ABSTRACT
Epigenetic regulator plays a pivotal role in breast cancer progression. We recently demonstrated that the epigenetic reader zinc finger MYND-type containing 8 (ZMYND8) mediates breast cancer progression and metastasis by activating hypoxia-inducible factor (HIF). This discovery provides new insights into the molecular mechanism underlying HIF activation and breast cancer progression.
Keywords: Hypoxia-inducible factor, epigenetic reader, gene regulation, breast cancer
Hypoxia is a hallmark of tumor microenvironment and drives breast tumor malignancy.1 Hypoxia-inducible factor (HIF), consisting of a α subunit (HIF-1α, HIF-2α, and HIF-3α) and a β subunit [HIF-1β and aryl hydrocarbon receptor nuclear translocator 2 (ARNT2)], is a master regulator of adaptive responses to hypoxia by inducing hundreds of downstream target genes.2 Our previous studies and others have shown that epigenetic regulators, including writers and erasers, are required for HIF-mediated transactivation.3 However, how the epigenetic reader modulates HIF transcriptional activity to promote breast tumor progression and metastasis is poorly understood.
We recently showed that the epigenetic reader zinc finger MYND-type containing 8 (ZMYND8) was upregulated by HIF-1 and HIF-2 in breast cancer cells.4 ZMYND8 was also highly expressed in human breast tumors and was correlated with poor clinical outcomes in patients with breast cancer, suggesting that ZMYND8 may have an oncogenic role in breast tumors. In vitro clonogenic and Boyden chamber cell migration and invasion assays revealed that ZMYND8 knockout (KO) significantly suppressed breast cancer cell colony formation, migration and invasion. Our in vivo orthotopic xenograft assay further showed that ZMYND8 KO significantly reduced breast tumor growth and metastasis to the lungs in severe combined immunodeficiency mice by increasing breast cancer cell death and decreasing tumor angiogenesis. HIF-1 and HIF-2 were required for ZMYND8-mediated breast tumor progression and metastasis in mice.
We elucidated the molecular mechanism by which ZMYND8 promotes breast cancer progression and metastasis.4 ZMYND8 physically interacted with HIF-1α and HIF-2α in breast cancer cells under hypoxia, and significantly increased HIF transcriptional activity. RNA sequencing analysis showed that about 62.7% of HIF target genes were enhanced by ZMYND8 in breast cancer cells under hypoxia. Chromatin immunoprecipitation with high-throughput DNA sequencing revealed that about 85.4% of HIF-1α peaks colocalized with ZMYND8 binding sites and about 92% of HIF-1 target genes showed a HIF-1α and ZMYND8 co-bound event at the hypoxia response elements (HREs) in breast cancer cells. These data indicate a global impact of ZMYND8 on HIF target gene expression in breast cancer cells.
ZMYND8 is known to bind acetyl lysine 14 of histone H3 (H3K14ac) and acetyl lysine 16 of histone H4 (H4K16ac).5,6 We found that hypoxia significantly increased occupancy of H3K14ac and H4K16ac at the HREs. Along with increased enrichment of H3K14ac and H4K16ac, ZMYND8 occupancy at the HREs was also elevated by hypoxia in breast cancer cells. Thus, these two histone modifications may recruit ZMYND8 to the HREs of HIF target genes in breast cancer cells.
Previous studies have indicated that RNA polymerase II is preloaded and paused at the promoter of HIF target genes and that release of paused RNA polymerase II is essential for HIF-mediated transactivation.7 We found that ZMYND8 triggered release of paused RNA polymerase II to promote the transcriptional elongation of HIF target genes in breast cancer cells by increasing phosphorylation of RNA polymerase II at serine 2 (S2P) at the promoter of HIF target genes. It was recently reported that ZMYND8 binds to the phosphorylated RNA polymerase II at serine 5 (S5P), but not RNA polymerase II-S2P,8 suggesting that a scaffold protein may bridge ZMYND8 and RNA polymerase II-S2P to induce the HIF-dependent transcriptional program. Bromodomain-containing protein 4 (BRD4) is known to induce RNA polymerase II-S2P.9 We found that BRD4 physically interacted with the MYND domain of ZMYND8 via its bromodomains and knockdown (KD) of BRD4 significantly decreased ZMYND8-mediated HIF target gene expression in breast cancer cells, indicating that ZMYND8 regulate the expression of HIF target genes through BRD4. We further showed that BRD4 was recruited by ZMYND8 to the HREs of HIF target genes in breast cancer cells. Taken together, ZMYND8 recruits BRD4 to the HREs of HIF target genes to induce RNA polymerase II-S2P, thereby promoting the transcriptional elongation of HIF target genes in breast cancer cells.
Lastly, we dissected the regulation of ZMYND8-BRD4 interaction in breast cancer cells.4 We found that ZMYND8 was acetylated at the lysine residues 1007 and 1034 within its MYND domain by the acetyltransferase p300 (EP300, best known as p300) and deacetylated by histone deacetylase (HDAC) as the treatment of a HDAC inhibitor trichostatin A increased ZMYND8 acetylation. Mutation of lysine 1007/1034 abolished ZMYND8 binding to BRD4 and ZMYND8-mediated HIF activation in breast cancer cells. Importantly, wild-type, but not lysine-acetylation mutant, ZMYND8 rescued the colony formation, migration and invasion abilities of ZMYND8 KD breast cancer cells in vitro, and partially restored reduced tumor growth and lung metastasis conferred by ZMYND8 KD in mice. Together, these data indicate that ZMYND8 acetylation is precisely controlled by p300 and HDAC and is required for HIF activation and breast cancer progression and metastasis.
