Abstract
The addition of the hepatitis C virus (HCV) protease inhibitors telaprevir and boceprevir to peginterferon alfa with ribavirin therapy has increased cure rates in HCV infection. Numerous other direct-acting antivirals (DAAs) are in advanced stages of development, including next-generation protease inhibitors, nonstructural protein (NS) 5A inhibitors, and nonnucleoside and nucleos(t)ide NS5B polymerase inhibitors. The classes have different potencies, different resistance mutation profiles, and different barriers to the emergence of resistance. A comprehensive table of resistance mutations for classes of DAAs is presented. Numerous combinations of DAAs with or without ribavirin have been evaluated in early studies of interferon alfa-free regimens, with results indicating that cure is indeed possible with such therapy and suggesting that identification of regimens that could produce cure in the majority of patients may occur within the foreseeable future. This article summarizes a presentation by David L. Wyles, MD, at the IAS-USA live continuing medical education activity held in New York in June 2012.
Hepatitis C virus (HCV) infection is characterized by a high rate of viral replication, the presence of an error-prone viral polymerase that acts twice in the replication cycle (converting positive to negative strand and negative to positive strand RNA), and absence of overlapping reading frames; these factors result in the generation of a large number of closely related viral variants (viral quasispecies) including drug-resistant variants. Infected cells have a turnover rate on the order of several weeks. However, the HCV replication unit is dynamic and is not integrated into host cell DNA. The absence of viral genome integration suggests that latent infection is highly unlikely. HCV replication occurs in the cytoplasm, and replication complexes turn over with a half-life on the order of 10 hours to 20 hours. These characteristics present a vulnerability that can be exploited to achieve eradication of the virus from infected persons through drug treatment. Antiviral resistance, however, may present challenges to development of effective direct-acting antiviral (DAA) regimens.
HCV Resistance to Telaprevir and Boceprevir
Initial studies of the nonstructural protein (NS) 3 HCV protease inhibitors (PIs) telaprevir and boceprevir, each used alone over 2 weeks, showed rapid emergence of resistant mutants. In patients with breakthrough viremia during the 2 weeks of treatment with telaprevir, for example, there was a nearly complete replacement of wildtype virus with drug-resistant variants. In patients who exhibited a continuous decline in viral load throughout the treatment period, resistant variants could nevertheless be found as a more prominent component of the viral quasispecies weeks to months after treatment had ended. In patients with HCV genotype 1a, prominent resistance mutations were the R155K/T and V36A/M substitutions. In patients with HCV genotype 1b, the A156V/T mutation was prominent. It was found that individual resistance mutations conferred a somewhat decreased replicative fitness compared with wild-type virus and were not associated with complete loss of antiviral activity of telaprevir or boceprevir. However, double mutants (eg, the R155K + V36M found in HCV genotype 1a) often exhibited increased fitness compared with single mutations and were associated with larger changes in antiviral 50% effective concentration (EC50).
In the PROVE (Protease Inhibitor for Viral Evaluation) 1 and 21,2 clinical trials with telaprevir in combination with peginterferon alfa and ribavirin, viral breakthrough occurred in approximately 7% of patients with HCV genotype 1a infection, compared with about 2% of those with genotype 1b infection; approximately 10% of patients with either genotype had relapse after cessation of HCV PI treatment. And, as shown in the boceprevir SPRINT-2 (Serine Protease Inhibitor Therapy 2)3 trial, the rate of emergence of resistance variants depended to a considerable degree on activity of peginterferon alfa in the individual patients. Patients having a decrease in HCV viral load greater than 1 log10 IU/mL during the 4-week lead-in period of peginterferon alfa with ribavirin therapy had very low rates of emergence of boceprevir-resistant mutants (< 5%) during subsequent triple therapy, whereas those with less than a 1 log10 IU/mL decrease in HCV RNA had higher rates (> 30%-45%).
