Abstract
Background
Genetic tools including constitutive and inducible promoters have been developed over the last few decades for strain engineering in Streptomyces. Inducible promoters are useful for controlling gene expression, however only a limited number are applicable to Streptomyces. The aim of this study is to develop a controllable protein expression system based on an inducible promoter using sugar inducer, which has not yet been widely applied in Streptomyces.
Results
To determine a candidate promoter, inducible protein expression was first examined in Streptomyces avermitilis MA-4680 using various carbon sources. Xylose isomerase (xylA) promoter derived from xylose (xyl) operon was selected due to strong expression of xylose isomerase (XylA) in the presence of d-xylose. Next, a xylose-inducible protein expression system was constructed by investigating heterologous protein expression (chitobiase as a model protein) driven by the xylA promoter in Streptomyces lividans. Chitobiase activity was detected at high levels in S. lividans strain harboring an expression vector with xylA promoter (pXC), under both xylose-induced and non-induced conditions. Thus, S. avermitilis xylR gene, which encodes a putative repressor of xyl operon, was introduced into constructed vectors in order to control protein expression by d-xylose. Among strains constructed in the study, XCPR strain harboring pXCPR vector exhibited strict regulation of protein expression. Chitobiase activity in the XCPR strain was observed to be 24 times higher under xylose-induced conditions than that under non-induced conditions.
Conclusion
In this study, a strictly regulated protein expression system was developed based on a xylose-induced system. As far as we could ascertain, this is the first report of engineered inducible protein expression in Streptomyces by means of a xylose-induced system. This system might be applicable for controllable expression of toxic products or in the field of synthetic biology using Streptomyces strains.
Electronic supplementary material
The online version of this article (10.1186/s12934-018-0991-y) contains supplementary material, which is available to authorized users.
Keywords: Xylose, Xylose isomerase promoter, Streptomyces avermitilis, Streptomyces lividans, Heterologous protein production
Background
Streptomyces strains are aerobic, gram-positive, mycelia-forming soil bacteria with high G+C content DNA. These strains have produced many useful compounds such as secondary metabolites [1, 2]. They are also used as industrial strains to produce various products such as commercial antibiotics and antifungals for therapeutic, environmental, and agricultural applications. Streptomyces strains also enable the secretion of various hydrolytic enzymes into culture medium; hence, they are useful for native and heterologous protein production as a secretion from culture medium [3, 4]. This secretion property might also be employed to eliminate endotoxin contamination to simplify purification processes, and to fold proteins precisely [5, 6].
Genetic tools have been developed for Streptomyces, and these tools have been employed for strain engineering and synthetic biology applications [7]. Promoters are one key factor for protein expression, and development of inducible and constitutive promoters has been reported over the last few decades for Streptomyces [3, 8]. Inducible promoters are useful for controlling gene expression in basic and applied studies. For example, the expression level of interesting genes can be changed at certain stages in order to investigate function, or for application in synthetic biology. Inducible promoters can also control the concentration of toxic products to limit the impact on cell growth. As yet, however, only a limited number of inducible promoters, such as PtipA and PnitA, have been developed for Streptomyces [9–11].
Inducible promoters by sugar inducers allow for the strict regulation of gene expression in target proteins using inexpensive input [12], and the promoters have been widely developed for protein expression in various species, including xylose-regulation systems for protein expression that include Pxyl (Bacillus subtilis [13], Clostridium perfringens [14], Brevibacillus choshinensis [15]), and PxylT (Lactococcus lactis [12]). Other systems using sugar inducers, such as arabinose and rhamnose, have been developed for Escherichia coli [16, 17]. Streptomyces strains have various specific permeases and are able to metabolize various carbon sources [18], however engineered inducible promoters by sugar inducers have not yet been widely applied for protein expression in Streptomyces.
The aim of this study is to develop an engineered inducible protein expression system for Streptomyces based on a sugar inducer. Xylose isomerase (xylA) promoter and xylose (xyl) operon were chosen in an initial attempt to investigate induced protein expression in a Streptomyces strain using various carbon sources. xylA promoter is a useful candidate for protein expression because actinomycetes strains are intrinsically good XylA protein producers under the control of xylA promoter [19]. xylA promoter from Actinoplanes missouriensis provides a system for overexpressing Cel6 protein in Streptomyces lividans, which is not inducible [20]. xylA promoter was also employed for genetic modification, such as efficient expression of cre gene for deletion of the 1.5 Mb region in Streptomyces avermitilis chromosome [21]. In the present study, an inducible protein expression system was designed under the control of xylA promoter with the components of XylR and d-xylose. Strictly regulated protein expression was finally achieved by d-xylose via constructed vector.
Results
Streptomyces avermitilis MA-4680 protein expression profiles for different carbon sources
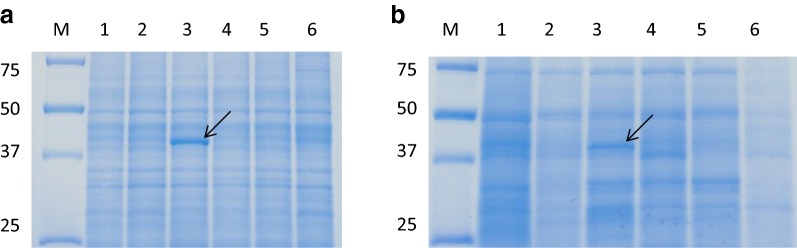
Inducible protein expression was first investigated in Streptomyces avermitilis MA-4680 using different carbon sources (glycerol, d-glucose, d-xylose, d-galactose, and d-fructose). Protein expression profiles of cell extracts and culture supernatants are shown for each carbon source (2.0%) in Fig. 1 (whole gels shown in Additional file 1: Figure S1). One protein was clearly detected above 37 kDa in the presence of d-xylose from both the cell extract and the culture supernatant (Fig. 1, arrows). On the other hand, this protein band was not detected in the presence of other carbon sources. The detected protein was identified as putative xylose isomerase (XylA, SAV_7182) by MALDI-TOF–MS analysis after the purification of cell extracts and culture supernatants. XylA expression was also detected for this strain in the presence of 1.0% d-xylose (Additional file 2: Figure S2). These results suggest that the expression of the xylA gene was induced in S. avermitilis by the addition of d-xylose.
