Abstract
Cell chemotaxis plays a pivotal role in normal development, inflammatory response, injury repair and tissue regeneration in all organisms. It is also a critical contributor to cancer metastasis, altered angiogenesis and neurite growth in disease. The molecular mechanisms regulating chemotaxis are currently being identified and key components may be pertinent therapeutic targets. Although these components appear to be mostly common in various cells, there are important differences in chemotactic signaling networks and signal processing that result in the distinct chemotactic behavior of mesenchymal cells compared to much better studied amoeboid blood cells. These differences are not necessarily predetermined based on cell type, but are rather chosen and exploited by cells to modify their chemotactic behavior based on physical constraints and/or environmental conditions. This results in a specific type of chemotactic migration in mesenchymal cells that can be selectively targeted in disease. Here, we compare the chemotactic behavior, signaling and motility of mesenchymal and amoeboid cells. We suggest that the current model of chemotaxis is applicable for small amoeboid cells but needs to be reconsidered for large mesenchymal cells. We focus on new candidate regulatory molecules and feedback mechanisms that may account for mesenchymal cell type-specific chemotaxis.
Keywords: Chemotaxis, Feedback regulation, Fibroblasts, Hydrogen peroxide, Signaling
List of abbreviations: GEFs, guanine nucleotide exchange factors; GPCRs, G-protein coupled receptors; LEGI, local excitation and global inhibition; MAP-kinase, mitogen-activated protein kinase; mTORC, mechanistic target of rapamycin complex; NOX, NADPH-oxidase; PTEN, phosphatase and tensin homolog; PI3-kinase, phosphatidylinositol-3-kinase; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PLA2, phospholipase A2; PDGF, platelet derived growth factor; РТР-1В, protein tyrosine phosphatase-1B; RTKs, receptor tyrosine kinases
Introduction
Directional migration of intramural cells is fundamental for tissue patterning and embryonic and postnatal development. In adults, directional movement of immune cells into sites of injury or infection is critical for the development of an inflammatory response, subsequent wound healing and tissue regeneration. The latter is provided by mesenchymal cells that move into damaged areas, produce connective tissue and maintain tissue homeostasis. These cells display a common fibroblast-like appearance and characteristic mode of migration. Hereafter, we focus on fibroblasts as the prototypical mesenchymal cell and standard experimental model. In addition, epithelial cancer cells disseminate by undergoing an epithelial-to-mesenchymal transition to acquire a mesenchymal migratory phenotype. Their directional migration is a hallmark of cancer metastasis and the pathogenesis of autoimmune diseases. This highlights the specificity and importance of the mesenchymal type of migration compared to the better understood amoeboid migration. For this reason, we focus on the mesenchymal-specific type of chemotaxis rather than common molecular mechanisms that have been exhaustively reviewed recently.1, 2, 3, 4, 5
Directional migration is based on the intrinsic propensity of cells to persistently move in one direction without turning.6 They can be ‘directed’ by various physical and chemical stimuli, such as light, temperature, substrate rigidity, matrix proteins, soluble substances, and so on. These stimuli form asymmetric patterns that are detected by cells and used as an external compass.7 Thus, this directed type of migration ultimately involves the conversion of asymmetrical external stimulants into internal gradients of signaling molecules that in turn influence the cytoskeleton that performs the motile responses.2, 5
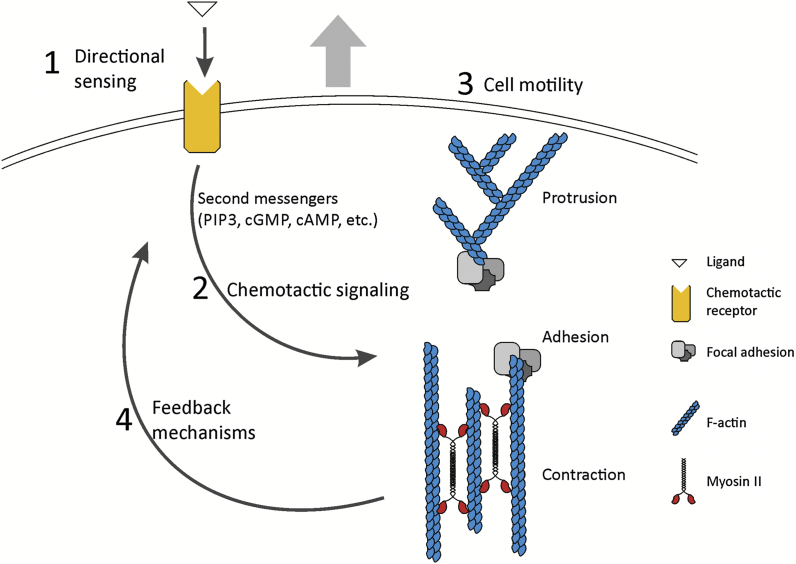
Chemotaxis is defined as directed migration of cells towards a source of soluble chemical agents (chemoattractants) that bind to surface receptors and stimulate cells to move. Chemotaxis involves 4 major components (Fig. 1). First, cells detect chemoattractants and determine the location of their source, a process known as directional sensing. Second, cells process this information and transmit it to the cytoskeleton by chemotactic signaling. Third, the cytoskeleton undergoes asymmetric redistribution and activation so that cells acquire a polarized morphology and increase cell motility. Finally, cells perceive spatiotemporal changes in external gradients and adapt to them through feedback mechanisms.