Overall, phase III telaprevir and boceprevir triple therapy trials in treatment-naive patients have shown that resistant variants are detected in 50% to 75% of patients not achieving sustained virologic response (SVR). Of the 10% to 15% with virologic failure (ie, excluding patients in whom therapy failed because of such factors as intolerance), greater than 90% have resistant variants as the predominant HCV species when viral breakthrough occurs. Viral breakthrough during treatment is associated with emergence of resistant variants conferring high-fold changes in sensitivity—eg, V36M plus R155K in HCV genotype 1a infection and A156T/V, T54S, and V55A in genotype 1b infection. Relapse after the end of treatment is associated with low-fold change variants, such as R155K or V36M alone in genotype 1a and T54A, A156S, or V170A in genotype 1b. Data on telaprevir resistant variants in patients not achieving SVR suggest that reversion to wild-type virus occurs in 96% of patients over 16 months off treatment,4 with less fit variants decaying more rapidly.
Population sequencing in patients from phase III trials of telaprevir and boceprevir showed that preexisting resistant variants were present in 5% to 7% of patients. Studies using ultradeep sequencing indicate that protease resistant variants are detectable at the 1% level in approximately 7% to 8% and include variants with substitutions at positions 155 and 156 that are associated with substantial loss of sensitivity. It is generally assumed that even when resistant variants cannot be detected, almost all single resistant variants are present prior to treatment at very low levels. An analysis of how preexisting resistant variants might affect clinical outcome from the boceprevir phase III trials suggested, again, that treatment response was dependent on whether patients had a good response to the 4-week lead-in treatment with peginterferon alfa and ribavirin. Overall, 66 (7%) of 980 patients had detectable resistant variants at baseline, including variants associated with treatment failure (V36M, R155K, T54A/S, and V55A; 65% of variants identified) and those that conferred reduced susceptibility to boceprevir but were not implicated in virologic failure (V36I/L, Q41H, V55I, V170M, and M175L; 35% of variants). Among patients who responded to peginterferon alfa, SVR rates were 80% overall, 78% in patients with the first group of variants, and 73% in patients with the second group of variants. Among patients with poor response to initial peginterferon alfa, SVR rates were approximately 30% overall, 0% in those with the first group of variants, and 50% in those with the second group. A total of 7 patients (<1% of the total population) did not respond to peginterferon alfa and had variants in the first group.
HCV resistance testing is clinically available, but given that the 2 available DAAs are completely cross-resistant (see Figure 1), results of testing have little clinical utility in making treatment decisions at this point. Baseline testing to determine the presence of resistant variants prior to therapy is also currently of little clinical value. It remains unclear what effect the identification of preexisting resistant variants has on treatment outcomes with the currently available PIs; telaprevir or boceprevir responses are still largely based on patient interferon-sensitivity. Available data suggest a very small proportion of treatment naive patients (<1%) will possess both baseline-resistant variants and be poor responders to interferon. Thus, there is no straightforward way to determine if patients with preexisting resistant variants should be spared telaprevir or boceprevir treatment.
Figure 1.

Mutations in hepatitis C virus (HCV) that impact susceptibility to HCV drugs approved by the US Food and Drug Administration (FDA) and to investigational drugs. The number on the bar refers to the amino acid position; the letter above the number refers to the wild-type amino acid and the letter(s) below the bar are relevant substitutions. Amino acid abbreviations: A, alanine; C, cysteine; D, aspartate; E, glutamate; F, phenylalanine; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; P, proline; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; W, tryptophan; Y, tyrosine. Bolded type under the amino acid position number represents the key mutations that are clearly associated with virologic failure and result in a resistant phenotype. ©IAS–USA 2012
* The majority of mutations seen in HCV genotype 1a are I170F/T substitutions in which I is the consensus amino acid. The majority of mutations seen in HCV genotype 1b are V170A/T, in which V is the consensus amino acid.
** Variants rarely seen in vivo and of unclear clinical significance. These variants are most frequently seen in combination with other resistance-associated mutations. In vitro, they do not confer substantial resistance to telaprevir (< 2-fold increase in median effective concentration [EC50]).
a Comprehensive resistance data have not been published. All mutations should be considered provisional.
b Indicates wildtype in HCV genotype 1a only.
c Indicates HCV genotype 1b only.
d Nonnucleoside polymerase inhibitor position designations according to Pauwels et al.7
With regard to preexisting resistant variants for other classes of DAAs in development, ultradeep pyrosequencing capable of identifying variants that account for as little as 0.5% of the viral population indicates that 10% of patients have a majority of preexisting variants resistant to nonnucleoside inhibitors of HCV nonstructural protein (NS)5B polymerase and 22.5% have a minority of resistant variants (in association with polymorphisms at the allosteric binding sites of the polymerase).5 Thus far, no preexisting resistant variants (S282T) to nucleos(t)ide NS5B polymerase inhibitors have been identified. Preexisting variants resistant to HCV NS5A inhibitors have been identified in approximately 16% of patients.
