Abstract
Left ventricular noncompaction (LVNC) is a distinct cardiomyopathy that is morphologically characterized by a two-layered myocardium, numerous prominent trabeculations, and deep intertrabecular recesses communicating with the left ventricular cavity. We present a case report regarding the identification of a new mutation in TNNI3 in a patient with LVNC using next-generation sequencing. A 13-year-old girl who had no family history of cardiac disease was hospitalized with dyspnea after exercise and electrocardiographic abnormalities during a school screening. Based on her clinical features, she was diagnosed with LVNC. Via genetic analysis, a TNNI3 heterozygous missense variant was identified in the proband. Although mutations in TNNI3 have been reported in patients with hypertrophic cardiomyopathy and restrictive cardiomyopathy, this is the first report of a mutation in this gene in a patient with LVNC.
<Learning objective: We identified a variant in TNNI3 in a patient with isolated left ventricular noncompaction using next-generation sequencing (NGS). Mutations in TNNI3 have been reported in patients with hypertrophic cardiomyopathy and restrictive cardiomyopathy. The use of NGS also results in the identification of multiple genetic variants of unknown significance to the investigated disease.>
Keywords: TNNI3, Arg192 His, Left ventricular noncompaction, Next-generation sequencing
Introduction
Left ventricular noncompaction (LVNC) is a recently classified form of inherited cardiomyopathy that is morphologically characterized by a two-layered myocardium, numerous prominent trabeculations, and deep intertrabecular recesses communicating with the left ventricular cavity. The clinical manifestation of LVNC is highly variable, ranging from no symptoms to a progressive deterioration that results in congestive heart failure, arrhythmia, thrombosis, and sudden cardiac death (SCD). To date, mutations in several genes have been reported in patients with LVNC, including the genes encoding tafazzin, LIM domain binding protein 3, α-dystrobrevin, lamin A/C, β-myosin heavy chain, α-cardiac actin, cardiac troponin T, PR domain containing 16, mindbomb E3 ubiquitin protein ligase 1, and sodium channel, voltage gated, type V alpha subunit [1], [2], [3]. However, the relatively small contribution of known mutations to the disease compared to the high proportion of familial cases suggests that other genes remain to be identified.
Next-generation sequencing (NGS) has been widely used recently because of its ability to investigate multiple genes at a reasonable cost. Meanwhile, mutations in TNNI3 have been reported in patients with hypertrophic cardiomyopathy (HCM) and restrictive cardiomyopathy (RCM), but there is no report of mutations in the gene in patients with LVNC. In this report, we identified a new mutation in TNNI3 in a patient with LVNC using NGS.
Case report
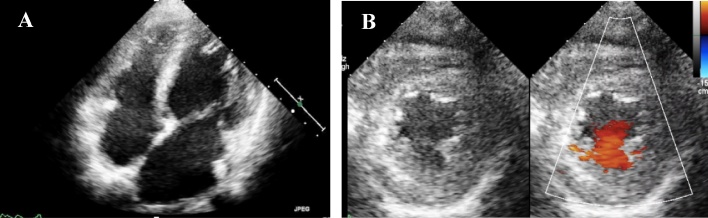
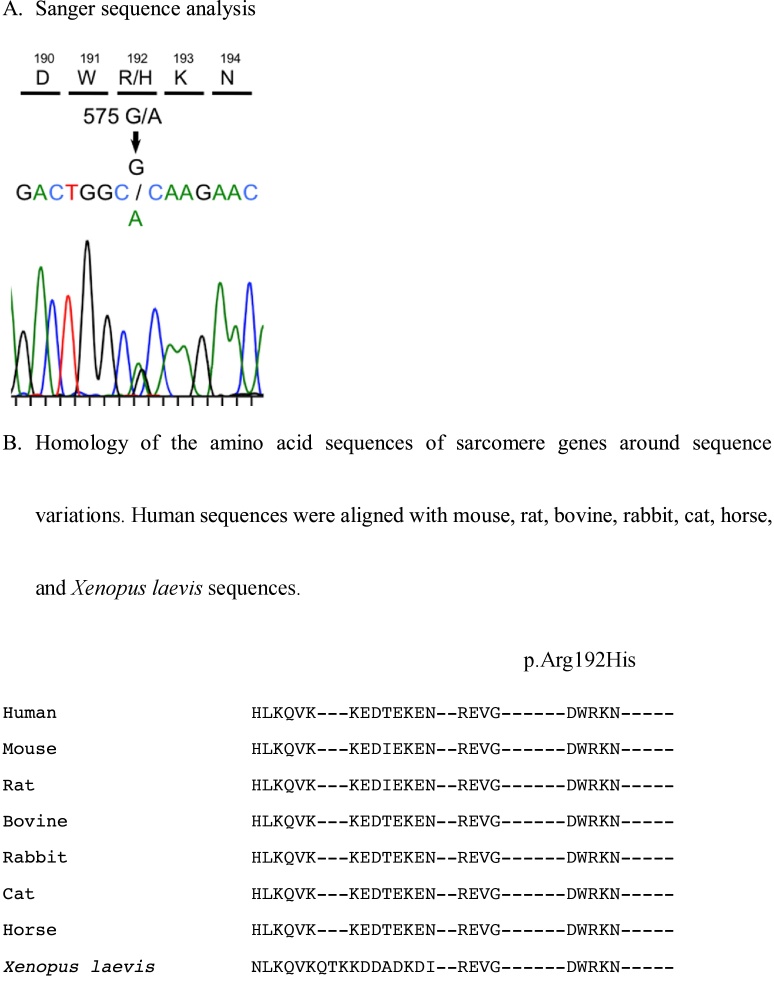
