Summary
Small non‐coding RNAs have emerged as possible biomarkers for various diseases including autoimmune diseases. A number of studies have demonstrated that the expression of specific microRNAs (miRNAs) is dysregulated in rheumatoid arthritis (RA). So far, all studies on miRNAs in RA patients have been performed using either microarray or reverse transcription–quantitative polymerase chain reaction (RT–qPCR) analyses. Compared to RT–qPCR and microarray analyses, next‐generation sequencing (NGS) allows the genome‐wide analysis of small RNAs and the differentiation between miRNAs that differ by a single nucleotide. The application of NGS to the analysis of small RNAs circulating in sera of RA patients has not been reported. This study provides a global overview of the circulating small RNAs in the sera of RA patients and healthy subjects and identifies differences between these groups using NGS. Several classes of small RNAs, including hY RNA‐derived fragments, tRNA‐derived fragments and miRNAs, were determined. Differentially expressed individual small RNAs were verified by RT‐qPCR. The levels of two miRNAs, miR‐223‐3p and miR‐16‐5p, were significantly lower in the sera from early RA patients than in those from established RA patients and healthy controls. In contrast, the serum level of miR‐16‐5p was higher in patients with established RA than in healthy control samples. These miRNAs may not only serve as biomarkers, but may also shed more light on the pathophysiology of RA.
Keywords: circulating small RNA, miRNAs, next generation sequencing, rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disorder that primarily affects the synovial tissue, leading to destruction of articular cartilage and bone. The precise cause of RA is still unknown, although both genetic and environmental factors have been shown to play a role. Serological parameters, e.g. anti‐citrullinated protein antibodies (ACPA), are important for diagnosing RA. Despite the excellent diagnostic value of ACPA and, albeit to a lesser extent, of rheumatoid factor, a substantial fraction of early RA (eRA) patients lack these serological markers. Moreover, these autoantibodies are not suitable to monitor treatment response. Identification of additional biomarkers for early RA diagnosis, for the prediction of disease outcome and for the monitoring of treatment response is among the most challenging issues in RA research.
Circulating nucleic acids have emerged as possible biomarkers of various diseases, including autoimmune diseases. Various types of small RNAs have been identified recently in body fluids, the best documented of which are microRNAs (miRNAs). The small RNAs differ in their size, structure, function and biogenesis. The vast majority (above 90%) of circulating miRNA is assembled in complexes with proteins that stabilize them 1, 2.
A number of studies have demonstrated that expression of specific miRNAs is dysregulated in RA. The levels of specific miRNA in plasma, serum, blood cells and fibroblasts showed correlations with the disease stage and activity, suggesting that they might serve as blood‐based biomarkers useful for diagnostic and therapeutic applications 3, 4, 5, 6. Both increased and decreased levels of miRNAs have been reported in RA, and in some cases the results are inconsistent 7.
Three techniques have been used to characterize miRNAs profiles; reverse transcription–quantitative polymerease chain reaction (RT–qPCR), microarray hybridization and next‐generation sequencing (NGS). All published studies on miRNAs in RA patients have been performed by either microarray hybridization or RT–qPCR. Compared to these techniques, NGS has several advantages, such as the global analysis of circulating (small) RNAs, including discovery of novel miRNAs, the ability to distinguish between miRNAs that differ by a single nucleotide (isomiRs), and it is more sensitive to rare small RNAs. Moreover, it provides absolute counts of small RNA abundance.
The objective of this study was to obtain a comprehensive overview of the abuncance of small RNAs in the sera of RA patients and healthy subjects, as well as differences between these groups.
Patients and methods
Patient samples
Three different sets of samples were used for this study. Set 1 consisted of four serum samples from healthy controls (CTR) and four serum samples from eRA patients and was used for NGS analysis. Set 2 consisted of 31 eRA samples, 15 established RA (estRA) samples and 30 CTR samples, and set 3 of freshly collected blood serum samples of six CTR subjects, three eRA and five estRA patients. Sets 2 and 3 were used to validate the NGS data with RT–qPCR analysis. The eRA and estRA patient sera were collected at the Erasmus Medical Center in Rotterdam and at the Radboud University Medical Center in Nijmegen. eRA patients were defined as patients who had arthritis in at least one joint with symptoms of < 1 year. Serum samples from CTR subjects were collected at the Sanquin Blood Bank in Nijmegen or at the Radboud University Medical Center. Demographic and clinical characteristics of the study participants are summarized in Supporting information, Table S1. All RA patients fulfilled the 2010 American College of Rheumatology/European League Against Rheumatism (ACR/EULAR) criteria 8 for the classification of RA. Patient sera were used in accordance with the code of conduct of research with human material in the Netherlands. All subjects gave informed consent and the study protocol was approved by local medical ethics committees.
RNA isolation, library preparation and deep sequencing
Small RNA was isolated from sera using the miRCURY RNA isolation kit (Biofluids; Exiqon, Vedbaek, Denmark), according to the manufacturer’s manual. A 100‐μl aliquot of each serum sample was used for RNA extraction. The TruSeq Small RNA sample preparation kit (Illumina, San Diego, CA, USA) was used to generate sequence libraries. The barcoded small RNA‐derived cDNA libraries from four eRA patients and from four CTR subjects were pooled and the quality and quantity of the cDNAs were determined by a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Sequencing of 51 cycles (single end read) was performed on a HiSeq2000 system (Illumina) using the 50 base pairs (bp) fragment sequencing protocol using a service provider (Imagif, CNRS, Paris, France).