Fig. 1.
SDS-PAGE analysis of S. avermitilis protein expression in a cell extracts and b culture supernatants: 2.0% 1. Glycerol, 2. d-glucose, 3. d-xylose, 4. d-galactose, 5. d-fructose, and 6. no sugar. M: molecular weight marker. Arrows indicate Xylose isomerase (XylA) protein
Genetic organization of xylose operon in S. avermitilis and mechanism of d-xylose metabolism
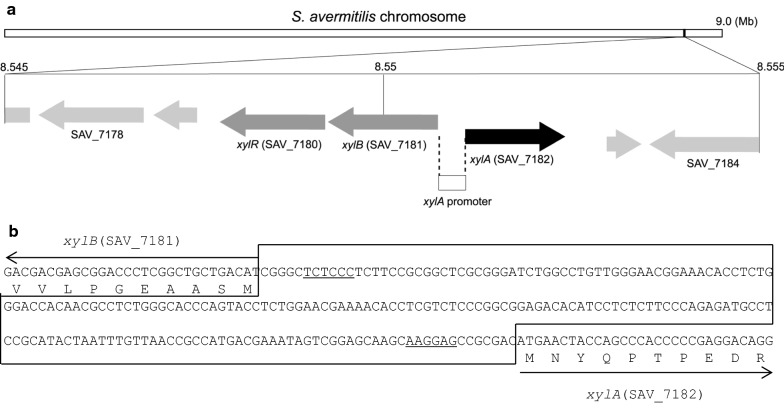
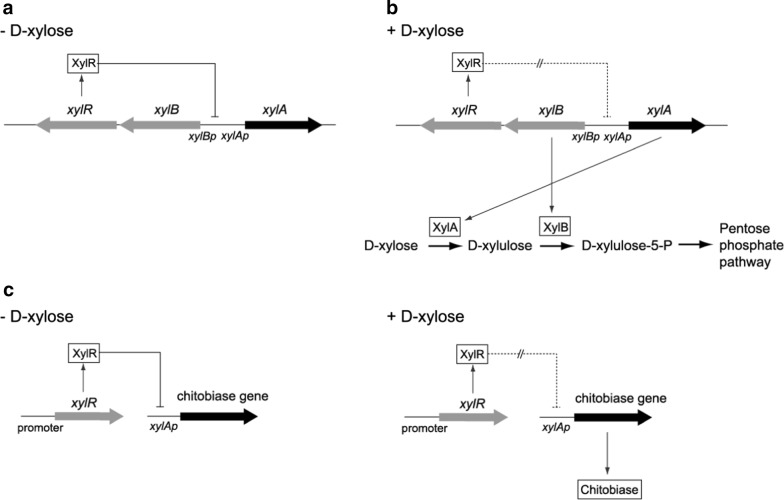
As mentioned above, S. avermitilis XylA was induced in the presence of d-xylose. The xylA gene is located in xylose (xyl) operon, which consists of xylA (SAV_7182; xylose isomerase), xylB (SAV_7181; xylulose kinase), and xylR (SAV_7180; xylose operon regulator) genes from the complete genome sequence of S. avermitilis MA-4680 [22] (Fig. 2). The xyl operon is related to d-xylose metabolism, which involves the transport of d-xylose, the isomerization of d-xylose to d-xylulose (mediated by XylA), and the phosphorylation of d-xylulose to d-xylulose-5-phosphate (mediated by XylB) (Fig. 3a, b). XylR acts as a repressor for xyl operon in S. lividans TK24, as reported in a deletion study of the xylR gene [23]. Genes driven by xyl promoters are induced in the presence of d-xylose [23, 24] (Fig. 3a, b). xylA promoter and xyl operon were selected in order to develop a xylose-dependent protein expression system in Streptomyces (Fig. 3c).
Fig. 2.
a Gene organization in the intergenic region containing the xyl operon of S. avermitilis MA-4680 from the complete genome sequence. b The nucleotide sequence of the intergenic promoter region of the divergently transcribled xylA and xylB genes of S. avermitilis. The putative ribosome binding sites from S. rubiginosus are shown as underlined sequences [24]
Fig. 3.
Genetic organization of Streptomyces xyl operon and d-Xylose metabolism. Schematic representation of putative gene regulation a in the absence of d-xylose and b in the presence of d-xylose. c Conceptual diagram of projected study
Chitobiase expression of the xylAp expression system in Streptomyces XC strain
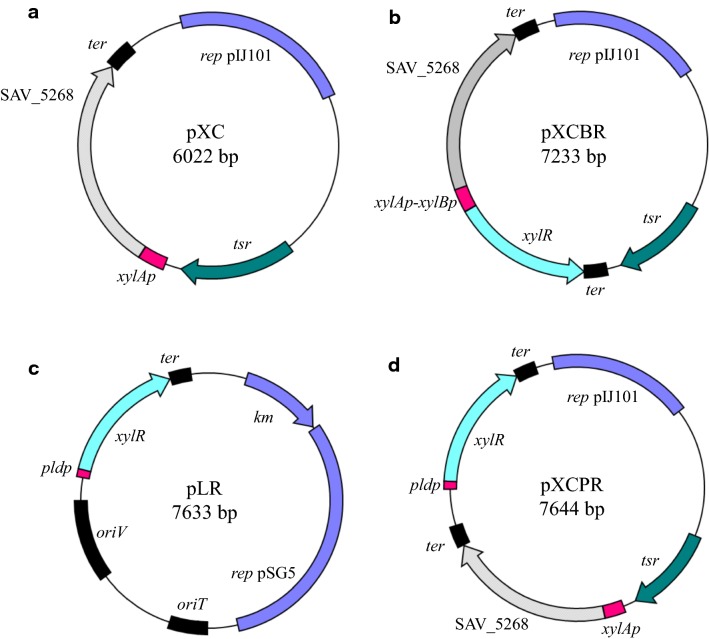
pXC vector was first constructed as a high-copy vector for protein expression under S. avermitilis xylA promoter without xylR gene in the same vector (Fig. 4a). The nagZ4 gene (SAV_5268), which encodes chitobiase in S. avermitilis, was employed as a reporter in order to investigate the system using the xylA promoter. The protein encoded by this gene has a putative signal peptide sequence, and the secretion of the protein was previously confirmed in culture supernatant (data not shown). This vector was transformed into S. lividans 1326 (named XC strain in Table 1).