In conclusion, ZMYND8 acetylation mediates breast cancer progression and metastasis by inducing the HIF-dependent transcriptional program (Figure 1). ZMYND8 is highly expressed in human breast tumors and predicts the worse outcome in breast cancer patients. Thus, our findings yield a potential biomarker and therapeutic target for the diagnosis and treatment of breast cancer.
Figure 1.
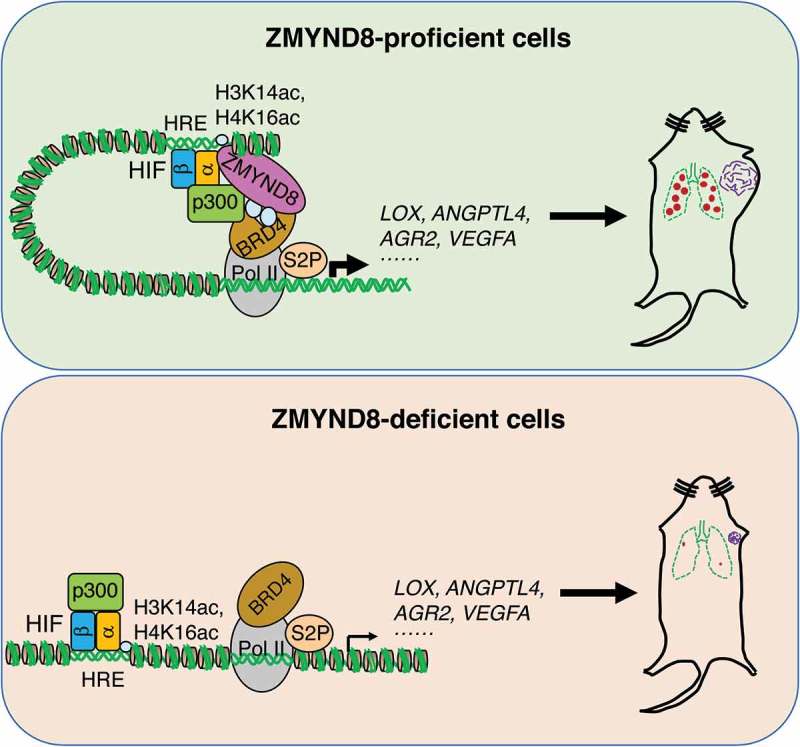
Zinc finger MYND-type containing 8 (ZMYND8) induces hypoxia-inducible factor (HIF)-dependent transcriptional programs that promote breast cancer progression. In ZMYND8-proficient breast cancer cells, ZMYND8 is acetylated by p300 (EP300, best known as p300) and recruits bromodomain containing protein 4 (BRD4) to the hypoxia response elements (HREs) to enhance RNA polymerase II-mediated elongation of HIF target genes, thereby promoting breast tumor growth and metastasis. In ZMYND8-deficient breast cancer cells, the recruitment of BRD4 to the HREs is blocked leading to impaired RNA polymerase II activation and HIF target gene expression, thereby suppressing breast tumor growth and metastasis. H3K14ac, acetyl lysine 14 of histone H3. H4K16ac, acetyl lysine 16 of histone H4. Pol II, RNA polymerase II. S2P, serine 2 phosphorylation.
Funding Statement
This work was supported by grants from Susan G. Komen (CCR16376227), NIH (R00CA168746), the Cancer Prevention & Research Institute of Texas (CPRIT, RR140036), Welch Foundation (I-1903-20160319), and the American Cancer Society/UTSW Simmons Cancer Center (ACS-IRG-02-196) to WL, and grants from NIH (R00NS078049, R35GM124693), CPRIT (RP170671), and Welch Foundation (I-1939-20170325) to YW. W.L. is a CPRIT Scholar in Cancer Research.
Disclosure of potential conflicts of interest
The authors report no conflicts of interest.
References
- 1.Vaupel P, Mayer A, Hockel M.. Tumor hypoxia and malignant progression. Methods Enzymol. 2004;381:.335–354. doi: 10.1016/S0076-6879(04)81023-1. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33(4):207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo W, Wang Y. Epigenetic regulators: multifunctional proteins modulating hypoxia-inducible factor-α protein stability and activity. Cell Mol Life Sci. 2018;75(6):1043–1056. doi: 10.1007/s00018-017-2684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Zhang B, Bao L, Jin L, Yang M, Peng Y, Kumar A, Wang JE, Wang C, Zou X, et al. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J Clin Invest. 2018;128(5):1937–1955. doi: 10.1172/JCI95089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong F, Chiu LY, Cox B, Aymard F, Clouaire T, Leung JW, Cammarata M, Perez M, Agarwal P, Brodbelt JS, et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015;29(2):197–211. doi: 10.1101/gad.252189.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savitsky P, Krojer T, Fujisawa T, Lambert JP, Picaud S, Wang CY, Shanle EK, Krajewski K, Friedrichsen H, Kanapin A, et al. Multivalent histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 2016;17(10):2724–2737. doi: 10.1016/j.celrep.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhry H, Schodel J, Oikonomopoulos S, Camps C, Grampp S, Harris AL, Ratcliffe PJ, Ragoussis J, Mole DR. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep. 2014;15(1):70–76. doi: 10.1002/embr.201337642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adhikary S, Sanyal S, Basu M, Sengupta I, Sen S, Srivastava DK, Roy S, Das C. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J Biol Chem. 2016;291(6):2664–2681. doi: 10.1074/jbc.M115.679985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]