Bioinformatics analyses
The raw Illumina reads were first demultiplexed and fastq files were generated. Both steps were performed using casava version 1.8.2 software. Adapter trimming was conducted using Cutadapt‐1.3 to produce reads in FastQ format. FastQC and FASTX tools were applied to check the read quality. Sequences with quality lower than phred = 30 were discarded from the analyses. Reads shorter than 18 nucleotides (nt) and with fewer than 10 counts were also discarded. A primary alignment was performed using Bowtie 2 against the human genome (hg38). Up to two mismatches were allowed during the alignment. Reference library sequences corresponding to mature human miRNAs, tRNAs and hY RNAs were downloaded from miRBase (http://www.mirbase.org; release 21), the genomic tRNA database (https://gtrnadb.ucsc.edu/) and GenBank (https://www.ncbi.nlm.nih.gov/nuccore/), respectively. Raw sequencing data are available on demand.
RT–qPCR
To validate the NGS data, the abundance of specific small RNAs in sera was analysed using the miR‐Q, RT–qPCR method 9. Small RNA was purified from sera using the miRCURY RNA isolation kit (Biofluids; Exiqon). The RevertAid M‐MuLV reverse transcriptase (ThermoFisher Scientific, Fremont, CA, USA) was employed to convert miRNA into cDNA, using primers with 3′‐ends complementary to the terminal six nucleotides at the 3′‐end of the small RNA of interest. The primer sequences are listed in Supporting information, Table S2. Subsequently, a qPCR was performed by using GoTaq©qPCR Master Mix (Promega, Madison, WI, USA) in combination with specific forward primers containing at least 16 nucleotides corresponding to the 5′‐end of the RNA of interest. All reactions were performed in duplicate, in parallel with no‐template controls. Two different methods for normalization were applied. The small RNA levels were normalized either based upon the serum volume used for RNA isolation, as equal volumes and the same isolation procedure were used for all samples, or the 5′ end of hY4 RNA fragment was used as endogenous control, as all three patient sets showed stable levels in sera. LinRegPCR software (version 2015.3) was used to determine amplification efficiency for each of the small RNA samples tested 10.
Statistical analysis
The edgeR algorithm (http://www.bioconductor.org/packages/3.3/bioc/html/edgeR.html version 3.12.0) 11 was applied to detect differential expression (DE) and to perform multi‐dimensional scaling (MDS) analysis. miRNAs were normalized by the trimmed mean of M‐values (TMM) normalization method in edgeR. To account for multiple testing and to control for the number of false positives, we performed Benjamini–Hochberg P‐value correction and the false discovery rate (FDR) was adjusted. Small RNAs with FDR < 0·05 and log2FC > 1 were considered as expressed differentially. Significance across more than two groups was assessed using analysis of variance (anova) (Kruskal–Wallis test). All statistical calculations were performed using GraphPad Prism 5 software. All P‐values were two‐sided and P‐values < 0·05 were considered statistically significant.
Results
Sample statistics and serum small RNA classification
To compare the profiles of small RNAs in sera of eRA patients and CTR, cDNA libraries from four samples of both groups (training set) were generated and sequenced using Illumina HiSeq 2000 with eight multiplexed samples in one sequencing lane. After the adapter trimming step, 6 669 112 high‐quality reads were obtained for all eight libraries together; range = 0·34–2·0 million for CTR and range = 0·3–1·2 million for the eRA samples, respectively. RNAs of 18–50 nucleotides long were selected for further analyses.
An analysis of the size distribution of the small RNAs revealed two main peaks, both for total number of reads and for the reads mapped to the human genome (hg38; Supporting information, Fig. S1). The highest read counts corresponded to RNA molecules of 30–32 nucleotides in length, while the second peak represented molecules of approximately 22 nts. The proportion of reads mapped to the human genome was significantly lower for eRA samples. In total, 84% of the CTR reads (Supporting information, Fig. S1a) and 41% of the eRA reads (Supporting information, Fig. S1b) could be mapped to the human genome.
Mapping of reads revealed that the majority of the human reads corresponded to three classes of small RNAs; hY RNAs, tRNAs and miRNAs (Supporting information, Fig. S2). Most of the human 30‐32 nts reads aligned to hY RNAs and tRNAs (Supporting information, Fig. S2a,b), whereas the 21–23 nts reads represented mainly miRNAs (Supporting information, Fig. S2c). The NGS results for individual samples are summarized in Fig. 1 and Supporting information, Table S3. These results show that the relative abundance of hY RNA‐ and tRNA‐derived molecules appeared to be reversed in the eRA and CTR samples, tRNAs being more abundant in the eRA samples and hY RNA being more abundant in the CTR samples. The non‐human reads, which include reads mapped to non‐human databases or reads which do not match to any sequence database, showed a less size‐restricted pattern, although the most abundant non‐human reads corresponded to molecules of 22–23 and 50 nts (Supporting information Fig,. S2d). The latter category may represent RNA fragments longer than 50 nts or failure of adapter removal.
Figure 1.
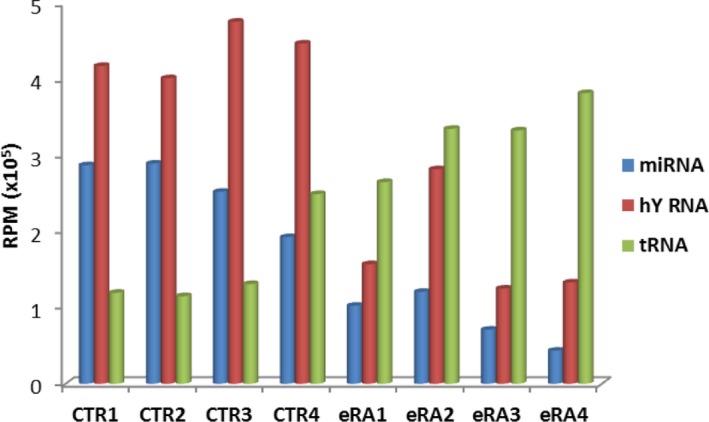
Small RNA abundance in sera of early rheumatoid arthritis (eRA) patients and control (CTR) individuals. The number of reads for miRNAs and fragments of hY RNAs and transfer RNAs (tRNAs) relative to reads mapped to the human genome is illustrated graphically for four CTR and four eRA samples. RPM = reads per million.