With regard to next-generation investigational PIs such as TMC-435 (simeprevir), it is known that 1 resistant variant (Q80K) is present in approximately 20% of patients at baseline (particularly common in patients with genotype 1a infection). In a phase II study, SVR rates in patients receiving a lower dose of simeprevir (75 mg po qd) were approximately 82% in those without the Q80K polymorphism versus 57% in those with it. At the higher dose (150 mg po qd), rates were somewhat closer, approximately 85% versus 67%.6 The higher dose is being tested in a phase III trial, which will include analysis to determine whether screening for the Q80K variant at baseline might have clinical utility in targeting patients for treatment.
Development of Interferon Alfa-Free Regimens
DAA Class Activity and Resistance
The currently identified mutations associated with resistance to telaprevir and boceprevir and investigational PIs, NS5A inhibitors, and nucleos(t)ide NS5B polymerase inhibitors are shown in Figure 1.
Among the current classes of HCV agents, the NS3 PIs have good potency (3-4 log10 IU/mL reductions in HCV RNA level) and established efficacy when used in combination with peginterferon alfa and ribavirin. However, they require several doses every day,have adverse effect profiles that can be improved on, have low to moderate barriers to resistance with cross-resistance, and have considerable drug interaction potential, since many are metabolized by or affect cytochrome P450 (CYP450) isoenzymes. NS5A inhibitors are very potent (4-5 log10 IU/mL reductions in HCV RNA level), can be dosed once daily, appear to have good adverse effect profiles, and provide broader HCV genotype coverage. However, they have a low barrier to resistance and also exhibit cross-resistance. Available data to date suggest that the nucleos(t)ide NS5B polymerase inhibitors are potent, and later-generation agents can be given once daily and have improved adverse effect profiles. These drugs have the highest barrier to resistance among the current classes, and thus appear to be strong candidates for inclusion in potential interferon alfa-sparing regimens. The nonnucleoside polymerase inhibitors have numerous target sites but modest potency. They also have the lowest barrier to resistance, because allosteric binding sites are not highly conserved.
To determine how best to use and combine these agents, modeling studies have been performed to provide a basis for rational design of interferon alfa-free regimens. Some of these models assume that all single and double mutants are present prior to exposure to DAAs, accounting for the rapid emergence of resistance with exposure to single drugs, and that triple mutants are selected by drug pressure within a day of starting treatment. It has thus been posited that a successful interferon alfa-free regimen would need to require the virus to develop 4 resistance mutations (ie, possess a 4-mutation barrier to resistance).
The NS5B polymerase has at least 4 allosteric binding sites and an active site. For each of these sites, inhibitors have been developed and a unique resistance profile identified. There is potential for cross-resistance at the palm site of the polymerase, against which 2 different classes of inhibitor have been developed. As noted, the nonnucleoside polymerase inhibitors are characterized by relatively low potency and a low barrier to resistance, with a wide variability of response being observed among patients receiving single-agent treatment. In a study of an investigational inhibitor (ANA598), the highest dose produced an approximately 1.5 log10 IU/mL reduction in HCV RNA level, with viral breakthrough occurring rapidly thereafter and the C316Y resistant variant being found in more than 50% of patients.
An example of the higher barrier to resistance among the nucleos(t)ide polymerase inhibitors is the effect of the resistant variant S282T on viral sensitivity and replicative fitness. This variant produces a low-fold change in EC50 (approximately 3-fold shift) and has replicative fitness that is 5% to 15% that of wild-type virus. The activity of NS5A inhibitors is indicated by a phase II study of one such investigational inhibitor (BMS-790052 or daclatasvir) that showed an SVR rate of 83% when used in combination with peginterferon alfa and ribavirin. When studied as a single agent, 4-log10 IU/mL decreases in HCV RNA level over the first several days are seen followed by viral breakthrough with a variety of resistance mutations. Some resistance mutations observed in patients with HCV genotype 1a infection were not observed in genotype 1b, apparently reflecting a difference in polymorphisms present at baseline in the 2 HCV genotypes. Those resistant variants that are common to the 2 genotypes have different effects on drug sensitivity. Thus, in the study of BMS-790052 alone, the resistant variants M28T, Q30H, L31V, and Y93H were identified in patients with genotype 1a infection and the variants L31V and Y93H were found in those with genotype 1b infection. There were dramatic differences in the effect on drug susceptibility with the common mutations by genotype: the L31V variant caused a 3000-fold change in genotype 1a but only a 28-fold change in genotype 1b, and the Y93H variant caused a 5000-fold change and a 24-fold change, respectively.