Fig. 4.
Schematic representation of xylose-induced expression vectors pXC, pXCBR, pLR, and pXCPR. oriT, origin of transfer; oriV, replication origin of E. coli from pK18mob (ATCC® 87095™); rep pIJ101 and rep pSG5, replicon of Streptomyces from pIJ101 [25] and pGM160 [36]; tsr, thiostrepton resistance gene; km, kanamycin resistance gene
Table 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant feature |
|---|---|
| Plasmid | |
| pIJ350 | Versatile vector for protein expression; thiostrepton resistance marker |
| pUC19 | Vector for cloning genes |
| pGM160 | A shuttle vector containing pSG5 ori |
| pK18mob | Cloning vector; oriT, oriV; kanamycin resistance marker |
| pXC | Vector for expression of chitobiase gene (SAV_5268) under the control of xylA promoter; thiostrepton resistance marker |
| pXCBR | Vector for expression of chitobiase gene (SAV_5268) under the control of xylA promoter and xylR gene under the control of xylB promoter; thiostrepton resistance marker |
| pLR | Vector containing pSG5 ori for expression of xylR gene under the control of pld promoter; kanamycin resistance marker |
| pXCPR | Vector for expression of chitobiase gene (SAV_5268) under the control of xylA promoter and xylR gene under the control of pld promoter; thiostrepton resistance marker |
| Strain | |
| Escherichia coli strain JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) e14− (mcrA−) supE44 relA1 Δ(lacproAB)/F’[traD36 proAB + lacI q lacZΔM15] |
| S. lividans 1326 | WT strain |
| XC | Streptomyces lividans strain harboring pXC vector |
| XCBR | Streptomyces lividans strain harboring pXCBR vector |
| XCr | Streptomyces lividans strain harboring pXC and pLR vectors |
| XCPR | Streptomyces lividans strain harboring pXCPR vector |
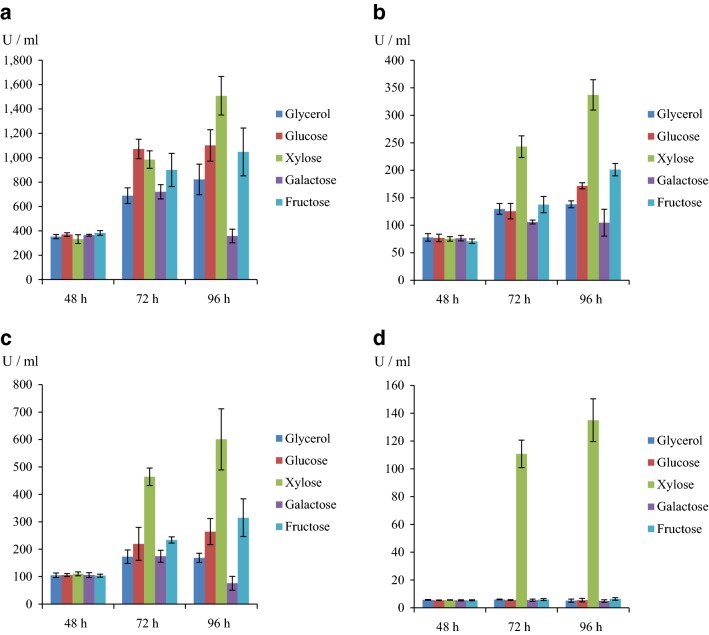
Xylose-induced chitobiase expression was investigated using the S. lividans XC strain. The strain was incubated for 48 h, at which time carbon sources (glycerol, d-glucose, d-xylose, d-galactose and d-fructose) were added to final concentration of 1.0%, and incubation continued for an additional 48 h (96 h total cultivation). This strain exhibited remarkable chitobiase activity at 48 h before addition of the carbon sources. After addition of carbon sources (glycerol, d-glucose, d-xylose, d-galactose and d-fructose) and further incubation to 96 h, activity was found to have increased 2.3, 3.0, 4.5, 1.0, and 2.7 times, respectively (Fig. 5a). The highest activity was detected in the presence of d-xylose at 96 h, and higher activity was detected with d-glucose compared to d-xylose at 72 h. The XC strain expressed chitobiase in the presence of all carbon sources. There is no evidence that xylA promoter is controlled by d-xylose, and xylA promoter appears to be constitutively expressed.
Fig. 5.
Chitobiase activity of a S. lividans XC, b S. lividans XCBR, c S. lividans XCr, and d S. lividans XCPR after 48, 72, and 96 h cultivation. After 48 h, carbon sources (glycerol, d-glucose, d-xylose, d-galactose, and d-fructose) were added to a final concentration of 1.0%, and cultivation was continued for 24 and 48 h (72 and 96 h total)
Chitobiase expression of the xylAp-XylR expression system in the XCBR strain
In order to develop a controllable expression system, pXCBR vector was constructed based on the S. avermitilis xylose (xyl) operon (Fig. 4b). pXCBR is a high-copy vector for chitobiase gene expression driven by the xylA promoter, and S. avermitilis xylR gene was introduced in the same vector. xylR gene is expressed under the control of xylB promoter in a common intergenic region between xylA (SAV_7182) and xylB (SAV_7181) genes derived from S. avermitilis chromosome. The expression vector was transformed into S. lividans 1326 (named XCBR strain in Table 1).