Multi‐dimensional scaling (MDS) analysis was applied to examine the samples for outliers and their relationship. In MDS plots, dimension 1 separates the RA samples from CTR samples, while dimension 2 corresponds to patient number. MDS analysis showed that CTR and RA samples were well grouped together for reads aligned to the human genome (Supporting information, Fig. S3a) and to miRNAs (Supporting information, Fig. S3c) and to a lesser extent for tRNA derived fragments (Supporting information, Fig. S3b). In general, the RA samples appear more heterogeneous than the CTR samples.
hY RNA‐derived fragments
The most abundant type of circulating small RNAs detected in sera of all samples was hY RNA. Y RNAs constitute a family of highly conserved small non‐coding RNAs, and in human cells four distinct Y RNAs (84–112 nts), hY1, hY3, hY4 and hY5 RNA have been found 12. Fragments of hY4 RNA were the most abundant hY RNA in all samples (Fig. 2a). Most hY RNA‐derived fragments are 30–32 nts long and correspond to both 5′‐ and 3′‐ends of the hY RNAs. However, 5′‐end fragments were much more abundant in comparison with 3′‐end fragments. Note that due to the size restriction of RNAs selected for sequencing (50 nts), our data do not provide information on the presence of full‐length hY RNAs. A comparison of CTR and eRA samples demonstrated lower read counts for hY4 and higher read counts for hY1 and hY5 RNA fragments in the eRA samples, but the differences were not significant.
Figure 2.
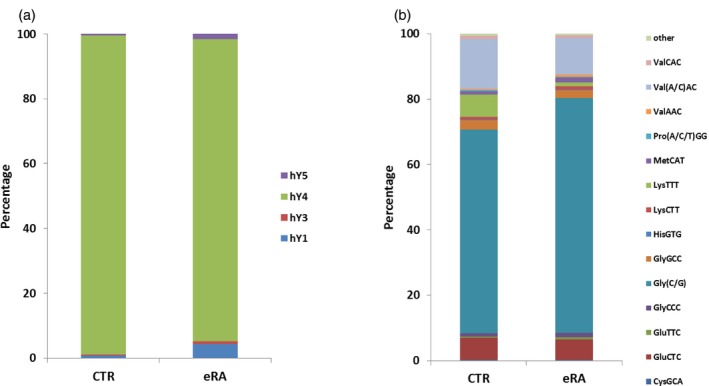
Relative abundance of distinct hY RNA‐ and transfer RNA (tRNA)‐derived fragments in early rheumatoid arthritis (eRA) and control (CTR) sera. The next‐generation sequencing (NGS) data were used to calculate the relative abundance of fragments from individual hY RNAs (a) or tRNAs (b) in sera from eRA patients and CTR individuals. Percentage of reads aligned to each type denotes the mean value for four serum samples.
tRNA‐derived fragments
The second class of abundant small RNAs in sera was tRNA, more specifically fragments derived from tRNAs (tRFs). Most of the tRFs are 29–33 nts in length and correspond to the 5′ ends of various tRNAs with cleavage sites located preferentially at the anti‐codon loops. Fragments corresponding to the internal parts and the 3′‐ends of tRNAs were also found, but with a much lower frequency than fragments derived from the 5′‐ends. The most abundant circulating tRNA fragments were derived from tRNA‐Gly‐(C/G)CC (average 67%), tRNA‐Val‐(A/C)AC (average 13%) and tRNA‐Glu‐CTC (average 6·6%, Fig. 2b, Supporting information, Table S4). The raw read frequencies for each tRF did not correlate with the number of tRNA genes per isotype (data not shown).
To determine whether tRFs were expressed differentially in sera of eRA patients and CTR, edgeR differential expression analysis was applied. The results showed that there was no significant difference between the levels of tRFs in eRA and CTR sera (FRD > 0·05; Supporting information, Table S5).
miRNA content
The third class of small RNAs for which a relatively large number of reads was obtained is miRNAs. An alignment to the miRBase database (version 21; containing 2588 human miRNAs) identified reads for 605 miRNAs throughout all samples together (Supporting information, Table S6). In each individual sample the number of miRNA reads varied from one to 426 883. Of the identified miRNAs, 82 miRNAs were detected in all samples. The 20 most abundant miRNAs identified in the eRA and CTR samples are listed in Table 1; miR‐486‐5p, miR‐92a‐3p, miR‐16‐5p and miR‐22‐3p were the most prominent. All these miRNAs are known to be expressed in blood cells, especially in erythrocytes 13, 14, 15.
Table 1.