A 13-year-old-girl who had no history of cardiac disease or SCD previously visited a hospital because of abnormalities in electrocardiography performed in a school screening. She had experienced dyspnea after exercise for a few years. Further, she had syncope during walking at 11 and 13 years of age. Although electroencephalography and magnetic resonance imaging of the skull were performed, abnormalities were not detected. The 12-lead electrocardiogram uncovered ST depression in leads II, III, and aVF and abnormalities of ST segments and T waves in the precordial lead. The two-dimensional echocardiogram revealed reduced systolic function, left atrium and ventricle enlargement, and impaired ventricular relaxation. The patient’s left ventricular ejection fraction, left ventricular end-diastolic dimension, and E/A and E/e′ ratios were 45%, 51 mm (z-score 2.58), 0.76, and 14.3, respectively. The ventricular end-diastolic posterior wall thickness was 7.6 mm (z-score 0.54). In addition, there were prominent trabeculations in the left ventricle, predominantly in the apex (Fig. 1A) and blood signals within the intertrabecular region (Fig. 1B). Parasternal short-axis echocardiogram at end-systole demonstrated the two-layered structure of thick noncompacted (NC) and thin compacted (C) layers; N/C ratio = 2.8 (Fig. 1B). Her B-type natriuretic peptide level was 291.7 pg/ml. Arrhythmia was not recorded on 24-h Holter monitoring. These findings were consistent with LVNC. Treatment with diuretics, β-blockers, angiotensin-converting enzyme inhibitors, and phosphodiesterase 3 inhibitors was started, and she was referred to our institution. The cardiac catheterization study showed that the mean pressures of pulmonary artery and the left pulmonary capillary wedge position were 21 and 13 mmHg, respectively. The left ventricular pressure was 88/20 mmHg. Endomyocardial biopsy uncovered hypertrophy with bizarre nuclei in myocytes, disarrangement of the cardiac muscle, and interstitial and perivascular fibrosis of the right ventricle. These findings were compatible with HCM. In the genetic analysis conducted using NGS (Table 1A), a TNNI3 heterozygous missense variant (NM_000363.4; c.575G>A, p.Arg192His) was identified in the proband (Fig. 2A, B). The patient’s cardiac function did not improve enough despite treatment of heart failure for 2 years in our institution. She had sudden cardiac death caused by ventricular fibrillation at the age of 15 years.
Fig. 1.
A. Apical four chamber echocardiogram of a patient with LVNC showing abnormal, highly trabeculated left ventricular myocardium with intratrabecular recesses. B. Parasternal short axis echocardiogram demonstrating the two-layered structure of noncompacted (NC) and compacted (C) layers. Color doppler echocardiogram demonstrating flow within the deep intertrabecular recesses in continuity with the left ventricular cavity.
LVNC, left ventricular noncompaction.
Table 1A.