These data highlight that there are differences between the HCV genotype 1 subtypes with regard to mutational and functional barriers to resistance. In another example, transition to the R155K variant requires only a single nucleotide change in genotype 1a, but requires 2 substitutions in genotype 1b. As shown in the study of the NS5A inhibitor, identical variants can have dramatically different effects on fold-change in susceptibility in genotype 1a versus genotype 1b.8
Promising investigational DAAs in phase II and III studies that are likely candidates for testing in interferon alfa-free regimens include the nucleoside polymerase inhibitor mericitabine and the nucleotide inhibitor GS-7977; the nonnucleoside inhibitor tegobuvir; the next-generation PIs simeprevir, BI-201335 (faldaprevir), and ritonavir-boosted danoprevir; and the NS5A inhibitor daclatasvir. Alisporivir, not a DAA, is a cyclophilin A inhibitor that interacts with HCV polymerase and NS5A. Although promising, this compound is on clinical hold because of an association with pancreatitis.
Initial Interferon Alfa-Free Studies
Early interferon alfa-free studies established that viral breakthrough is universal when combinations with low barriers to resistance, such as a nonnucleoside polymerase inhibitor plus a PI, are used, with addition of ribavirin often preventing early breakthrough and prolonging response. On the other hand, the higher resistance barrier combination of a PI with a nucleoside polymerase inhibitor resulted in a continuous antiviral response during short-term treatment, with no on-treatment viral breakthroughs observed.
The first trial to show that patients could achieve cure with an interferon alfa-free regimen was recently reported by Lok and colleagues.9 This small trial compared the DAA combination of the NS5A inhibitor daclatasvir and the PI asunaprevir (2-drug regimen) with the 2 agents in combination with peginterferon alfa and ribavirin (4-drug regimen) in subjects who had not responded to peginterferon alfa and ribavirin. Breakthrough occurred in 6 of the 11 dual-therapy patients during treatment and in 1 after treatment had stopped, with 4 being cured (Figure 2). All 10 patients receiving the 4-drug regimen achieved cure. It is of interest that both patients with HCV genotype 1b in the dual therapy arm achieved cure, suggesting a higher barrier to resistance in this genotype. The patient relapsing at the end of treatment had a preexisting R155K PI-resistant variant, with breakthrough being marked by emergence of the NS5A-resistant variant Q30E. Breakthrough was associated with a higher baseline viral load and was characterized by emergence of resistance to both drug classes.
Figure 2.