Top 20 most abundant miRNA in sera of rheumatoid arthritis (RA) patients and control (CTR) individuals
| Human miRNAs | CTR sera* (%) | RA sera* (%) | Possible source of miRNA† |
|---|---|---|---|
| hsa‐miR‐486‐5p | 54 ± 9 | 46 ± 12 | RBC 16 |
| hsa‐miR‐92a‐3p | 14 ± 2 | 13 ± 3 | RBC, platelets, T cells, B cells, RASF 16, 17, 18 |
| hsa‐miR‐16‐5p | 4·8 ± 2·2 | 2·1 ± 1·4 | RBC, platelets, granulocytes, leucocytes 17, 18 |
| hsa‐miR‐22‐3p | 2·3 ± 0·7 | 3·8 ± 1·5 | Not known |
| hsa‐miR‐451a | 2·3 ± 0·3 | 1·4 ± 0·6 | RBC 17, 18 |
| hsa‐miR‐486‐3p | 2·2 ± 0·3 | 1·3 ± 1·5 | Granulocytes 19 |
| hsa‐miR‐191‐5p | 2·1 ± 0·4 | 2·6 ± 2·6 | Neutrophils, monocytes 20 |
| hsa‐miR‐26a‐5p | 1·4 ± 0·7 | 2·1 ± 1·7 | Platelets, T cells, B cells 17 |
| hsa‐miR‐423‐5p | 1·4 ± 0·8 | 1·8 ± 0·8 | Not known |
| hsa‐miR‐25‐3p | 0·9 ± 0·1 | 0·40 ± 0·04 | RBC, granulocytes 17 |
| hsa‐miR‐181a‐5p | 0·7 ± 0·2 | 1·0 ± 0·3 | Neutrophils, RASF 18, 21 |
| hsa‐miR‐30e‐5p | 0·7 ± 0·3 | 0·4 ± 0·1 | RASF 21 |
| hsa‐miR‐3184‐3p | 0·6 ± 0·5 | 2·1 ± 1·5 | Not known |
| hsa‐let‐7f‐5p | 0·6 ± 0·3 | 1·3 ± 1·4 | RBC, platelets, granulocytes 17 |
| hsa‐miR‐126‐5p | 0·5 ± 0·2 | 1·5 ± 1·2 | Platelets, RASF 18, 22 |
| hsa‐miR‐30d‐5p | 0·5 ± 0·2 | 0·4 ± 0·2 | Not known |
| hsa‐miR‐223‐3p | 0·5 ± 0·1 | 0·2 ± 0·2 | Granulocytes, platelets 17, 23 |
| hsa‐miR‐10b‐5p | 0·49 ± 0·07 | 2·9 ± 1·3 | RASF 18 |
| hsa‐miR‐423‐3p | 0·4 ± 0·1 | 0·3 ± 0·1 | Not known |
| hsa‐miR‐10a‐5p | 0·32 ± 0·05 | 1·7 ± 1·1 | Not known |
*Percentage of total number of miRNA reads.
†RBC, red blood cells; RASF, rheumatoid arthritis synovial fibroblasts.
A comparison of the levels of miRNAs in eRA and CTR sera by edgeR analysis demonstrated that six differentially expressed miRNAs passed the Benjamini–Hochberg correction; three were found at higher levels (miR‐10a, miR‐10b and miR‐320a) in eRA and the other three at reduced levels (miR‐223‐3p, miR‐25‐3p and miR‐16‐5p) in eRA sera in comparison with CTR samples (Table 2; log2FC > 1, FDR < 0·05).
Table 2.
miRNAs differentially expressed in eRA patients’ sera vs control (CTR) samples
| Log2FC* | Log2CPM† | P‐value | FDR‡ | |
|---|---|---|---|---|
| hsa‐miR‐10b‐5p | 2·02 | 14·01 | 6·3E–05 | 0·0011 |
| hsa‐miR‐10a‐5p | 1·83 | 13·16 | 7·0E–05 | 0·0011 |
| hsa‐miR‐320a | 1·90 | 11·16 | 0·00036 | 0·0038 |
| hsa‐miR‐223‐3p | –2·03 | 11·58 | 0·00049 | 0·0039 |
| hsa‐miR‐25‐3p | –1·30 | 13·02 | 0·0056 | 0·03 |
| hsa‐miR‐16‐5p | –1·48 | 15·44 | 0·023 | 0·03 |
*Log2FC is log2 fold change; positive values indicate elevated and negative values reduced levels in eRA samples, respectively.
†Log2CPM: log2 read count per million;
‡FDR = false discovery rate.
Validation of NGS data using RT–qPCR
To validate the NGS data, the 5′‐end of hY4 RNA and the six differentially expressed miRNAs were selected for analysis with independent patient (eRA, n = 31 and estRA, n = 15) and control samples (n = 30; set 2). The levels of these RNAs were determined by RT–qPCR, using the miR‐Q approach, which has been developed for small miRNA‐sized RNA molecules 9. For four of the miRNAs (miR‐10a, miR‐10b, miR‐320a and miR‐25‐3p) the Ct values appeared to be too low for reliable conclusions and thus the altered levels of these miRNAs in eRA could not be confirmed. In agreement with the NGS data, the relative levels of the 5′‐end of hY4 RNA molecules did not show differences between CTR and eRA samples. The levels of miR‐223‐3p and miR‐16‐5p were found to be significantly lower in eRA sera in comparison with CTR and estRA samples (Fig. 3a), confirming our NGS data. Although miR‐223‐3p levels did not differ between estRA and CTR samples, the level of miR‐16‐5p was significantly higher in estRA compared with CTR samples (P = 0·014). The levels of these miRNAs did not correlate with age, disease activity score 28 in joints (DAS28) and C‐reactive protein (CRP) levels, except for the ESR, which appeared to correlate with serum miR‐223‐3p levels in eRA (P = 0·034).
Figure 3.
