ABSTRACT
BBX proteins are a family of zinc finger transcription factors that are versatile regulators of plant development. The 32 BBX proteins in Arabidopsis are subdivided into five structural groups based on their domain structure. Members of group IV play important and diverse roles in light-regulated development. The N-terminal B-box domains mediate DNA binding and transcriptional regulation. The C-terminal region determines the functional diversity of the structurally similar group IV members as reported in our recent study investigating the basis of functional diversification between BBX21 and BBX24. We also found that multi-layered regulation of HY5 by the BBX proteins leads to a diverse repertoire of developmental effects. Here we provide a comprehensive structure-function analysis of the group IV BBX proteins.
KEYWORDS: B-box, BBX21, BBX24, HY5, light signaling, photomorphogenesis, post-transcriptional regulation
BBX proteins constitute a family of Zinc-finger transcription factors that are highly conserved across the plant kingdom. BBX proteins characteristically possess one or two B-box motifs in their N-terminal region, in some cases together with a C-terminal CCT domain. The 32-membered BBX family in Arabidopsis is categorized into five different structural groups based on the number and types of motifs present.1 They are largely known for their regulatory roles in light-mediated developmental processes such as photomorphogenesis. Interestingly, closely related BBX proteins having comparable structural features have been found to impart opposite effects on development. A prominent example is the members of structural group IV. ELONGATED HYPOCOTYL 5 (HY5) is a central downstream regulator of light-mediated developmental processes and acts as one of the most important factors that promote photomorphogenesis.2 Six among the eight members of group IV (BBX20-25) regulate photomorphogenesis in a HY5-dependent manner (Fig. 1). Intriguingly, BBX20, BBX21, BBX22 and BBX23 are positive regulators of photomorphogenesis whereas BBX24 and BBX25 are negative regulators.3,4,5,6,7,8 In our recent study, by taking the examples of BBX21 and BBX24, we have shown that the opposite functions of these proteins are determined by their diverse C-terminal regions.9 Our findings also indicated that multi-layered regulation of HY5 by closely related BBX proteins is the key in mediating their antagonistic actions. This work is aimed at analysing the mechanistic basis of the opposite roles played by BBX21 and BBX24 and discussing some insights that can potentially propel questions for the future.
Figure 1.
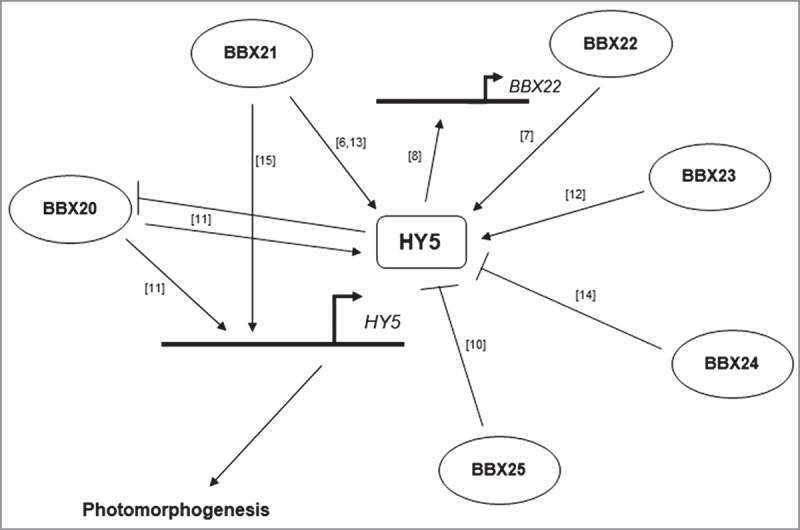
A model showing the HY5-dependency of group IV BBX proteins in the regulation of photomorphogenesis. Arrowed lines and flat-headed lines respectively indicate positive and negative regulation.
Essential roles for B-boxes in BBX21 and BBX24
Intact B-boxes are crucial for the functioning of BBX proteins and their domain topology is highly conserved in plants.1 Mutations in the B-box motifs of BBX21, BBX22, BBX24 and BBX25 resulted in disruption of their ability for protein-protein interactions with HY5.3,4,6,10 BBX21 has also been shown to bind the HY5 promoter and positively regulate the transcription of HY5.11 In a recent study, it was shown that disruption of the second B-box, but not the first one, completely abolishes the function of BBX21.12 The zinc-ligating Asp-75 in the second B-box of BBX21 was shown to be crucial for its function. A mutation in this residue (D75A) abrogates its ability to bind on the T/G box of HY5 promoter, probably by disrupting the structure of the whole B-box. This abolishes the subsequent transcriptional activation of HY5.3,12 The zinc-ligating residues in B-box2 are largely conserved between BBX21 and BBX24. However, BBX21, but not BBX24, transcriptionally regulates HY5.9,11,12
We have used chimeric domain swap lines to delineate the basis of this functional difference between BBX21 and BBX24.9 The residues up to the 98th and 101st amino acid respectively in BBX24 and BBX21 (which include their B-boxes) constitute the N-terminal region while the C-terminal region represents the residues after these (Fig. 2a). Interestingly, overexpressing the chimeric protein with the N-terminal regions of BBX24 and the C-terminal regions of BBX21 (BB24C21) could transcriptionally upregulate HY5 similar to BBX21 overexpressors whereas overexpression of the chimera BB21C24 did not have any effect on the mRNA levels of HY5.9 Assuming that BB24C21 chimera can bind to the HY5 promoter in a similar way to BBX21 and considering the essentiality of B-box2 in its DNA binding ability, it is possible that the C-terminal region of BBX21 has an important regulatory effect on the functioning of B-box2. Comparing the abilities of the chimeric proteins BB24C21 and BB21C24 to bind the HY5 promoter might help to resolve this question.