Figure 1.
General organization of chemotactic migration. The four major modules of chemotactic migration are depicted. Directional sensing occurs at the level of chemoattractant (ligand) binding to a chemotactic receptor and transmembrane signal transduction at the level of second messengers. Then, chemotactic signaling transmits information to the cytoskeleton, where it targets three major components of cell motility: protrusions, adhesions and contractility. Finally, feedback mechanisms regulate sensitivity to external cues and mediate adaptive responses to changes in their gradients. See the text for details.
The lower eukaryotic amoeba Dictyostelium is a commonly used model organism for studying chemotaxis (www.dictybase.org).1, 8 It has provided considerable insight into chemotactic behavior, mechanisms of signal reception and transduction inside cells.2 Importantly, these mechanisms tend to be generally conserved in many cell types of higher eukaryotes.9 They are almost identical in amoeba-like neutrophils of higher vertebrates, which helps to explain neutrophil biology and immune function in higher organisms.
Fibroblasts are typically mesenchymal cells. Consistent with physiological function,10, 11 they clearly differ from amoeboid cells in chemotactic and motile behavior.12, 13, 14 However, they similarly perceive and process chemotactic stimuli, and move using common machinery and principles of motility as discussed below.
Here, we compare fibroblasts to amoeboid cells through the four components of chemotaxis depicted in Fig. 1. The major differences are summarized in Table 1 and are detailed further in the text. We conclude that differences in chemotactic signaling and, particularly, in feedback mechanisms are most likely to account for differences in the chemotactic behavior of these cells. Although fibroblasts have been found to lack the feedback loops that are present in amoeboid cells, we suggest that they possess a separate type of feedback that functions in the cytosol, which is why it has not been detected with commonly used membrane probes. We further suggest that hydrogen peroxide may mediate this feedback downstream of chemotactic receptors and activated Rac1.
Table 1.
Major chemotactic differences between amoeboid and mesenchymal cells. GPCRs, G-protein-coupled receptors; RTKs, receptor tyrosine kinases; PLA2, phospholipase A2.