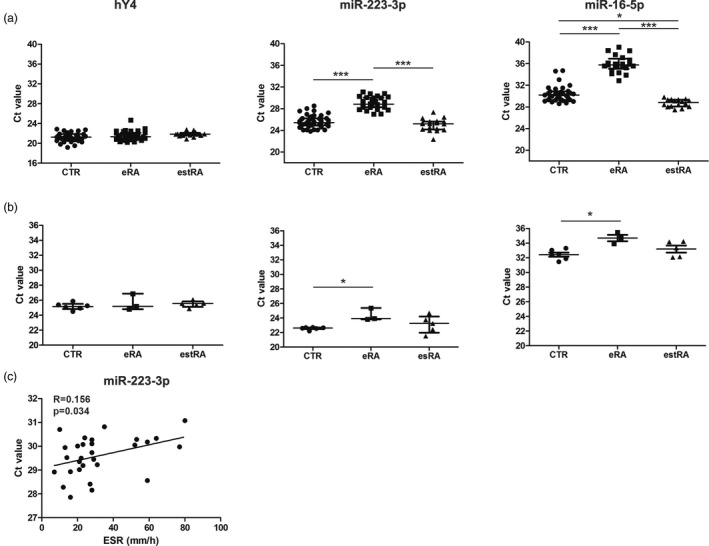
Reverse transcription–quantitative polymerase chain reaction (RT–qPCR) analysis of the 5′‐end of hY4 RNA, miR‐223‐3p and miR‐16‐5p in rheumatoid arthritis (RA) and control (CTR) sera. Small RNAs were isolated from frozen sera and the presence of the 5′‐end fragment of hY4 RNA, miR‐223‐3p and miR‐16‐5p was determined by the miR‐Q approach (a). miR‐Q analyses of the same small RNAs in fresh sera from eRA and estRA patients and from CTR individuals (b). Spearman’s correlation between serum miR‐223‐3p levels in eRA patients and ESR (c). Spearman’s coefficient (r) is shown along with its significance (P‐value). The Mann–Whitney two‐sample tests were performed to examine the differences between RA and CTR (*P < 0·05; ***P < 0·001); n.s. = non‐significant.
Influence of pre‐analytical variability on serum samples
Previous studies indicated that there is a general systematic influence of the storage conditions and/or specimen processing procedures on small RNA patterns of biofluids and that the specific effect of the storage conditions has to be verified for each small RNA separately 13, 14, 15. To determine whether a pre‐analytical step such as freezing/thawing affected the outcome of small RNA measurements in serum samples, small RNAs were isolated immediately after blood withdrawal (set 3) and the levels of hY4, miR‐223‐3p and miR‐16‐5p were determined. 16, 17, 18, 19, 20, 21, 22, 23 Similar to the observations made with the frozen samples, lower levels of miR‐223‐3p and miR‐16‐5p were observed in eRA compared to CTR samples (Fig. 3b; P = 0.04 and P = 0.06 for miR‐223‐3p and miR‐16‐5p, respectively). Moreover, there was no significant difference for the 5′‐end of hY4 RNA, miR‐223‐3p and miR‐16‐5p between CTR and estRA samples (Fig. 3b).
Discussion
The results of this study provide a global overview of circulating small RNAs in the sera of RA patients. To our knowledge, this is the first study in which the profiles of circulating small RNAs in sera of RA patients were analysed and compared with those of healthy controls using the NGS approach. We found a high degree of diversity among the circulating small RNAs in all tested samples, although the profiles were dominated by fragments derived from hY4 RNA, tRNA‐Gly, tRNA‐Glu and tRNA‐Val, and by miR‐486‐5p, which together comprised 51 and 71% of all reads mapped to hg38 for eRA and controls, respectively. The abundance of these RNAs in serum/plasma is similar to observations made in independent studies 24, 25. However, no significant differences in the levels of these most abundant small RNAs were found between eRA and healthy control samples. In contrast, for two less abundant miRNAs, miR‐223‐3p and miR‐16‐5p both NGS and RT–qPCR data revealed decreased levels in sera of eRA patients. Both miRNAs have been reported previously to be associated with RA. In one study the miR‐16‐5p level was decreased in the sera of naive RA patients in comparison with estRA, while it was comparable with that in samples of healthy subjects 26. Our data demonstrated significantly lower levels of miR‐16‐5p in the sera of patients with eRA in comparison with CTR and estRA samples in two separate cohorts of frozen and fresh samples. The differences in treatment could be one possible explanation for discrepancy between our results and others. In our study eRA patients in both cohorts were treated with either methotrexate (MTX), prednisone or both. Another potential explanation is the quantification method used to measure miRNA levels. It has been shown previously that quantification of serum miRNAs is influenced significantly by different normalization approaches; let‐7a was used as a stable reference miRNA for normalization 27. Indeed, let‐7a is among the most stable reference genes, although Niimoto et al. 28 reported that let‐7a was up‐regulated significantly in the interleukin (IL)‐17 producing CD4+ T cells from RA patients. We have used two different approaches to normalize the data; the small RNA levels were normalized based either upon the serum volume used for RNA isolation or the 5′ end of the hY4 RNA fragment was used as endogenous control.
In another study, elevated levels of miR‐16‐5p were found in peripheral blood mononuclear cells from RA patients in comparison with healthy controls, and the levels correlated with active disease in RA patients 4. The concentration of miR‐16‐5p in RA synovial fluid was significantly higher than that in osteoarthritis (OA) synovial fluid 29. The differences between the biological materials analysed might, at least, in part explain the discrepancy.
Interestingly, it has been shown previously that chloramidine (Cl‐amidine), a pan‐protein arginine deiminase (PAD) inhibitor, up‐regulates miR‐16‐5p expression in mice 30. In agreement with this, Cl‐amidine treatment decreased the expression of the miR‐16‐5p targets, such as cyclins D1, E1 and the cell proliferation marker Ki67, in epithelial cells. As an increase in PAD enzyme activity in sera of RA patients compared to controls has been reported 31, it is tempting to speculate that PAD activation during disease development leads to a decrease of miRNA‐16‐5p expression in blood and/or synovial fluid cells and increased cell proliferation rates, e.g. during chronic immune activation and pannus formation. It remains to be established whether variation in PAD activation affects miR‐16‐5p expression in blood cells or synovium from RA patients.