Chitobiase activity was investigated in the XCBR strain. Incubation and addition of carbon sources were carried out as described above. This strain exhibited a certain amount of chitobiase activity before addition of the carbon sources (Fig. 5b). With the addition of glycerol, d-glucose, d-xylose, d-galactose, and d-fructose, activity at 96 h was found to have increased 1.8, 2.2, 4.5, 1.4, and 2.8 times, respectively. d-xylose induced slightly higher chitobiase activity than other carbon sources at 72 and 96 h. These results suggest that the xylA promoter was slightly regulated by XylR expression in the intergenic region, but such regulation was insufficient when using the region of divergent promoters.
Chitobiase expression of the xylAp-XylR expression system in the XCr and XCPR strains
Different sets of basic vector and promoter were employed to construct pLR and pXCPR (Fig. 4c, d). The pLR vector is based on the medium copy number plasmid pSG5, while the pXCPR vector is based on the high copy number plasmid pIJ101 [25]. Streptomyces cinnamoneum phospholipase D (PLD) promoter was selected to control xylR gene expression in pLR and pXCPR vectors [26]. pXC (vector for expression of chitobiase gene, Fig. 4a) and pLR vectors were transformed into S. lividans 1326 (named XCr strain in Table 1). pXCPR vector was transformed into S. lividans 1326 (named XCPR strain in Table 1).
XCr and XCPR strains were investigated for chitobiase expression using the various carbon sources as described above. The XCr strain exhibited moderate chitobiase activity at 48 h. The addition of 1.0% glycerol, d-glucose, d-xylose, d-galactose, and d-fructose increased activity 1.6, 2.5, 5.4, 0.7, and 3.0 times at 96 h, respectively (Fig. 5c), suggesting that induction is weak in the XCr strain. The XCPR strain, on the other hand, exhibited inducible expression of chitobiase (Fig. 5d). Chitobiase activity was low in this strain before addition of the carbon sources. The activity was 24-fold higher after addition of d-xylose at 96 h, while the activity remained low after the addition of other carbon sources. These results indicate that the XCPR strain strictly regulates the xylA promoter with d-xylose. Chitobiase activity was also investigated in the XCPR strain with the different d-xylose concentrations from 0.0 to 2.0% at 48, 72, and 96 h (Additional file 3: Figure S3). The difference in activity between 1.0 and 2.0% was found to be slight, and growth was barely affected with different d-xylose concentrations from 0.0 to 2.0%.
Discussion
An inducible promoter would be useful in Streptomyces for such applications as controllable expression of heterologous protein, or as a genetic tool for expressing interesting genes at certain stages [7, 8, 10]. The aim of this study is to develop a controllable gene expression system for Streptomyces strains based on inducible promoter related with carbon sources. A candidate inducible promoter was first selected by analyzing induced proteins with different carbon sources, and an inducible protein expression system was constructed driven by the candidate promoter.
Protein induction was observed with different carbon sources in S. avermitilis MA-4680. S. avermitilis is particularly well-known for avermectin production [27], and the complete genome sequence of this strain has already been reported [22]. In the analysis of induced proteins using various carbon sources as inducer, XylA protein was found to be well expressed with d-xylose in the cell extract and the culture supernatant (Fig. 1). Extracellular secretion of XylA in some Streptomyces species has been reported [19], but this is fairly uncommon because it does not contain a putative secreted signal sequence. The XylA detected in culture supernatants might have been released from the cell due to cell wall permeability, or by partial lysis of the cells [19].
The xylA gene exists in the xyl operon from the complete genome sequence of S. avermitilis MA-4680 (Fig. 2) [22]. The S. avermitilis xyl operon consists of xylA (SAV_7182), xylB (SAV_7181), and xylR (SAV_7180) genes. In comparison, S. coelicolor A3 (2) [28] and S. lividans 1326 [29] show the same gene organization such as xylA (SCO1169 or SLI_1446), xylB (SCO1170 or SLI_1447), and xylR (SCO1171 or SLI_1448) genes (Additional file 4: Figure S4). On the other hand, the organization of the xylA, xylB and xylR locus in S. griseus (NBRC 13350) [30] is a little different, such as xylR (SGR_1069), xylA (SGR_1070), and xylB (SGR_1071) genes (Additional file 4: Figure S4). xyl gene regulatory mechanism by regulatory protein differs among species. The regulator functions as an activator in E. coli, but as a repressor in Bacillus [31–33]. Regulatory protein (XylR) acts as a repressor for xyl operon in Streptomyces TK24, as reported in a deletion study of the xylR gene [23]. In the present study, the xylA promoter and xyl operon were selected in order to develop a xylose-dependent protein expression system in Streptomyces (Fig. 3c).
A set of protein expression vectors was constructed based on S. avermitilis xylA promoter and the xyl operon (Fig. 4). Extracellular chitobiase activity was investigated in S. lividans strain harboring these vectors. S. lividans is known as an attractive host for heterologous protein production because it has a low level of endogenous proteolytic activity, and plasmid-based expression systems have been established [4, 5]. Expression of extracellular chitobiase from S. avermitilis was examined as a model. S. lividans also contains a putative chitobiase gene in its chromosome (SLI_3133), however S. lividans wild-type strain tested under the same conditions as in the present study displayed no intrinsic chitobiase activity.
In the strain harboring pXC vector without S. avermitilis xylR gene in the same vector, activity was detected under both xylose-induced and non-induced conditions (Figs. 4a, 5a), indicating that the XC strain is not capable of controlling protein expression with d-xylose. xylR gene functioning as a repressor also exists in the S. lividans chromosome, but intrinsic XylR might be insufficient for regulation of the S. avermitilis xylA promoter, presumably because of the high-copy number of the xylA promoter in the vector as opposed to the expression level of xylR gene in the chromosome.
From these results, vectors were constructed containing the S. avermitilis XylR component in order to repress xylA promoter activity in the absence of d-xylose. In the XCBR vector, the common intergenic region between xylA and xylBR genes was employed for the expression of S. avermitilis xylR gene (Figs. 2 and 4b). Common intergenic regions exist in several bacteria species, such as between xylR and xylAB genes in Bacillus subtilis and between xylR and xylBA genes in Clostridium, although gene organization is different in Streptomyces [13, 14]. Divergent promoters in these regions are easily employed to control xylR gene for vector construction of an inducible target protein expression in these strains [13, 14]. In the present study, the intergenic region of Streptomyces xyl operon was employed to express the xylR gene by placing the gene under the control of xylB promoter (Fig. 4b). Although protein induction occurred using the constructed vector, the induction ratio was weak in the Streptomyces strain (XCBR strain in Fig. 5b).
