articles. Oncotarget. 2017;8:66402–66413. doi: 10.18632/oncotarget.18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Abd N.E., Fawzy N.A., El-Sheikh S.M., Soliman M.E. Circulating miRNA-122, miRNA-199a, and miRNA-16 as biomarkers for early detection of hepatocellular carcinoma in Egyptian patients with chronic hepatitis C virus infection. Mol. Diagn. Ther. 2015;19:213–220. doi: 10.1007/s40291-015-0148-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhan Y., Zheng N., Teng F., Bao L., Liu F., Zhang M., Guo M., Guo W., Ding G., Wang Q. MiR-199a/b-5p inhibits hepatocellular carcinoma progression by post-transcriptionally suppressing ROCK1. Oncotarget. 2017;8:67169–67180. doi: 10.18632/oncotarget.18052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou J., Lin L., Zhou W., Wang Z., Ding G., Dong Q., Qin L., Wu X., Zheng Y., Yang Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Shin V.Y., Siu J.M., Cheuk I., Ng E.K., Kwong A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br. J. Cancer. 2015;112:1751–1759. doi: 10.1038/bjc.2015.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukigi M., Bilim V., Yuuki K., Ugolkov A., Naito S., Nagaoka A., Kato T., Motoyama T., Tomita Y. Re-expression of miR-199a suppresses renal cancer cell proliferation and survival by targeting GSK-3β. Cancer Lett. 2012;315:189–197. doi: 10.1016/j.canlet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Duan Z., Choy E., Harmon D., Liu X., Susa M., Mankin H., Hornicek F. MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol. Cancer Ther. 2011;10:1337–1345. doi: 10.1158/1535-7163.MCT-11-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minna E., Romeo P., De Cecco L., Dugo M., Cassinelli G., Pilotti S., Degl’Innocenti D., Lanzi C., Casalini P., Pierotti M.A. miR-199a-3p displays tumor suppressor functions in papillary thyroid carcinoma. Oncotarget. 2014;5:2513–2528. doi: 10.18632/oncotarget.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichimi T., Enokida H., Okuno Y., Kunimoto R., Chiyomaru T., Kawamoto K., Kawahara K., Toki K., Kawakami K., Nishiyama K. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int. J. Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 30.Qiao C., Yuan Z., Li J., He B., Zheng H., Mayer C., Li J., Xiao X. Liver-specific microRNA-122 target sequences incorporated in AAV vectors efficiently inhibits transgene expression in the liver. Gene Ther. 2011;18:403–410. doi: 10.1038/gt.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G., Dong X., Tian W., Lu Y., Hu J., Liu Y., Yuchi J., Wu X. Evaluation of miR-122-regulated suicide gene therapy for hepatocellular carcinoma in an orthotopic mouse model. Chin. J. Cancer Res. 2013;25:646–655. doi: 10.3978/j.issn.1000-9604.2013.11.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu T., Chen Q., Huang Y., Huang Q., Jiang L., Guo L. Low microRNA-199a expression in human amniotic epithelial cell feeder layers maintains human-induced pluripotent stem cell pluripotency via increased leukemia inhibitory factor expression. Acta Biochim. Biophys. Sin. (Shanghai) 2012;44:197–206. doi: 10.1093/abbs/gmr127. [DOI] [PubMed] [Google Scholar]
- 33.Gentner B., Schira G., Giustacchini A., Amendola M., Brown B.D., Ponzoni M., Naldini L. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 34.Rüegger S., Großhans H. MicroRNA turnover: when, how, and why. Trends Biochem. Sci. 2012;37:436–446. doi: 10.1016/j.tibs.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Sanei M., Chen X. Mechanisms of microRNA turnover. Curr. Opin. Plant Biol. 2015;27:199–206. doi: 10.1016/j.pbi.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Della Peruta M., Badar A., Rosales C., Chokshi S., Kia A., Nathwani D., Galante E., Yan R., Arstad E., Davidoff A.M. Preferential targeting of disseminated liver tumors using a recombinant adeno-associated viral vector. Hum. Gene Ther. 2015;26:94–103. doi: 10.1089/hum.2014.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng W., Liu T., Wan X., Gao Y., Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279:2047–2059. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu R., Liu C., Zhang D., Liu B., Chen X., Rycaj K., Jeter C., Calhoun-Davis T., Li Y., Yang T. miR-199a-3p targets stemness-related and mitogenic signaling pathways to suppress the expansion and tumorigenic capabilities of prostate cancer stem cells. Oncotarget. 2016;7:56628–56642. doi: 10.18632/oncotarget.10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X.L., Jia Q., Lv L., Deng T., Gao J. Tumorspheres derived from HCC cells are enriched with cancer stem cell-like cells and present high chemoresistance dependent on the Akt pathway. Anticancer. Agents Med. Chem. 2015;15:755–763. doi: 10.2174/1871520615666150202111721. [DOI] [PubMed] [Google Scholar]
- 40.Friedrich J., Seidel C., Ebner R., Kunz-Schughart L.A. Spheroid-based drug screen: considerations and practical approach. Nat. Protoc. 2009;4:309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 41.Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller-Klieser W., Kunz-Schughart L.A. Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Morrison B.J., Steel J.C., Morris J.C. Sphere culture of murine lung cancer cell lines are enriched with cancer initiating cells. PLoS ONE. 2012;7:e49752. doi: 10.1371/journal.pone.0049752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.