Figure 2.
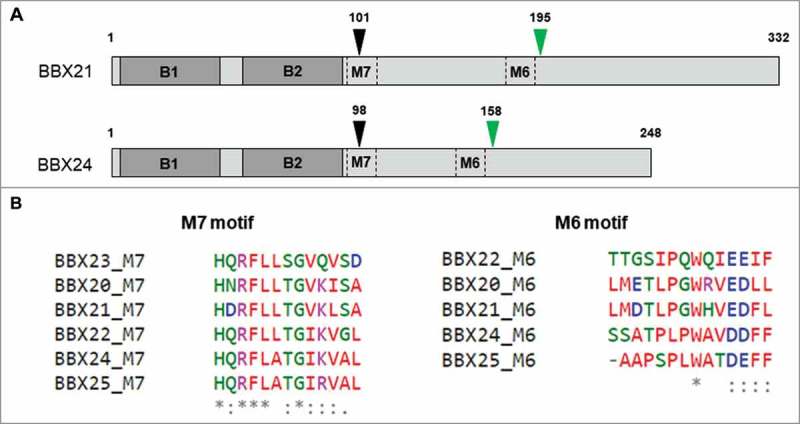
(A) Schematic representation of full length BBX21 and BBX24 proteins. B1 and B2 represent the first and second B-box. Motif 7 and Motif 6 are denoted by M6 and M7. The black arrowhead indicates the position from where the C-terminal regions were swapped to make BB21C24 and BB24C21. The green arrowhead indicates the position from where the regions containing M6 motif were swapped. (B) Multiple sequence alignment of M7 and M6 motifs in the C-terminal regions of BBX20-BBX25 using Clustal Omega.
Importance of C-terminal regions in functional diversity of group IV BBX proteins
Evolutionary diversification of C-terminal regions seems largely responsible for the functional diversity of BBX proteins within the same structural groups. We have shown that the C-terminal regions dictate the opposite roles of BBX21 and BBX24. As mentioned above, when the C-terminal regions were interchanged, BBX21 functionally behaved like BBX24 and vice versa.9 Similarly, it was shown in a previous study that sequences C-terminal to the B-boxes of CO/BBX1 were required for flowering time regulation in COL1/BBX2.13 The C-terminal regions of group IV BBX proteins possess two conserved motifs of unknown function, namely Motif 6 (M6) and Motif 7 (M7). Sequence analysis with 214 full length BBX proteins across the plant kingdom suggested that M7 motif is highly conserved (98%) whereas M6 motif is relatively less conserved (64%) among the group IV members.1 Our experiments with chimeric lines suggested that M6 motif might play role in the functional determination as the C-terminal region of BBX21 lost its ability to promote photomorphogenesis when its M6 motif was swapped with that of BBX24 (Fig. 2a). It also indicated that the highly conserved M7 motif is less important in functional determination (Fig. 2b).9 The M6 motifs of BBX20 and BBX21 are similar to the conserved M6 sequence whereas the M6 motifs of BBX22, BBX24 and BBX25 are only partially conserved (Fig. 2b).1 BBX23 does not possess an M6 motif. Since only BBX20 and BBX21, not the other members of the group, can regulate HY5 at transcriptional levels, it would be interesting to examine if the extent of sequence conservation in the M6 motif has any relation with the ability of the protein to transcriptionally regulate HY5.