From the result of protein expression using the XCBR strain, different sets of promoter and vector were employed for suitable expression of the S. avermitilis xylR gene to optimize xylose-induced expression. S. cinnamoneum phospholipase D (PLD) promoter was employed for expression of the XylR regulatory protein. The promoter is constitutive, and examples have been reported to express heterologous genes under the control of the promoter in S. lividans [3, 34]. Moreover, two different vectors were employed. The pLR vector (Fig. 4c) is based on a medium copy number plasmid, and the pXCPR vector (Fig. 4d) is based on a high copy number plasmid. Xylose-induced expression was investigated using XCr strain (harboring pXC and pLR vectors; Fig. 4a, c), and XCPR strain (harboring pXCPR vector; Fig. 4d). Different protein expression behaviors were observed between these two strains. The XCr strain showed regulation of chitobiase expression at low levels (Fig. 5c), while the XCPR strain exhibited strictly regulated expression at 72 and 96 h (Fig. 5d). XylR expression from pLR (medium copy number, Fig. 4c) might be insufficient to regulate the xylA promoter in the XCr strain, but XylR expression was sufficient using pXCPR (high copy number, Fig. 4d) in the XCPR strain. As seen in the XC strain, intrinsic XylR may be insufficient for regulation of this system (Fig. 5a). Results show that strictly regulated xylose-induced expression is achieved by the combination of xyl operon components (xylA promoter and XylR) for d-xylose metabolism in Streptomyces.
The engineering of a strictly regulated xylose-induced system has only just begun, and further study is needed for practical applications using d-xylose, such as controlling the expression of toxic recombinant proteins, or gene expression in the field of synthetic biology. Moreover, the d-xylose induced system might be employed using mixed sugars such as d-glucose. Gene expression was induced in the presence of 1% (w/v) d-xylose and d-glucose in S. rubiginosus, although the expression was lower than in the presence of d-xylose alone [24]. The constructed system in the present study may be applicable in such a case.
Conclusions
In this study, strictly regulated protein expression was achieved, based on a xylose-induced system. First, the induction of XylA in a model Streptomyces strain was found to be greater with d-xylose than with other carbon sources. Next, xylose-induced protein expression system was developed using constructed vectors containing xylA promoter and XylR components. As far as we could ascertain, this is the first report of engineered inducible protein expression in Streptomyces by means of a xylose-induced system. This system might be applicable with Streptomyces for controllable expression of toxic products or in the field of synthetic biology.
Methods
Bacterial strains in this study
Streptomyces avermitilis MA-4680 (NBRC 14893) and S. lividans 1326 (NBRC 15675) were purchased from National Institute of Technology and Evaluation (NITE, Chiba, Japan). Escherichia coli JM109 (Takara, Shiga, Japan) was used as the host for DNA manipulation (Table 1).
SDS analysis of S. avermitilis MA-4680 induced protein in the presence of different carbons
Streptomyces avermitilis MA-4680 was cultured on inorganic salt starch agar plates. A single colony was pre-cultured in TSB medium (17 g/l pancreatic digest of casein, 3 g/l papaic digest of soybean meal, 2.5 g/l glucose, 5 g/l sodium chloride, and 2.5 g/l dipotassium phosphate, Becton, Dickinson and Company, Sparks, MD, USA) for 48 h at 28 °C, then inoculated into modified TSB medium (17 g/l pancreatic digest of casein, 3 g/l papaic digest of soybean meal, 5 g/l sodium chloride, and 2.5 g/l dipotassium phosphate) in a 500 ml baffled flask in the presence of 1.0 or 2.0% various carbon sources (glycerol, d-glucose, d-xylose, d-galactose, and d-fructose) for 96 h at 28 °C. Cells were harvested by centrifugation and re-suspended in 20 mM potassium phosphate buffer (pH 7.0). Suspensions were sonicated and centrifuged. Cell extracts were analyzed by e-PAGEL (15%) (ATTO, Tokyo, Japan). Culture supernatants (200 μl) were desalted with Bio-Spin®6 Tris Columns (Bio-Rad, Hercules, CA, USA), then lyophilized. Samples were dissolved by SDS sample buffer (0.125 M Tris–HCl (pH 6.8), 4% (w/v) SDS solution, 20% (w/v) glycerol, 0.01% (w/v) BPB, 0.12 M 2-mercaptoethanol) and loaded onto e-PAGEL (15%). Culture supernatants and cell extracts were concentrated using Amicon® Ultra (Merck Millipore, Co Cork, Ireland). From the samples, fractions including XylA protein were separated via HiTrap Q column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). After the fractions were concentrated using Amicon® Ultra, XylA protein was purified by SDS-PAGE and analyzed by MALDI-TOF–MS (Bruker Daltonics, Leipzig, Germany).