| Parameter | Amoeboid cells | Mesenchymal cells | |
|---|---|---|---|
| Morphology | Size | 5–10 μm | 50–150 μm |
| Native function | Catching bacteria or finding other cells | Reparing connective tissue at the region of wound | |
| Polarization time | 30–60 s | 30–50 min | |
| Speed | 10–20 μm/min | 0.25–1 μm/min | |
| Shape | Non-spread cells, constantly changing shape, usually a single protrusion | Spread cells, fairly constant shape, multiple protrusions | |
| Protrusions | Large relative to cell size: filopodia, lamellipodia and blebs | Local relative to cell size: microspikes, filopodia, lamellipodia, rarely blebs | |
| Adhesiveness | Weak | Strong | |
| Contractility | Weak | Strong | |
| Chemotactic behavior | Starved cells | Not polarized and immobile | Often intrinsically polarized but immobile |
| Uniform stimulation | Not polarized and immobile; stochastic protrusions and random migration possible | Intrinsic cues direct polarization and migration | |
| Gradient stimulation | Rapid polarization and chemotaxis along the gradient | Intrinsic polarization counteracts external gradients | |
| Directional sensing | Simultaneously recognize different chemoattractants in the wide range of concentrations; detect steep and shallow gradients | Recognize single gradients in midpoint concentration of chemoattractant; detect only steep gradients | |
| Changing gradient direction | Fast repolarization and turning | Slow and partial reorientation | |
| Chemotactic signaling | Chemoattractants | Microbial products, pathogen- or damage-associated molecular patterns, components of complement system, cAMP | Growth factors, extracellular matrix proteins |
| Receptors | GPCRs | RTKs | |
| Receptor-associated G-proteins | Trimeric Gi and G12/13, the βγ-complex is important | Small Ras GTPase | |
| Major signaling modules | PI3 kinase/PIP3, PLA2, cGMP/cAMP, mTORC2 | PI3 kinase/PIP3, PLA2, mTORC2, MAP-kinases, Src-family tyrosine kinases | |
| Cytoskeleton regulation | Small GTPases Rac, Cdc42 and Rho | Small GTPases Rac, Cdc42 and Rho | |
| Amplification step | Second messenger level (i.e. PIP3) | ? | |
| Cytoskeleton | Actin microfilaments | Dynamic actin cytoskeleton, no stress fibers | Localized actin dynamics, developed stress fibers |
| Microtubules | ? | Involved in polarization | |
| Intermediate filaments | ? | ? | |
| Feedback | PAK1/PIXα-dependent Cdc42 activation and PIP3-dependent Rac1 activation (ref.4) | ? | |
Chemotactic behavior of mesenchymal cells relative to amoeboid cells
According to the overall morphology and manner of movement, cells are conventionally divided into two general types: amoeboid and mesenchymal cells. Among them, free-living amoeba Dictyostelium, intramural immune neutrophils, and connective tissue fibroblasts are considered to be prototypical cells. They have clearly distinct biology and functions that to a great extent explain their morphology and behavior. Other cells, such as endothelial, epithelial, neural, smooth muscle, primordial germ and progenitor cells, as well various cancer cells, can be broadly classified into one of the two above types by their mode of migration and chemotactic behavior. However, the mesenchymal type of migration is a hallmark of many pathological states associated with aberrant cell migration. Nonetheless, the mechanisms that regulate mesenchymal cell migration are much less known or understood than those of amoeboid cells. This lack of knowledge markedly limits our ability to target these cellular components in disease.
Amoeboid cells, such as fast moving neutrophils and Dictyostelium, are most commonly used in studies of chemotaxis. These cells have similar motile and chemotactic behavior, are highly responsive to chemoattractants and sensitive to changes in their gradients.15, 16 They are very small (10–20 μm) and can persistently move to a source of chemoattractants with high speeds of up to 20 μm/min (i.e., one cell size a minute). Because they constantly change shape, their type of migration is called amoeboid.15, 17
Amoeba Dictyostelium inhabits the soil and relies completely on chemotaxis to fulfill nutritional and reproductive needs.8 It feeds on bacteria, which it chases by chemotaxis. When food runs out, the amoebae begin to differentiate and aggregate into a single fruit body.18 They move up the gradient of cyclic AMP, which they produce and expel at the rear of the cell body to help followers navigate into the assembly. Perhaps the most useful property of the amoeba for researchers is its simple genome, which has been sequenced and used to identify molecular players in chemotactic migration.19
Neutrophils are the model of choice for studying mammalian chemotactic migration.16, 20 Defense is their primary function in an organism. They hunt invading bacteria by chemotaxis and mediate inflammatory responses. Consistently, the major chemoattractants for neutrophils are bacterially derived substances, such as formyl-methionyl-leucyl-phenylalanine (fMLP) and lipopolysaccharide (LPS), various inflammatory cytokines, and components of a complement system, such as С5α.3 Neutrophils distinguish between different chemoattractants to fulfill their tasks. They are directed by several chemotactic gradients, which they recognize in a preferential manner to move interstitially for long distances.21 Additionally, both neutrophils and amoebae have to react promptly to changes in prey location. They both possess an effective adaptive system, which provides a capacity for rapid responses in speed and direction of movement.3