List of the analyzed genes associated with inherited cardiac disease.
| ABCC9, ACTC1, ACTN2 AKAP9, ANK2, BAG3, BMPR1A, CACNA1C, CACNB2, CALR3, CAPN3, CAV3, DES, DMD, DSC2, DSG2, DSP, ELN, EMD, GAA, GATA4, GLA, GPD1L, HCN4, JUP, KCNE1, KCNE2, KCNE3, KCNH2, KCNJ2, KCNQ1, KRAS, LAMP2, LDB3, LMNA, MIB1, MYBPC3, MYH11, MYH6, MYH7, MYL2, MYL3, MYLK, MYOZ2, NKX2-5, NRAS, PKP2, PLN, PRDM16, PRKAG2, PTPN11, RAF1, RPS7, RYR2, SCN1B, SCN3B, SCN4B, SCN5A, SGCD, SMAD3, SNTA1, SOS1, STARD3, TAZ, TBX5, TGFBR1, TGFBR2, TMEM43, TNNC1, TNNI3, TNNT2, TPM1, VCL |
Fig. 2.
A. Sanger sequence analysis. B. Homology of the amino acid sequences of sarcomere genes around sequence variations. Human sequences were aligned with mouse, rat, bovine, rabbit, cat, horse, and Xenopus laevis sequences.
Discussion
In this case, it was difficult to distinguish LVNC from HCM or RCM. The echocardiographic findings, prominent trabeculations and blood signals within the intertrabecular region in the left ventricle, were compatible with LVNC. On the other hand, the histological finding, bizarre nuclei in myocytes, disarrangement of the cardiac muscle, and interstitial and perivascular fibrosis, were compatible with HCM. However, the echocardiogram revealed left ventricular wall thickness was not hypertrophic. The cardiac catheterization data showed increased left ventricular end-diastolic pressure, which was consistent with restrictive hemodynamics. However, her left ventricular systolic function was depressed. This was not afforded to typical RCM. Based on these findings, we diagnosed this case as LVNC.
TNNI3, localized on chromosome 19q13.4, is composed of eight exons, and it encodes cardiac troponin I (cTnI), which is expressed in cardiac muscle. It is well known that the major sensor of intracellular Ca2+ levels is the troponin complex, which consists of the subunits cTnI, troponin C, and troponin T. Their primary function is to control the interaction between thick and thin filaments during muscle contraction and relaxation. cTnI has an inhibitory effect, which is reversed by troponin C after the binding of Ca2+, which subsequently introduces conformational changes in the entire troponin complex, leading to muscle contraction. The mechanisms by which cTnI mutations affect the contractility of the sarcomere were recently reviewed in depth by Tardiff [4]. The TNNI3 Arg192His mutation in the troponin complex may introduce alterations in Ca2+ affinity and protein–protein interactions, which may ultimately lead to the development of cardiomyopathy.
cTnI was first identified as a disease-causing gene in patients with HCM by Kimura et al. in 1997 [5]. Mutations in TNNI3 have been reported in patients with HCM and RCM. Interestingly, the identification of cTnI as the disease-causing gene in a large family with affected relatives presenting with either RCM or HCM, made it apparent that transitional forms of the conditions appear even among individuals carrying the same variant. The Arg192His variant in TNNI3, which was identified in patients with HCM and RCM, has been reported by Rai et al. and Yang et al. (Table 1B) [6], [7]. They also reported that HCM and RCM with restrictive physiology, both featuring the clinical expression of mutant TNNI3, are associated with worse clinical onset and disease. Our patient carrying the same mutation, who exhibited diastolic dysfunction and whose endomyocardial biopsy revealed HCM-like features, similarly struggled to recover from cardiac dysfunction despite full treatment for heart failure. The clinical presentation of LVNC is highly variable, and Towbin proposed at least eight different phenotypes of the disease [8]. The restrictive LVNC phenotype was defined by enlargement of one or both atria relative to ventricles of normal or small size with evidence of impaired diastolic filling in the absence of significant valvular heart disease. This definition is compatible with our case, and her Arg192His mutation may cause the diversity of cardiomyopathies such as HCM, RCM, and LVNC.
Table 1B.
Known variant identified as pathogenic in this study.
| Variant | dbSNP | Minor allele frequency |
Disease databases |
||
|---|---|---|---|---|---|
| ExAC | HGVD | ClinVar | HGMD | ||
| TNNI3_c.575G>A (p.Arg192His) | rs104894729 | – | – | Pathogenic/Likely pathogenic | CM030288 (Disease-causing mutation) |
| Familial restrictive cardiomyopathy 1 | Cardiomyopathy, restrictive | ||||
| Primary familial hypertrophic cardiomyopathy | |||||
| Cardiomyopathy | |||||
HGVD, Human Genetic Variation Database.