A number of studies also demonstrated an association of miR‐223‐3p with RA. Anti‐tumour necrosis factor (TNF)‐α/disease‐modifying anti‐rheumatic drug (DMARD) combination therapy of RA patients resulted in significant up‐regulation of serum miRNAs, including both miR‐223‐3p and miR‐16‐5p. The changes observed in miR‐223‐3p and miR‐16‐5p expression correlated significantly with the changes observed for clinical parameters, such as DAS28, and for inflammatory parameters, such as CRP or ESR 32. In the case of frozen samples, we observed no difference for the miR‐223‐3p level and a higher level of miRNA‐16‐5p in sera from estRA patients, who were treated with DMARDs, in comparison with CTR samples. The miR‐16‐5p levels were similar for estRA and CTR when fresh blood was used to isolate small RNAs. These data indicate that changes in miRNA levels should be interpreted with care if the samples of the different groups are not collected and stored by the same procedures.
miR‐223‐3p is expressed more highly in the synovium from of RA patients than in osteoarthritis synovium due to the increased number of miR‐223‐3p‐positive cells 33. Higher miR‐223‐3p levels were also observed in T cells from RA patients 34, 35 and in microvesicles from RA synovial fluid 36. In contrast to our results, the concentrations of miR‐223‐3p were reported to be increased significantly in plasma of RA patients in another study 37. This discrepancy might be explained by the fact that we analysed serum samples. It has been shown recently that plasma samples are commonly contaminated by platelets and a single freeze/thaw cycle of plasma increases the number of platelet‐derived microparticles dramatically, contaminates the extracellular miRNA pool and affects the levels of miRNA detection 38. Serum samples become depleted of platelets during the clotting process. MiR223‐3p is expressed in cells of the myeloid lineage and its levels are relatively high in polymorphonuclear neutrophils. Therefore, these cells can be an important source of circulating miR‐223‐3p 17. The analysis of leucocyte/platelet markers, such as CD45 and ITGA2B, might shed more light on this possibility 22.
Although there were no significant differences between the relative quantities of the individual hY RNA and tRNA fragments, our NGS data revealed a higher proportion of reads mapped to tRNA in eRA samples and higher frequency of reads mapped to hY RNA in the CTR samples. These differences might be explained by either preferential release or higher turnover rates of certain types of circulating small RNAs. Similar differences in serum abundance of tRNA‐ and Y RNA‐derived small RNAs have been observed in head and neck squamous cells carcinoma and breast cancer patients when compared to healthy individuals 39, 40. tRNA‐derived small RNAs are involved in inhibition of apoptosis and promotion of survival of injured cells 41. Y RNA‐derived small RNAs influence DNA replication, cell proliferation, stress response and apoptosis 42. Further studies are required to shed more light on this possibility and to reveal potential (patho)physiological effects of the circulating fragments of these RNAs.
We also observed a higher proportion of non‐human reads in eRA samples compared with healthy individuals. DNA from a wide variety of bacteria has been detected previously in the synovium and serum of RA patients and was shown to be associated with different forms of arthritis 43, 44, 45. We detected the presence of circulating small RNAs derived from different bacterial species. Small RNAs from fungi and yeasts were also found. Systematic, prospective studies will be required to verify these data.
This study has one notable limitation. To identify small RNA biomarker candidates, we used a small sample size for the NGS analyses. As a consequence, NGS results need to be validated by an independent, less laborious method and a larger sample size. Indeed, only a fraction of the candidates identified by NGS were confirmed by RT–qPCR.
In conclusion, the results of our study suggest that circulating miR‐223‐3p and miR‐16‐5p can be used as a biomarker for eRA and provide further support for the association of miR‐16‐5p and miR‐223‐3p with RA. Previously, miR‐223 was proposed as a marker of disease activity and both miR‐16 and miR‐223 as possible predictors of disease outcome in eRA 26. Nevertheless, the clinical value of these potential biomarkers still needs to be established. In this respect, it is also important to note that miR‐16 was linked to oncosuppression and was identified as a promising biomarker in diagnosing cancers such as gastric, breast and prostate cancers 17, 22, 38. As a consequence, the interpretation of quantitative data on these molecules in a clinical setting might be somewhat complex.
Disclosure
The authors declare no conflicts of interest.
Author contributions
All authors were involved in drafting of the article or revising it critically and all authors approved the final version to be published. M. D. has full access to all of the data in the study and takes responsibility for the integrity of the data and accuracy of the data analysis. Study concept and design: M. D. and G. P.; acquisition of data: M. D., J. B. and G. P.; analysis and interpretation of data: M. D., J. B., R. T. and G. P.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Length distribution of reads mapped to hg38 and total reads of small RNA isolated from sera of control (CTR) and early rheumatoid arthritis (eRA) patients. (a) Length distribution of reads obtained from four serum samples of CTR subjects. (b) Length distribution of reads obtained from four serum samples of eRA patients. The y‐axis shows the number of reads for a given small RNA length (in nucleotides, nt). Bars represent mean values. Different colours of bars denote total reads (blue) and reads which aligned to hg38 (red).
Fig. S2. Size distribution of different classes of small RNAs. Length distribution of reads obtained from serum samples of control (CTR) (upper graphs) and early rheumatoid arthritis (eRA) (lower graphs) samples. (a) hY RNA‐derived molecules; (b) transfer RNA (tRNA)‐derived molecules; (c) miRNAs; (d) non‐human reads.
