ABSTRACT
Studies in single cell transcriptomics have significantly expanded our understanding of tumor biology, including recent analyses in head and neck squamous cell carcinoma. Here, we focus on the role of a partial epithelial-to-mesenchymal (EMT) program in these tumors, with discussion of its dynamics, regulation, and implications for diagnostic and therapeutic approaches.
KEYWORDS: head and neck cancer, single cell sequencing, EMT, metastasis
Decades of research has begun uncovering programs that drive tumor growth, invasion, and metastasis, with significant progress based on whole exome and genome sequencing approaches.1 While large-scale datasets across hundreds of patients have proven invaluable, insight from these approaches is limited by use of bulk tissues. As a result, these techniques do not allow gene expression profiles for distinct cell types to be distinguished, leaving open the question of whether an upregulated or downregulated gene is arising from malignant cells versus the surrounding stroma or immune infiltrates. This is particularly problematic in cases where cancer cells express programs also associated with stroma (e.g. mesenchymal genes) or immune infiltrates (e.g. inflammatory signatures). In addition, bulk approaches do not allow the description of distinct malignant subpopulations with diverse gene expression and cellular states, thereby missing intra-tumoral heterogeneity. However, increasing evidence suggests intra-tumoral heterogeneity among malignant cells, and their interactions, are important for diverse aspects of tumor biology and patient survival.
Recent improvements in single cell transcriptomics provide a method for the detailed characterization of gene expression states in individual cells. These approaches can be applied to tissue samples, providing a direct, unbiased analysis of intra-tumoral heterogeneity and offering clarity over long-standing questions in oncology. Our previous single cell work in human glioma has highlighted expression heterogeneity that underlies cellular hierarchies of cancer stem cells and differentiation,2-4 while studies in melanoma have shed light on drug resistance programs and immune responses.5 However, until recently, detailed single cell studies of epithelial tumors and the process of metastasis were lacking.
A persistent challenge in oncology relates to understanding the mechanisms by which epithelial cells may lose their attachments to the underlying extracellular matrix, invade the basement membrane, and ultimately metastasize. Epithelial-to-mesenchymal transition (EMT) – which occurs during normal development and wound healing – may be co-opted in human tumors6 to drive tumor spread. However, the reliance on animal models and individual EMT marker genes has limited the ability to confidently identify and characterize EMT in human tumors, with its significance in metastasis still unproven. Furthermore, many studies have failed to detect EMT, or argue that EMT is not required for metastasis, leading to significant controversy in oncology.
To address these challenges, we recently utilized single cell RNA-seq to characterize head and neck squamous cell carcinoma (HNSCC) – one of the more common epithelial tumors.7 Through unbiased analysis of the malignant cells we identified an EMT-like expression program. This program included several classical EMT markers (such as podoplanin (PDPN), vimentin (VIM), and integrin alpha-5 (ITGA5)), many extracellular matrix genes traditionally associated with EMT (matrix metalloproteinases, integrins and laminins), and Transforming Growth Factor Beta (TGFβ) Induced (TGFBI), consistent with the role of TGFβ signaling in EMT (Fig. 1). Apart from direct proof for the presence of an EMT-like process in human malignant cells, this work highlighted four important points that may clarify the role of EMT in cancer.
Figure 1.
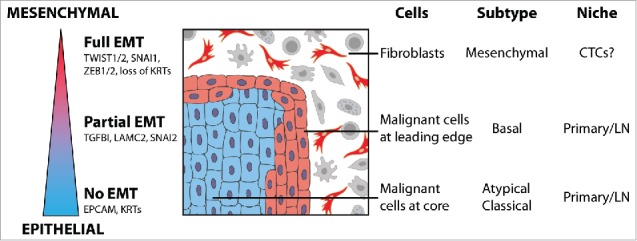
Partial epithelial-to-mesenchymal transition plays a role in head and neck cancer. Epithelial tumors, including head and neck squamous cell carcinoma, may exist along a spectrum of epithelial-to-mesenchymal transition (EMT), including a partial EMT (p-EMT). Individual cells in tumors, the cancer genome atlas (TCGA) subtypes, and tumor niches demonstrate distinct states along this spectrum.
First, EMT-like cells maintained expression of epithelial markers (e.g. multiple cyto-keratins) and did not express most of the transcription factors considered to be EMT master regulators, with the exception of SNAI2 (Snail2, also known as Slug). Interestingly, recent studies suggest that SNAI2 may be the first transcription factor upregulated in EMT,8 raising the possibility that this program is a partial EMT (p-EMT), rather than full EMT. In addition, in vitro analyses suggested that cells dynamically transition between p-EMT and non-p-EMT states, with TGFβ acting as a potent stimulus for this transition. Thus, consistent with prior studies,6 EMT in oncology may be more of a spectrum than suggested in classical EMT models from developmental biology (Fig. 1). Accordingly, analysis of “full-EMT” markers in cancer may be misleading and could underlie some of the discrepancies in the field.