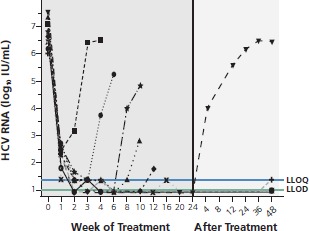
Virologic response to the investigational drugs daclatasvir and asunaprevir (interferon alfa-free regimen). Viral breakthrough occurred in 6 of 11 patients during dual therapy and 1 patient relapsed after therapy. The patient with relapse had a preexisting R155 hepatitis C virus (HCV) gene variant, with relapse marked by emergence of a Q30E variant. LLOQ indicates lower limit of quantification; LLOD, lower limit of detection. Adapted from Lok et al.9
The ability of a 2-drug regimen to achieve cure in genotype 1b patients was supported by a study in Japanese patients, who almost exclusively have genotype 1b infection. In this trial, the combination of daclatasvir and asunaprevir produced cure in 77% of more than 40 genotype 1b patients who had not responded to peginterferon alfa with ribavirin or who could not receive peginterferon alfa.10
The findings in the study reported by Lok and colleagues tend to support the notion that a 4-mutation barrier is needed for cure in genotype 1a patients, given the fact that most of these patients had breakthrough and that the dual-drug regimen did not provide such a barrier. It is less clear whether the regimen satisfies this theoretical requirement in patients with genotype 1b infection, in whom it nevertheless appeared to be successful in achieving cure. Another issue highlighted by this trial is whether continued administration of dual therapy in patients with viral breakthrough leads to emergence of additional resistance mutations. Patients with breakthrough on dual therapy continued treatment with the addition of peginterferon alfa and ribavirin. In one of these patients, a reduction in viral load occurred with the addition of peginterferon alfa and ribavirin, followed by another breakthrough, with the patient having 6 months of cumulative exposure to a failing regimen including a PI and NS5A inhibitor. PI resistance due to variants at the 168 position evolved from wild-type at baseline to D168Y and D168A during dual therapy. Both of these variants are associated with high fold changes in drug susceptibility (525-fold and 30-fold respectively) but very low relative fitness (23% and 1% respectively). Continued evolution during 4-drug therapy led to emergence of the D168T variant, which is associated with a high fold change in drug susceptibility (170-fold) and increased replicative fitness (160% that of wild-type). Such findings suggest that failing DAAs should be stopped if viral breakthrough occurs, to prevent continued evolution of resistance.
More Recent Interferon Alfa-Free Studies
With regard to more recent studies of interferon alfa-free regimens, the results of the Gilead Sciences 0120 study with the 3 DAAs plus ribavirin was reported at the 2012 EASL (European Association for the Study of the Liver) conference. Sulkowski and colleagues evaluated quadruple therapy including a PI, NS5A antagonist (2 dosage levels), nonnucleoside polymerase inhibitor, and ribavirin.11 Patients in the high-dose NS5A antagonist group (arm 2) were randomized to 12 weeks or 24 weeks of therapy if HCV RNA level was undetectable at 2 weeks (very rapid virologic response [vRVR]). All other patients received 24 weeks of treatment. Patients without vRVR had peginterferon alfa added to their treatment. Patients were also analyzed according to variants in IL28B, a gene in which the CC variant is predictive of response to peginterferon alfa with ribavirin. As shown in Figure 3, there was less difference between vRVR rates according to whether patients had IL28B CC or non-CC genotypes with the stronger regimen containing the higher NS5A antagonist dose. Viral breakthrough was more common in patients with genotype 1a HCV and was characterized by triple-class resistance variants in the majority of cases. Preliminary SVR data for patients with vRVR showed 4-week SVR in nearly all arm-2 patients after the 12-week treatment course (Figure 4). The data showed 12-week SVR in 80%. In the vRVR group getting 24 weeks of treatment, 4-week SVR was observed in all patients with available data and 12-week SVR was observed in most. Overall, 87% of patients without vRVR had viral suppression with the addition of peginterferon alfa.
Figure 3.
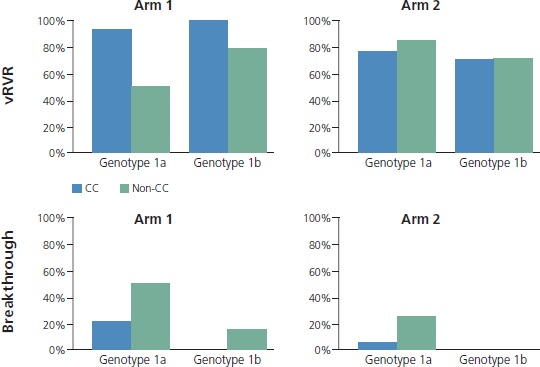
Peginterferon alfa-free therapy with 4-drug combination of a protease inhibitor, nonstructural protein 5A antagonist (2 dosage levels), nonnucleoside polymerase inhibitor, and ribavirin. Very rapid virologic response (vRVR) rates shown in genotype 1a and 1b patients receiving therapy. Arm 2 patients received the highest dose of nonstructural protein 5A antagonist. Patients were also analyzed according to IL28B variant (CC or non-CC) [top]. Triple-class resistance was seen in most of 1a breakthroughs [bottom]. Adapted with permission from Sulkowski et al.11
Figure 4.