Fig. S3. Multi‐dimensional scaling (MDS) plot of abundance levels of small RNAs in sera of early rheumatoid arthritis (eRA) patients and control (CTR) subjects. MDS analysis of the sequence data clusters the samples based on similarities; subjects with similar small RNA expression are represented by points close on the other. The leading log‐fold‐change (logFC) is the average of the largest absolute log‐fold changes between each pair of RNA samples. (a) Small RNAs aligned to hg38 genome. (b) Transfer RNA (tRNA)‐derived small RNA. (c) miRNAs. The filtering criteria are read counts > 10 and two mismatches.
Table S1. Demographic and clinical characteristics of the patients with rheumatoid arthritis and healthy subjects
Table S2. Complete list of primers used in the study
Table S3. List of sequence reads identified in sera of early rheumatoid arthritis (eRA) patients and control (CTR) subjects
Table S4. Percentage of different transfer RNA (tRNA) derived fragment (tRFs) in sera of rheumatoid arthritis (RA) patients and controls (CTR)
Table S5. The abundance of tRNA‐derived fragments in sera of early rheumatoid arthritis (eRA) patients versus control (CTR) samples
Table S6. miRNAs identified by next‐generation sequencing (NGS). miRNA raw read counts and normilised read counts (per million reas) are shown for each sample
Acknowledgements
We thank Wilbert Boelens (Radboud University, Nijmegen, the Netherlands) for critical reading of the manuscript and helpful suggestions and Jolanda Luime and Erik Lubberts (University Medical Center Rotterdam, the Netherlands) for providing serum samples.
References
- 1. Arroyo JD, Chevillet JR, Kroh EM et al Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 2011;108:5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chevillet JR, Kang Q, Ruf IK et al Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA 2014;111:14888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li J, Wan Y, Guo Q et al Altered microRNA expression profile with miR‐146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther 2010;12:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR‐146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther 2008;10:R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stanczyk J, Pedrioli DM, Brentano F et al Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum 2008;58:1001–9. [DOI] [PubMed] [Google Scholar]
- 6. Stagakis E, Bertsias G, Verginis P et al Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR‐21 regulates aberrant T cell responses through regulation of PDCD4 expression. Ann Rheum Dis 2011;70:1496–506. [DOI] [PubMed] [Google Scholar]
- 7. Salehi E, Eftekhari R, Oraei M, Gharib A, Bidad K. MicroRNAs in rheumatoid arthritis. Clin Rheumatol 2015;34:615–28. [DOI] [PubMed] [Google Scholar]
- 8. Aletaha D, Neogi T, Silman AJ et al Rheumatoid arthritis classification criteria. Arthritis Rheum 2010;2010(62):2569–81. [DOI] [PubMed] [Google Scholar]
- 9. Sharbati‐Tehrani S, Kutz‐Lohroff B, Bergbauer R, Scholven J, Einspanier R. miR‐Q: a novel quantitative RT‐PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol Biol 2008;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruijter JM, Ramakers C, Hoogaars WM et al Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009;37:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lerner MR, Boyle JA, Hardin JA, Steitz JA. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 1981;211:400–2. [DOI] [PubMed] [Google Scholar]
- 13. Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLOS ONE 2011;6:e20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras‐Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 2011;57:833–40. [DOI] [PubMed] [Google Scholar]
- 15. Backes C, Leidinger P, Altmann G et al Influence of next‐generation sequencing and storage conditions on miRNA patterns generated from PAXgene blood. Anal Chem 2015;87:8910–6. [DOI] [PubMed] [Google Scholar]
- 16. Pritchard CC, Kroh E, Wood B et al Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res 2012;5:492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Teruel‐Montoya R, Kong X, Abraham S et al MicroRNA expression differences in human hematopoietic cell lineages enable regulated transgene expression. PLOS ONE 2014;9:e102259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de la Rica L, Urquiza JM, Gómez‐Cabrero D et al Identification of novel markers in rheumatoid arthritis through integrated analysis of DNA methylation and microRNA expression. J Autoimmun 2013;41:6–16. [DOI] [PubMed] [Google Scholar]
- 19. Bianchi E, Bulgarelli J, Ruberti S et al MYB controls erythroid versus megakaryocyte lineage fate decision through the miR‐486‐3p‐mediated downregulation of MAF. Cell Death Differ 2015;22:1906–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edelstein LC, McKenzie SE, Shaw C, Holinstat MA, Kunapuli SP, Bray PF. MicroRNAs in platelet production and activation. J Thromb Haemost 2013;11:340–50. [DOI] [PubMed] [Google Scholar]
- 21. Ward JR, Heath PR, Catto JW, Whyte MK, Milo M, Renshaw SA. Regulation of neutrophil senescence by microRNAs. PLOS ONE 2011;6:e15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaudewitz D, Skroblin P, Bender LH et al Association of microRNAs and YRNAs with platelet function. Circ Res 2016;118:420–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plé H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLOS ONE 2012;7:e50746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Mote P, Martin DI. 5'‐YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics 2013;45:990–8. [DOI] [PubMed] [Google Scholar]