The allelic frequency of all detected variants was determined using the Exome Aggregation Consortium (ExAC) database and the Human Genetic Variation Database (HGVD), which contain data for 1208 Japanese individuals. All variants with a minor allele frequency (MAF) of ≥0.01% among the ExAC and HGVD population were filtered out. As the prevalence of LVNC is reported to be 0.014% [9], [10], we chose an initial MAF filter of 0.01%. We obtained functional or/and segregation analysis data on previously reported variants from both the human genome mutation database (HGMD) and the ClinVar disease mutation database. To evaluate the pathogenicity of the remaining variants, we utilized seven different silico in silico predictive algorithms: FATHMM, SIFT, PROVEAN, Align GVGD, MutationTaster2, PolyPhen2, and CADD. The all detected mutations and CADD phred score of the detected mutation after filtering are shown in the Supplemental file. Variants predicted deleterious/pathogenic by at least 5 of the 7 in silico algorithms were considered likely pathogenic. As a result, we found only one variant.
To further explore the potential relationship between genotype and phenotype, long-term follow-up studies are needed. It is essential to investigate the natural history of the condition among affected individuals carrying identical mutations and compare these findings with the disease expression associated with different mutations and disease genes.
Conclusion
We identified a variant in TNNI3 in a patient with isolated LVNC using NGS. Applying NGS technology, we were able to simultaneously analyze 73 cardiomyopathy-related genes, apart from identifying the mutation in TNNI3. Although mutations in TNNI3 have been reported in patients with HCM and RCM, this is the first report of a mutation in this gene in a patient with LVNC.
Our results reflect a substantial increase in the yield obtained using modern NGS techniques. Nevertheless, the use of NGS also results in the identification of multiple genetic variants of unknown significance to the investigated disease, and further investigations are required to confirm their significance.
Conflict of interest
The authors have no conflict of interest concerning this article.
Acknowledgments
None.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jccase.2018.04.001.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Ichida F., Tsubata S., Bowles K.R., Haneda N., Uese K., Miyawaki T. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation. 2001;103:1256–1263. doi: 10.1161/01.cir.103.9.1256. [DOI] [PubMed] [Google Scholar]
- 2.Vatta M., Mohapatra B., Jimenez S., Sanchez X., Faulkner G., Perles Z. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol. 2003;42:2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 3.Kenton A.B., Sanchez X., Coveler K.J., Makar K.A., Jimenez S., Ichida F. Isolated left ventricular noncompaction is rarely caused by mutations in G4.5, alpha-dystrobrevin and FK binding protein-12. Mol Genet Metab. 2004;82:162–166. doi: 10.1016/j.ymgme.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Tardiff J.C. Thin filament mutations: developing an integrative approach to a complex disorder. Circ Res. 2011;108:765–782. doi: 10.1161/CIRCRESAHA.110.224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura A., Harada H., Park J.E., Nishi H., Satoh M., Takahashi M. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–382. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 6.Rai T.S., Ahmad S., Ahluwalia T.S., Ahuja M., Bahl A., Saikia U.N. Genetic and clinical profile of Indian patients of idiopathic restrictive cardiomyopathy with and without hypertrophy. Mol Cell Biochem. 2009;331:187–192. doi: 10.1007/s11010-009-0157-7. [DOI] [PubMed] [Google Scholar]
- 7.Yang S.W., Chen Y., Li J., Yin J., Qin Y.M., Andelfinger G. Clinical characteristics and genetic analysis of three pediatric patients with idiopathic restrictive cardiomyopathy. Zhonghua Xin Xue Guan Bing Za Zhi. 2013;41:304–309. [PubMed] [Google Scholar]
- 8.Towbin J.A., Lorts A., Jefferies J.L. Left ventricular non-compaction cardiomyopthy. Lancet. 2015;386:813–825. doi: 10.1016/S0140-6736(14)61282-4. [DOI] [PubMed] [Google Scholar]
- 9.Jenni R., Oechslin E.N., van der Loo B. Isolated ventricular non-compaction of the myocardium in adults. Heart. 2007;93:11–15. doi: 10.1136/hrt.2005.082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh R., Thomson K.L., Ware J.S., Funke B.H., Woodley J., McGuire K.J. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference sanples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.