Second, the mesenchymal program previously described by the Cancer Genome Atlas (TCGA) and thought to be expressed by cancer cells was shown to be expressed by non-malignant mesenchymal cells, namely fibroblasts and myocytes. Since these are “true” mesenchymal cells – while malignant cells appear to have only a partial mesenchymal profile – the non-malignant mesenchymal profiles overshadowed the malignant ones. Accordingly, tumors with a high frequency of non-malignant mesenchymal cells had high expression of mesenchymal programs in bulk profiles, leading to their classification as a mesenchymal subtype, regardless of the state of the malignant cells. This highlights the limitations of bulk tissue analyses and the caution required when interpreting bulk profiles. Notably, other studies suggest that a similar conclusion can be made in colon cancer,9 raising the possibility that mesenchymal programs described in diverse cancer types have been confounded by the presence of non-malignant mesenchymal cells.
Third, p-EMT was found in a subset of patient samples, suggesting that the controversy around EMT may also reflect tumor-specific biology. P-EMT was found in seven tumors corresponding to the “malignant basal” HNSCC subtype (formerly divided into basal and mesenchymal due to non-malignant cells1), and in this subtype the p-EMT state was predictive of invasion and metastasis. However, three tumors corresponding to the atypical and classical HNSCC subtypes lacked the p-EMT program, and analysis of TCGA data further supported the lack of p-EMT in those HNSCC subtypes. Thus, the underlying genetics and molecular subtype of a tumor may determine the presence and significance of EMT in human tumors (Fig. 1). Notably, atypical and classical subtype tumors metastasize at comparable rates to the malignant-basal subtype tumors, warranting further studies of their metastatic process. A form of EMT might still exist in those tumors, but could be rare and hence not captured in the current analysis. Alternatively, EMT-independent mechanisms may govern their spread, such as collective cell migration of cells.10
Fourth, although p-EMT was correlated with the presence of lymph node metastasis, direct comparison of such metastases to their primary tumors showed highly similar expression patterns, both with respect to p-EMT and to other genes. Thus, no “metastasis signature” could be identified. If expression of particular genes (e.g. p-EMT) facilitates metastasis, then once the metastases are formed their expression appears to revert back to the levels in the primary tumor, consistent with the idea that EMT is followed by the opposing mesenchymal-epithelial transition (MET).6,10 This highlights another challenge in studying the role of EMT in cancer and calls for further analysis of circulating tumor cells (CTCs) (Fig. 1). Future studies investigating these major questions, as well as single cell studies of other human tumors will be essential: A detailed understanding of intra-tumoral heterogeneity is likely to identify novel diagnostic biomarkers, such as those of p-EMT, as well as potential therapeutic targets.
Acknowledgments
This work was supported by research grants from the Zuckerman STEM Leadership Program, and the Benoziyo Endowment Fund for the Advancement of Science (I.T.).
References
- 1.Cancer Genome Atlas N Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129. PMID:25631445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al.. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. doi: 10.1126/science.1254257. PMID:24925914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al.. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–13. doi: 10.1038/nature20123. PMID:27806376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hoverstadt V, Escalante LE, Saw ML, Rodman C, et al.. Decoupling genetics, lineages and tumor micro-environment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017;355(6332). doi: 10.1126/science.aai8478. PMID:28360267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al.. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–96. doi: 10.1126/science.aad0501. PMID:27124452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. PMID:27368099. [DOI] [PubMed] [Google Scholar]
- 7.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al.. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171(7):1611-1624 e24. doi: 10.1016/j.cell.2017.10.044. PMID:29198524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dijk D Nainys J, Sharma R, Kathail P, Carr AJ, Moon KR, Mazutis L, Wolf G, Krishnaswamy S, Pe'er D. MAGIC: A diffusion-based imputation method reveals gene-gene interactions in single-cell RNA-sequencing data. BioRxiv Pre-print. 2017. [Google Scholar]
- 9.Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, et al.. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49(5):708–18. doi: 10.1038/ng.3818. PMID:28319088. [DOI] [PubMed] [Google Scholar]
- 10.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168(4):670–91. doi: 10.1016/j.cell.2016.11.037. PMID:28187288. [DOI] [PMC free article] [PubMed] [Google Scholar]