Construction of pXC vector
A chitobiase expression vector pXC carrying a xylose isomerase promoter (xylAp) was constructed as follows:
Plasmid pXC carries the Streptomyces avermitilis xylAp, followed by the coding sequence for SAV_5268 (nagZ4 gene) and the Streptomyces cinnamoneum PLD terminator (pldt) on pIJ350 [35]. To construct this plasmid, xylAp (the selected region lacks xylB RBS from xylA-xylB divergent promoter region in Fig. 2) and pldt fragments were amplified via PCR using primer pairs XIApF-XIpR and PLDtF-PLDtR, respectively (Table 2). These two pairs included 27-bp overlapping sequences containing multi-cloning sites that allowed them to anneal with one another, so that the products worked as primers to amplify a 0.4 kb joined fragment. Plasmid pIJ350 was digested with PstI, the ends of which were blunted by KOD DNA polymerase (Toyobo, Osaka, Japan) and then ligated with a 0.4-kb amplified fragment containing xylAp and pldt. The resultant vector was subsequently digested with Aor51HI and EcoT22I and ligated with a sav5268-containing fragment, which was amplified by PCR using primers SAV5268F and SAV5268R (Table 2) and then digested with EcoT22I. Thus constructed, the plasmid was then named pXC.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| XIApF | 5′-CTTCCGCGGCTCGCGGGATCTGGC |
| XIpR | 5′-GCATGCATCTAGAAGCTTGAATTCGCTAGCGCTGCGGCTCCTTGCTTGCTCCG |
| PLDtF | 5′-AGCGAATTCAAGCTTCTAGATGCATGCGACGACTGAGCGCCCGGACG |
| PLDtR | 5′-ATTTCCGGTCGGTTCGGGGCCAGCGCAT |
| PLDpF | 5′-ATATATGGTACCGGCTCCCGGGAGCTGATAGC |
| PLDpR | 5′-GCATGCATCTAGAAGCTTGAATTCGCTAGCGCTGCATCCTTAAACGAAGTAAC |
| PLDtR_KpnI | 5′-ATATATGGTACCATTTCCGGTCGGTTCGGGGCCAGCGCAT |
| SAV5268F | 5′-ATGAGACAGCACCACAGAACGCC |
| SAV5268R | 5′-ACATGCATGCCTACGCACCGGGCCAGGGAA |
| XylAp_XylBp_NdeI | 5′-ATATATCATATGGCTCTCCCTCTTCCGCGGCTCGCGGGATCTG |
| XylRF_NdeI | 5′-ATATATCATATGACCGCACCGCTGCACGA |
| PLDtR_PstI | 5′-ATATATCTGCAGATTTCCGGTCGGTTCGGGGCCAGCGCAT |
| XylRF | 5′-AATTATAGCGCTATGACCGCACCGCTGCACGA |
| SG5F_BstBI | 5′-TTTTTTCGAACGCGTCGTCGTCGACGGCCT |
| SG5R_BstBI | 5′-TTTTTTCGAAGATCACGAGGTCACTCCGTC |
| PsXylRF_EcoRI | 5′-ATAATTGAATTCGGCTCCCGGGAGCTGATAGC |
| PtXylRR_HindIII | 5′-ATGCATAAGCTTATTTCCGGTCGGTTCGGGGC |
| XylRR | 5′-ATATATATGCATCTACCGGTGCGTCGCCGC |
Overlap sequences are underlined
Aor51HI restriction sites are shown in bold
Construction of pXCBR and pXCPR vectors
The inducible chitobiase expression vectors pXCBR and pXCPR, carrying a xylose isomerase promoter (xylA promoter) and a xylR gene under the control of different promoters, were constructed as follows:
Plasmid pXCBR contains the xylR gene under control of xylB promoter in a common intergenic region between xylA and xylB genes. To construct this plasmid, the xylAp-sav5268-pldt fragment was amplified by PCR using the primer pair (XylAp_XylBp_NdeI and PLDtR_KpnI primers) and pXC vector as a template (Table 2). The xylR-pldt fragment was also amplified by PCR using the primer pair (XylRF_NdeI and PLDtR_PstI primers) and pXCPR vector as a template. The xylAp-sav5268-pldt fragment was digested with NdeI and KpnI. The xylR-pldt fragment was digested with NdeI and PstI. These fragments were ligated and inserted into the PstI/KpnI restriction sites of pIJ350, yielding plasmid pXCBR.
Plasmid pXCPR contains the xylR gene under control of the S. cinnamoneum phospholipase D (PLD) promoter [26]. To construct this plasmid, pldp and pldt fragments were amplified via PCR using primer pairs PLDpF-PLDpR and PLDtF-PLDtR_KpnI (Table 2). Amplified pldp and pldt fragments were joined by 27-bp overlapping sequences, and the fragment was amplified via PCR using the primer pair PLDpF-PLDtR_KpnI. The amplified fragment and pUC19 were digested with KpnI and ligated together. The resultant vector was digested with Aor51HI and EcoT22I and ligated with the xylR-containing fragment amplified via PCR using the primer pair XylRF-XylRR and S. avermitilis genome as a template. The resultant plasmid was referred to as pPR. The XylR expression cassette pldp-xylR-pldt was obtained from pPR as a KpnI digested fragment. This fragment was inserted into the KpnI site of pXC, yielding the plasmid pXCPR.
Construction of pLR vector
The pLR vector contains pSG5 origin and was constructed as follows:
pSG5 origin was amplified by PCR using the primer pair (SG5F_BstBI and SG5R_BstBI primers) and pGM160 as a template [36]. The amplified fragment and pK18mob vector were digested with BstBI and ligated together [37]. The resultant vector was subsequently digested with EcoRI and HindIII and was ligated with the XylR expression cassette pldp-xylR-pldt, which was amplified by PCR using PsXylRF_EcoRI and PtXylRR_HindIII primers (Table 2) and digested with EcoRI and HindIII. The constructed plasmid was designated pLR.
Transformation, culture, and expression of chitobiase in S. lividans
Streptomyces lividans XC, XCBR, and XCPR were obtained by introducing plasmids pXC, pXCBR, and pXCPR into S. lividans 1326 via protoplast transformation [38]. S. lividans XCr was obtained by introducing plasmid pLR into S. lividans XC. Single colonies of each transformant were pre-cultured in modified TSB medium for 3 days at 28 °C. Seed cultures were then transferred into the TSB medium, and cultivation was performed at 28 °C for 48 h. After 48 h, various carbon sources (glycerol, d-glucose, d-xylose, d-galactose, and d-fructose) were added to the medium to a final concentration of 1.0%, and cultivation was continued at 28 °C for 24 and 48 h (72 and 96 h total). Supernatants were harvested by centrifugation. In order to investigate chitobiase expression in XCPR strain at different d-xylose concentrations, seed cultures were transferred into the modified TSB medium, and cultivation was performed at 28 °C for 48 h. After 48 h, d-xylose was added to final concentrations of 0.0, 0.5, 1.0, and 2.0%. Cultivation was continued at 28 °C for 24 and 48 h (72 and 96 h total).