- 25. Tosar JP, Gámbaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Assessment of small RNA sorting into different extracellular fractions revealed by high‐throughput sequencing of breast cell lines. Nucleic Acids Res 2015;43:5601–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Filková M, Aradi B, Senolt L et al Association of circulating miR‐223 and miR‐16 with disease activity in patients with early rheumatoid arthritis. Ann Rheum Dis 2014;73:1898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem 2015;61:1333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Niimoto T, Nakasa T, Ishikawa M, Okuhara A et al MicroRNA‐146a expresses in interleukin‐17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet Disord 2010;11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murata K, Yoshitomi H, Tanida S et al Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther 2010;12:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Witalison EE, Cui X, Causey CP, Thompson PR, Hofseth LJ. Molecular targeting of protein arginine deiminases to suppress colitis and prevent colon cancer. Oncotarget 2015;6:36053–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Basu PS, Majhi R, Ghosal S, Batabyai SK. Peptidyl‐arginine deiminase: an additional marker of rheumatoid arthritis. Clin Lab 2011;57:1021–5. [PubMed] [Google Scholar]
- 32. Castro‐Villegas C, Pérez‐Sánchez C, Escudero A et al Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti‐TNFα. Arthritis Res Ther 2015;17:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shibuya H, Nakasa T, Adachi N et al Overexpression of microRNA‐223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod Rheumatol 2013;23:674–85. [DOI] [PubMed] [Google Scholar]
- 34. Fulci V, Scappucci G, Sebastiani GD et al miR‐223 is overexpressed in T‐lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol 2010;71:206–11. [DOI] [PubMed] [Google Scholar]
- 35. Lu MC, Yu CL, Chen HC, Yu HC, Huang HB, Lai NS. Increased miR‐223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin‐like growth factor‐1‐mediated interleukin‐10 production. Clin Exp Immunol 2014;177:641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim ND, Lochhead RB, Schmit R, Kohler MJ, Luster AD. Microvesicle‐associated hsa‐miR‐223‐3p is elevated in rheumatoid synovial fluid compared with osteoarthritis synovial fluid (abstract). Arthritis Rheumatol 2015;67 (Suppl:10). https://acrabstracts.org/abstract/microvesicle-associated-hsa-mir-223-3p-is-elevated-in-rheumatoid-synovial-fluid-compared-with-osteoarthritis-synovial-fluid/ (accessed 07 August 2018). [Google Scholar]
- 37. Ormseth MJ, Solus JF, Vickers KC, Oeser AM, Raggi P, Stein CM. Utility of select plasma microRNA for disease and cardiovascular risk assessment in patients with rheumatoid arthritis. J Rheumatol 2015;42:1746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mitchell AJ, Gray WD, Hayek SS et al Platelets confound the measurement of extracellular miRNA in archived plasma. Sci Rep 2016;6:32651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Victoria Martinez B, Dhahbi JM, Nunez Lopez YO et al Circulating small non‐coding RNA signature in head and neck squamous cell carcinoma. Oncotarget 2015;6:19246–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DI. Deep sequencing of serum small RNAs identifies patterns of 5' tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer 2014;6:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raina M, Ibba M. TRNAs as regulators of biological processes. Front Genet 2014;5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kowalski MP, Krude T. Functional roles of non‐coding Y RNAs. Int J Biochem Cell Biol 2015;66:20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gérard HC , Wang Z , Wang GF et al Chromosomal DNA from a variety of bacterial species is present in synovial tissue from patients with various forms of arthritis. Arthritis Rheum 2001;44:1689–97. [DOI] [PubMed] [Google Scholar]
- 44. Martinez‐Martinez RE, Abud‐Mendoza C, Patiño‐Marin N, Rizo‐Rodríguez JC, Little JW, Loyola‐Rodríguez JP. Detection of periodontal bacterial DNA in serum and synovial fluid in refractory rheumatoid arthritis patients. J Clin Periodontol 2009;36:1004–10. [DOI] [PubMed] [Google Scholar]
- 45. Schumacher HR Jr, Arayssi T, Crane M et al Chlamydia trachomatis nucleic acids can be found in the synovium of some asymptomatic subjects. Arthritis Rheum 1999;42:1281–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Length distribution of reads mapped to hg38 and total reads of small RNA isolated from sera of control (CTR) and early rheumatoid arthritis (eRA) patients. (a) Length distribution of reads obtained from four serum samples of CTR subjects. (b) Length distribution of reads obtained from four serum samples of eRA patients. The y‐axis shows the number of reads for a given small RNA length (in nucleotides, nt). Bars represent mean values. Different colours of bars denote total reads (blue) and reads which aligned to hg38 (red).
Fig. S2. Size distribution of different classes of small RNAs. Length distribution of reads obtained from serum samples of control (CTR) (upper graphs) and early rheumatoid arthritis (eRA) (lower graphs) samples. (a) hY RNA‐derived molecules; (b) transfer RNA (tRNA)‐derived molecules; (c) miRNAs; (d) non‐human reads.
Fig. S3. Multi‐dimensional scaling (MDS) plot of abundance levels of small RNAs in sera of early rheumatoid arthritis (eRA) patients and control (CTR) subjects. MDS analysis of the sequence data clusters the samples based on similarities; subjects with similar small RNA expression are represented by points close on the other. The leading log‐fold‐change (logFC) is the average of the largest absolute log‐fold changes between each pair of RNA samples. (a) Small RNAs aligned to hg38 genome. (b) Transfer RNA (tRNA)‐derived small RNA. (c) miRNAs. The filtering criteria are read counts > 10 and two mismatches.
Table S1. Demographic and clinical characteristics of the patients with rheumatoid arthritis and healthy subjects
Table S2. Complete list of primers used in the study
Table S3. List of sequence reads identified in sera of early rheumatoid arthritis (eRA) patients and control (CTR) subjects
Table S4. Percentage of different transfer RNA (tRNA) derived fragment (tRFs) in sera of rheumatoid arthritis (RA) patients and controls (CTR)
Table S5. The abundance of tRNA‐derived fragments in sera of early rheumatoid arthritis (eRA) patients versus control (CTR) samples
Table S6. miRNAs identified by next‐generation sequencing (NGS). miRNA raw read counts and normilised read counts (per million reas) are shown for each sample