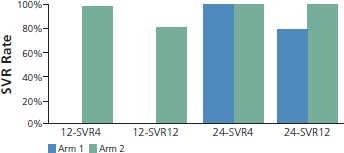
Sustained virologic response (SVR) in patients with very rapid virologic response (vRVR) on 4-drug regimen that did not include peginterferon alfa. Patients in the high-dose nonstructural protein 5A antagonist group (arm 2) who achieved vRVR were randomized to 12 weeks (12-SVR) or 24 weeks (24-SVR) of therapy. Overall, 87% (26/30) of patients without vRVR had viral suppression with the addition of peginterferon alfa. Adapted with permission from Sulkowski et al.11
Another study reported by Poordad and colleagues at the 2012 EASL conference12 examined the triple combination of a ritonavir-boosted PI (ABT-450, at 2 dosage levels), a nonnucleoside polymerase inhibitor (ABT-333), and ribavirin for 12 weeks. This combination was given to treatment-naive patients and to prior null responders to peginterferon alfa who had genotype 1 infections. In treatment-naive patients, SVR rates 12 weeks after treatment were 95% in the higher-dose boosted PI group and 93% in the lower-dose group, and 47% in the prior nonresponders. Among the treatment-naive patients, 1 in each group stopped treatment within 2 weeks due to intolerance or nonadherence; thus, all patients completing treatment had a 12-week SVR. Reasons for the poor treatment outcomes in the prior nonresponders remain unclear; none had the IL28B CC genotype. These patients exhibited an initial decline in viral load, but many had viral breakthrough during treatment or had relapse after treatment, with failure being associated with dual class resistance.
The ELECTRON study examined 12 weeks of treatment with the nucleotide polymerase inhibitor GS-7977 (sofosbuvir) plus ribavirin in patients with genotype 1 HCV who were treatment naive or were prior nonresponders to peginterferon alfa with ribavirin, and in patients with genotype 2 or 3 virus who were either treatment naive or had prior treatment failures.13,14 The trial also examined the polymerase inhibitor alone in treatment-naive patients with genotype 2 or 3 HCV, and an additional arm assessed the combination of GS-7977, ribavirin, and peginterferon alfa in treatment-naive patients with genotype 2 or 3 virus. Suppression was observed in all groups during the 12 weeks of treatment, consistent with other observations indicating that virtually no patients receiving the nucleotide polymerase inhibitor exhibited viral breakthrough during treatment. However, shortly after stopping treatment, different relapse patterns emerged. Four-week SVR rates following GS-7977 plus ribavirin were 100% in the treatment-naive genotype 2 or 3 group, 88% in the treatment-naive genotype 1 group, 80% in the treatment-experienced genotype 2 or 3 group, and only 11% (1 of 9 patients) in the genotype 1 prior null responders. SVR rates at 4 weeks were 60% in the treatment-naive genotype 2 or 3 group receiving the polymerase inhibitor without ribavirin. Population sequencing and deep sequencing studies in 5 of the 8 patients with relapse in the genotype 1 null responder group showed no evidence of the S282T polymerase resistance variant. That observation leaves open the question of how relapse occurred in these patients in the absence of identified resistance mutations.
Finally, another study included 24 weeks of treatment with the nucleotide polymerase inhibitor GS-7977 and the NS5A inhibitor daclatasvir with or without ribavirin, in 3 groups of treatment-naive patients with genotype 1 infection.15 One group started NS5A inhibitor therapy a week after starting polymerase inhibitor treatment and one started both concurrently, with neither group receiving ribavirin; the third group started all 3 drugs together. Four-week SVR rates were 100% in all 3 groups. Further follow-up will help determine whether the apparent lack of need for ribavirin with this DAA combination is borne out in this setting.
Conclusion
HCV variants that are resistant to DAAs emerge in patients who experience virologic failure. Multiclass resistance emerges in patients on interferon alfa-free regimens that contain DAAs, with the possible exception of resistance to nucleoside polymerase inhibitors. The long-term impact of emergence of resistant variants is not known at this point. It is known that such variants decay over time, but it remains unclear whether reexposure to the same drug classes will result in rapid reemergence of resistance. Progress in devising interferon alfa-free regimens is rapid and can be expected to continue in this manner for the next several years. Hope remains that interferon-free regimens can be crafted for the vast majority of HCV patients within the near future.