Measurement of chitobiase activity
Culture supernatants were measured for chitobiase assay. Chitobiase activity was measured using 4-Nitrophenyl N-acetyl-β-d-glucosaminide (Sigma-Aldrich, St. Louis, MO, USA) as a substrate. An assay was performed according to an established method [39]. One unit of enzyme activity was defined as the amount of enzyme that releases 1 μmol of 4-nitrophenol per minute. Assays were repeated three times.
Additional files
Additional file 1: Figure S1. SDS-PAGE gels of S. avermitilis protein expression profiles for (A) cell extracts and (B) culture supernatants: 2.0% 1. Glycerol, 2. D-glucose, 3. D-xylose, 4. D-galactose, 5. D-fructose, and 6. no sugar. M: molecular weight marker. Arrows indicate Xylose isomerase (XylA) protein.
Additional file 2: Figure S2. SDS-PAGE gels of S. avermitilis protein expression profiles for (A) cell extracts and (B) culture supernatants: 1.0% 1. Glycerol, 2. D-glucose, 3. D-xylose, 4. D-galactose, 5. D-fructose, and 6. no sugar. M: molecular weight marker. Arrows indicate Xylose isomerase (XylA) protein.
Additional file 3: Figure S3. (A) Chitobiase activity and (B) optical density (600 nm) of S. lividans XCPR after 48, 72, and 96 h cultivation. After 48 h, D-xylose was added to a final concentration of 0.0, 0.5, 1.0, and 2.0%. Cultivation was continued for 24 and 48 h (72 and 96 h total).
Additional file 4: Figure S4. Gene organization in the xyl operon of S. avermitilis MA-4680, S. coelicolor A3 (2), S. lividans 1326, and S. griseus (NBRC 13350) [22, 28, 29, 30].
Authors’ contributions
YN, NK, CO, HI, and MS designed research; YN, NK and AU performed research; YN, NK and AU analyzed data; and YN and NK wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to Akiko Matsuyama for technical assistance.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its additional files].
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by a Special Coordination Fund for Promoting Science and Technology, Creation of Innovative Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuji Noguchi, Email: yuji.noguchi@nagase.co.jp.
Norimasa Kashiwagi, Email: nkashiwagi@port.kobe-u.ac.jp.
Atsuko Uzura, Email: atsuko.uzura@nagase.co.jp.
Chiaki Ogino, Email: ochiaki@port.kobe-u.ac.jp.
Akihiko Kondo, Email: akondo@kobe-u.ac.jp.
Haruo Ikeda, Email: ikeda@ls.kitasato-u.ac.jp.
Masahiro Sota, Email: masahiro.sota@nagase.co.jp.
References
- 1.Hwang KS, Kim HU, Charusanti P, Palsson BØ, Lee SY. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol Adv. 2014;32:255–268. doi: 10.1016/j.biotechadv.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, Yang H, Shin HD, Li J, Du G, Chen J. Recent advances in recombinant protein expression by Corynebacterium, Brevibacterium, and Streptomyces: from transcription and translation regulation to secretion pathway selection. Appl Microbiol Biotechnol. 2013;97:9597–9608. doi: 10.1007/s00253-013-5250-x. [DOI] [PubMed] [Google Scholar]
- 4.Vrancken K, Anné J. Secretory production of recombinant proteins by Streptomyces. Future Microbiol. 2009;4:181–188. doi: 10.2217/17460913.4.2.181. [DOI] [PubMed] [Google Scholar]
- 5.Binnie C, Cossar JD, Stewart DI. Heterologous biopharmaceutical protein expression in Streptomyces. Trends Biotechnol. 1997;15:315–320. doi: 10.1016/S0167-7799(97)01062-7. [DOI] [PubMed] [Google Scholar]
- 6.Anné J, Vrancken K, Van Mellaert L, Van Impe J, Bernaerts K. Protein secretion biotechnology in Gram-positive bacteria with special emphasis on Streptomyces lividans. Biochem Biophys Acta. 2014;1843:1750–1761. doi: 10.1016/j.bbamcr.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 7.Medema MH, Breitling R, Takano E. Synthetic biology in Streptomyces bacteria. Methods Enzymol. 2011;497:485–502. doi: 10.1016/B978-0-12-385075-1.00021-4. [DOI] [PubMed] [Google Scholar]
- 8.Myronovskyi M, Luzhetskyy A. Native and engineered promoters in natural product discovery. Nat Prod Rep. 2016;33:1006–1019. doi: 10.1039/C6NP00002A. [DOI] [PubMed] [Google Scholar]
- 9.Takano E, White J, Thompson CJ, Bibb MJ. Constructions of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene. 1995;166:133–137. doi: 10.1016/0378-1119(95)00545-2. [DOI] [PubMed] [Google Scholar]
- 10.Herai S, Hashimoto Y, Higashibata H, Maseda H, Ikeda H, Omura S, Kobayashi M. Hyper-inducible expression system for Streptomycetes. Proc Natl Acad Sci. 2004;101:14031–14035. doi: 10.1073/pnas.0406058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto M, Hashimoto Y, Saitoh Y, Kumano T, Kobayashi M. Development of nitrilase promoter-derived inducible vectors for Streptomyces. Biosci Biotechnol Biochem. 2016;80:1230–1237. doi: 10.1080/09168451.2016.1148577. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi A, Jamet E, Commissaire J, Renault P, Langella P, Azevedo V. A xylose-inducible expression system for Lactococcus lactis. FEMS Microbiol Lett. 2004;239:205–212. doi: 10.1016/j.femsle.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Bhavsar AP, Zhao X, Brown ED. Development and Characterization of a Xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nariya H, Miyata S, Kuwahara T, Okabe A. Development and characterization of a xylose-inducible gene expression system for Clostridium perfringens. Appl Environ Microbiol. 2011;77:8439–8441. doi: 10.1128/AEM.05668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Urzo N, Martinelli M, Nenci C, Brettoni C, Telfold JL, Maione D. High-level intracellular expression of heterologous proteins in Brevibacillus choshinensis SP3 under the control of a xylose inducible promoter. Microb Cell Fact. 2013;12:12. doi: 10.1186/1475-2859-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wegerer A, Sun T, Altenbuchner J. Optimization of an E. colil-rhamnose-inducible expression vector: test of various genetic module combinations. BMC Biotechnol. 