Multi-layered regulation of HY5 by BBX21
Our data suggest that in addition to the transcriptional regulation, BBX21 regulates HY5 also at post-transcriptional level.9 The physical interaction between BBX21 and HY5 might be important for this post-transcriptional regulation.3 The significance of this interaction in the modulation of photomorphogenesis has not been adequately addressed so far. BBX21 overexpressing lines which exhibit very strong phenotypes showed only two-fold upregulation in HY5 expression. It seems quite unlikely that the intense effect of BBX21 overexpression on levels of expression of HY5 target genes, anthocyanin accumulation levels etc. is merely a result of a two-fold increase in HY5 expression.9,11 The fact that 35S:HY5 lines do not phenocopy 35S:BBX21 lines suggests that mere overexpression of HY5 at transcript levels might not be sufficient for significant regulatory impact on its downstream targets.14 Additional post-transcriptional regulation might contribute, perhaps in a more prominent manner, to regulate HY5 activity on its downstream targets. Concordantly, in addition to the disruption of its ability to bind on HY5 promoter, the D75A mutation in the second B-box of BBX21 also results in a complete loss of HY5 interaction, suggesting that this mutation attenuates both the transcriptional and post-transcriptional regulation of HY5 by BBX21. In contrast, the D84A mutation in the second B-box, disrupts the physical interaction with HY5, but still retains partial ability to promote photomorphogenesis.3,12 This mutation might therefore be informative in separating the transcriptional and post-transcriptional dependency by HY5 on BBX21. While most of these observations are pointing towards the possible promotion of the action of HY5 by BBX21 at protein level, in contrast, BBX21 regulates ABA signalling by forming non-DNA binding heterodimers with HY5 and hence inhibiting its binding on ABI5 promoter.15 Together, more studies are needed to elucidate the several mechanisms by which BBX21 post-transcriptionally regulates HY5 to control various developmental processes like photomorphogenesis and ABA signalling in diverse ways.
Conclusion
In summary, the diverse C-terminal regions of closely related BBX proteins are crucial for their functional divergence. The M6 motif in the C-terminal region might play influential role in the functional determination of group IV BBX proteins. The loosely conserved nature of this motif within the group might introduce functional differences among these proteins. BBX21 and BBX24 oppositely modulate photomorphogenesis through their antagonistic regulation of HY5 at different levels. BBX24 regulates HY5 at protein level whereas BBX21 regulates HY5 both at transcriptional and post-transcriptional levels. We propose that the post-transcriptional regulation of HY5 by BBX21 play a major role in the modulation of photomorphogenesis, potentially being more important than its well-characterized transcriptional regulation of HY5.
Funding Statement
Science and Engineering Research Board, Department of Science and Technology, India. Department of Biotechnology, India. Deutsche Forschungsgemeinschaft, Germany.
Disclosure statement
Authors disclose no potential conflicts of interest.
Acknowledgments
SD would like to thank Department of Biotechnology (Ramalingaswami Fellowship, IYBA, BT/PR19193/BPA/118/195/2016) and SERB (EMR/2016/000181), Government of India for funding. H.J acknowledges the funding by DFG (JO 1409/1-1). PY and NJ respectively acknowledge DBT and DST-INSPIRE, Govt. of India for their PhD fellowships.
References
- 1.Crocco CD, Botto JF. BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene. 2013;531:44–52. doi: 10.1016/j.gene.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 2.Gangappa SN, Botto JF. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol Plant. 2016;9:1353–65. doi: 10.1016/j.molp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007;19:3242–55. doi: 10.1105/tpc.107.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008;20:2324–38. doi: 10.1105/tpc.108.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CS, Li YH, Chen LT, Chen WC, Hsieh WP, Shin J, Jane WN, Chou SJ, Choi G, Hu JM, et al.. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008;54:205–19. doi: 10.1111/j.1365-313X.2008.03401.x. [DOI] [PubMed] [Google Scholar]
- 6.Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell. 2013;25:1243–57. doi: 10.1105/tpc.113.109751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei C-Q, Chien C-W, Ai L-F, Zhao J, Zhang Z, Li KH, Burlingame AL, Sun Y, Wang Z-Y. The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J Genet Genomics. 2016;43:555–63. doi: 10.1016/j.jgg.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XY, Huai JL, Shang FF, Xu G, Tang WJ, Jing YJ, Lin R. A PIF1/PIF3-HY5-BBX23 Transcription Factor Cascade Affects Photomorphogenesis. Plant Physiol. 2017;174:2487–500. doi: 10.1104/pp.17.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Job N, Yadukrishnan P, Bursch K, Datta S, Johansson H. Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 2018;176:2963–76. doi: 10.1104/pp.17.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangappa SN, Holm M, Botto JF. Molecular interactions of BBX24 and BBX25 with HYH, HY5 HOMOLOG, to modulate Arabidopsis seedling development. Plant Signal Behav. 2013;8:37–41. doi: 10.4161/psb.25208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci U S A. 2016;113:7655–60. doi: 10.1073/pnas.1607687113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu D, Jiang Y, Li J, Holm M, Deng XW. The B-box domain protein BBX21 promotes photomorphogenesis. Plant Physiol. 2018;176:2365–75. doi: 10.1104/pp.17.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S-K, Park H-Y, Jang YH, Lee JH, Kim J-K. The sequence variation responsible for the functional difference between the CONSTANS protein, and the CONSTANS-like (COL) 1 and COL2 proteins, resides mostly in the region encoded by their first exons. Plant Sci. 2013;199–200:71–8. doi: 10.1016/j.plantsci.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 14.Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–22. doi: 10.1016/S1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 15.Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M. Convergence of Light and ABA signaling on the ABI5 promoter. PLoS Genet. 2014;10:e1004197. doi: 10.1371/journal.pgen.1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]