References
- 1.McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360(18):1827-1838. [DOI] [PubMed] [Google Scholar]
- 2.Hezode C, Forestier N, Dusheiko G, et al. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360(18):1839-1850. [DOI] [PubMed] [Google Scholar]
- 3.Poordad F, McCone JJr, Bacon BR, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan JC, De Meyer S, Bartels DJ, et al. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. J Hepatol. 2011;54(Suppl 1):S4. [DOI] [PubMed] [Google Scholar]
- 5.Margeridon S, Pogam SL, Liu TF, et al. No detection of variants bearing NS5B S282T mericitabine (MCB) resistance mutation in DAA treatment-naïve HCV genotype 1 (G1)-infected patients using ultradeep pyrosequencing (UDPS). Hepatology. 2011;54(S1):532A.21574174 [Google Scholar]
- 6.Lenz O, Fevery B, Vijgen L, et al. TMC435 in combination with peginterferon alpha-2a/ribavirin in treatment-naive patients infected with HCV genotype 1: virology analysis of the PILLAR study. [Abstract 1329.] 62nd Annual Meeting of the American Association for the Study of Liver Disease (AASLD) November 4-8, 2011; San Francisco, CA. [Google Scholar]
- 7.Pauwels F, Mostmans W, Quirynen LM, et al. Binding-site identification and genotype profiling of hepatitis C virus polymerase inhibitors. J Virol. 2007;81(13):6909-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridell RA, Wang C, Sun JH, et al. Geno-typic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology. 2011;54(6):1924-1935. [DOI] [PubMed] [Google Scholar]
- 9.Lok AS, Gardiner DF, Lawitz E, et al. Preliminary study of two antiviral agents for hepatitis C genotype 1. N Engl J Med. 2012;366(3):216-224. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki F, Ikeda K, Toyota J, et al. Dual oral therapy with the NS5A inhibitor daclatasvir (BMS-790052) and NS3 protease inhibitor asunaprevir (BMS-650032) in HCV genotype 1b-infected null responders or ineligible/intolerant to peginterferon. [Abstract 14.] 47th Annual Meeting of the European Association for the Study of the Liver (EASL) April 18-22, 2012; Barcelona, Spain. [Google Scholar]
- 11.Sulkowski M, Rodriguez-Torres M, Lawitz E, et al. Interim sustained virologic response rates in treatment-naïve HCV genotype 1a and 1b patients treated for 12 or 24 weeks with an interferon-free all-oral Quad regimen. [Abstract 1421.] 47th Annual Meeting of the European Association for the Study of the Liver (EASL) April 18-22, 2012; Barcelona, Spain. [Google Scholar]
- 12.Poordad F, Lawitz E, Kowdley KV, et al. A 12-week interferon-free regimen of ABT-450/r + ABT-333 + ribavirin achieved SVR12 in more than 90% of treatment-naïve HCV genotype-1-infected subjects and 47% of previous non-responders. [Abstract 1399.] 47th Annual Meeting of the European Association for the Study of the Liver (EASL) April 18-22, 2012; Barcelona, Spain. [Google Scholar]
- 13.Gane EJ, Stedman CA, Hyland RH, et al. ELECTRON: once daily PSI-7977 plus RBV in HCV GT1/2/3. [Abstract 1113.] 47th Annual Meeting of the European Association for the Study of the Liver (EASL) April 18-22, 2012; Barcelona, Spain. [Google Scholar]
- 14.Gane E, Stedman C, Anderson J, et al. 100% Rapid virologic response for PSI-7977 + ribavirin in genotype 1 null responders (ELECTRON): early viral decline similar to that observed in genotype 1 and genotype 2/3 treatment-naïve patients. [Abstract 54LB.] 19th Conference on Retroviruses and Opportunistic Infections (CROI) March 5-8, 2012; Seattle, WA. [Google Scholar]
- 15.Sulkowski MS, Gardiner D, Lawitz E, et al. Potent viral suppression with the all-oral combination of daclatasvir (NS5A inhibitor) and GS-7977 (nucleotide NS5B inhibitor), +/- ribavirin, in treatment-naive patients with chronic HCV GT1, 2, or 3 (100% SVR gt1, 91% gt2). [Abstract 1422.] 47th Annual Meeting of the European Association for the Study of the Liver (EASL) April 18-22, 2012; Barcelona, Spain. [Google Scholar]