2008;8:2. doi: 10.1186/1472-6750-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertram R, Schlicht M, Mahr K, Nothaft H, Saier MH, Jr, Titgemeyer F. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2) J Bacteriol. 2004;186:1362–1373. doi: 10.1128/JB.186.5.1362-1373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhosale SH, Rao MB, Deshpande VV. Molecular and industrial aspects of glucose isomerase. Microbiol Rev. 1996;60:280–300. doi: 10.1128/mr.60.2.280-300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li JX, Zhao LM, Wu RJ, Zheng ZJ, Zhang RJ. High-level overproduction of Thermobifida enzyme in Streptomyces lividans using a novel expression vector. Int J Mol Sci. 2013;14:18629–18639. doi: 10.3390/ijms140918629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sasaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 23.Young HG, Kim WC, Joo GJ, Kwak YY, Shin JH, Roh DH, Park HD, Rhee IK. Deletion of xylR gene enhances expression of Xylose Isomerase in Streptomyces lividans TK24. J Microbiol Biotechnol. 2008;18:837–844. [PubMed] [Google Scholar]
- 24.Wong HC, Ting Y, Lin HC, Reichert F, Mymbo K, Watt KW, Toy PL, Drummond RJ. Genetic organization and regulation of the xylose degradation genes in Streptomyces rubiginosus. J Bateriol. 1991;173:6849–6858. doi: 10.1128/jb.173.21.6849-6858.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kendall KJ, Cohen SN. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988;170:4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino C, Negi Y, Matsumiya T, Nakaoka K, Kondo A, Kuroda S, Tokuyama S, Kikkawa U, Yamane T, Fukuda H. Purification, characterization, and sequence determination of phospholipase D secreted by Streptoverticillium cinnamoneum. J Biochem. 1999;125:263–269. doi: 10.1093/oxfordjournals.jbchem.a022282. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda H, Kazuo SY, Omura S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol. 2014;41:233–250. doi: 10.1007/s10295-013-1327-x. [DOI] [PubMed] [Google Scholar]
- 28.Bentley SD, Chater KF, Cerdeno-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Globe A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murohy L, Oliver K, O’Neil S, Rabbinovitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 29.Cruz-Morales P, Vijgenboom E, Iruegas-Bocardo F, Girard G, Yáñez-Guerra LA, Ramos-Aboites HE, Pernodet JL, Anné J, van Wezel GP, Barona-Gómez F. The genome sequence of Streptomyces lividans 66 reveals a novel tRNA-dependent peptide biosynthetic system within a metal-related genomic island. Genome Biol Evol. 2013;5:1165–1175. doi: 10.1093/gbe/evt082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rygus T, Scheler A, Allmansberger R, Hillen W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus megaterium encoded regulon for xylose utilization. Arch Microbiol. 1991;155:535–542. doi: 10.1007/BF00245346. [DOI] [PubMed] [Google Scholar]
- 32.Scheler A, Rygus T, Allmansberger R, Hillen W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus licheniformis encoded regulon for xylose utilization. Arch Microbial. 1991;155:526–534. doi: 10.1007/BF00245345. [DOI] [PubMed] [Google Scholar]
- 33.Song S, Park C. Organization and regulation of the d-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J Bacteriol. 1997;179:7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noda S, Miyazaki T, Miyoshi T, Miyake M, Okai N, Tanaka T, Ogino C, Kondo A. Cinnamic acid production using Streptomyces lividans expressing phenylalanine ammonia lyase. J Ind Microbiol Biotechnol. 2011;38:643–648. doi: 10.1007/s10295-011-0955-2. [DOI] [PubMed] [Google Scholar]
- 35.Kieser T, Hopwood DA, Wright HM, Thompson CJ. pIJ101, a multicopy broad host range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185:223–238. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- 36.Muth G, Nussbaumer B, Wohlleben W, Pühler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in streptomycetes. Mol Gen Genet. 1989;219:341–348. doi: 10.1007/BF00259605. [DOI] [Google Scholar]
- 37.Schӓfer A, Tauch A, Jӓger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 38.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical streptomyces genetics. Norwich: John Innes Centre; 2000. [Google Scholar]
- 39.Kim K, Ji HS. Effect of chitin sources on production of chitinase and chitosanase by Streptomyces griseus HUT 6037. Biotechnol Bioprocess Eng. 2001;6:18–24. doi: 10.1007/BF02942245. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. SDS-PAGE gels of S. avermitilis protein expression profiles for (A) cell extracts and (B) culture supernatants: 2.0% 1. Glycerol, 2. D-glucose, 3. D-xylose, 4. D-galactose, 5. D-fructose, and 6. no sugar. M: molecular weight marker. Arrows indicate Xylose isomerase (XylA) protein.
Additional file 2: Figure S2. SDS-PAGE gels of S. avermitilis protein expression profiles for (A) cell extracts and (B) culture supernatants: 1.0% 1. Glycerol, 2. D-glucose, 3. D-xylose, 4. D-galactose, 5. D-fructose, and 6. no sugar. M: molecular weight marker. Arrows indicate Xylose isomerase (XylA) protein.
Additional file 3: Figure S3. (A) Chitobiase activity and (B) optical density (600 nm) of S. lividans XCPR after 48, 72, and 96 h cultivation. After 48 h, D-xylose was added to a final concentration of 0.0, 0.5, 1.0, and 2.0%. Cultivation was continued for 24 and 48 h (72 and 96 h total).
Additional file 4: Figure S4. Gene organization in the xyl operon of S. avermitilis MA-4680, S. coelicolor A3 (2), S. lividans 1326, and S. griseus (NBRC 13350) [22, 28, 29, 30].
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its additional files].