In the absence of stimuli, the amoeboid Dictyostelium and neutrophil cells have a rounded morphology and remain stationary.22 When stimulated in a uniform manner (i.e., the entire membrane is equally accessible to chemoattractant), the cells start growing protrusions stochastically throughout the plasma membrane. Despite being activated, the cells do not move because they cannot temporally link successive protrusions at a single site on the membrane, thus the cell body is not displaced in a single direction. Occasionally, random displacement does occur due to a predisposition of new protrusions to grow at the base of previous ones, a phenomenon known as correlated random walk.23, 24
When a chemoattractant comes from a particular source (i.e., in a gradient fashion), successive protrusions develop on the side of the membrane facing the stimulus. They are biased to this side through local upregulation of chemotactic signaling and activation of the protrusive machinery. As a result, cells persistently move in one direction with increased speed. If the gradient changes direction, cells respond by either biasing new protrusions to this new direction and turning gradually or by growing a new protrusion and turning abruptly. The choice depends on the strength of the external gradient, such that shallow gradients (i.e., weak stimulation) cannot effectively counteract polarization and only induce gradual changes toward the new direction. In contrast, steep gradients are sufficiently strong to repolarize the cytoskeleton and induce new formations.23, 24
Polarization is the key event in directional motility. It supports continuous growth of pseudopods at the cell front and localizes contractile actomyosin bundles to the rear. Whereas the cytoskeleton of unstimulated Dictyostelium and neutrophil cells is inconspicuous and prone to remodeling, the cytoskeleton of fibroblasts is prominent and developed.14 Thus, amoeboid cells have much more plasticity than fibroblasts. Shallow chemotactic gradients induce marked polarization in amoeboid cells, which can easily repolarize if a gradient changes direction. This feature allows immune leukocytes or free-living amoebae to travel long distances and gather precisely at the source of a pathogen or pheromone.
Fibroblasts are much larger and slower cells.25 They move in a manner clearly distinct from amoeboid cells, but in a manner that is characteristic of most mesenchymal cells, which has been referred to as the mesenchymal type of migration.16, 26 Fibroblasts are highly adhesive and spread 5-10 times larger than neutrophils. Their typical speeds are approximately 0.25–1 μm/min,16, 27 which means that they require an hour to cover a distance of the cell length. This brings into question what mechanisms these cells use to maintain their direction of movement.
Chemotactic behavior and regulation in fibroblasts is consistent with their functions, which are distinct from amoeboid cells. Fibroblasts act to maintain connective tissue homeostasis and mediate wound healing.28 In a homeostatic state at low or no stimulation, fibroblasts remain randomly polarized and move slowly while directed by proteins of the extracellular matrix (a process known as haptotaxis). When undergoing wound healing, fibroblasts move into the wound by chemotaxis and mediate repair and matrix remodeling. When in the wound, fibroblasts are activated by a high absolute concentration of PDGF. As a result, cells switch from persistent migration to proliferation. Their migration is directed by chemoattractants, primarily platelet-derived growth factor (PDGF), which is released upon damage and inflammation.28 Platelets, which aggregate in wounded areas, produce extremely steep gradients of PDGF. Fibroblasts do not need to locate the source of PDGF precisely; they just migrate in the general direction of the wound while producing matrix proteins. Once in the wound, fibroblasts are exposed to a shallow-gradient, high-concentration of PDGF (greater than 5 ng/ml). They cannot effectively navigate in such conditions and switch from migration to proliferation.29 Overall, this behavior is consistent with the initial recruitment of these cells to wounded areas, followed by an increase in the number of cells, to accelerate wound healing.10
Additionally, fibroblasts are highly adhesive and sensitive to protein composition and extracellular matrix rigidity. They easily respond to the matrix by localized changes in integrin-mediated signaling and cytoskeleton reorganization.30 When stabilized, these differences become prominent between distant parts of these large, flat cells. In this manner, internal asymmetry of the cytoskeleton and overall cell polarity is established even in the absence of chemoattractants. Together with other internal factors (see Ref. 31), these mechanisms determine the intrinsic polarity of fibroblasts.
In contrast to amoeboid cells, fibroblasts lack certain mechanisms that help cell behavior adapt to altered stimulation. They lack feedback circuits that are used by amoeboid cells to amplify weak chemotactic signals,32 which helps to explain why fibroblasts are poorly sensitive to shallow chemotactic gradients. In addition, they only respond by chemotaxis to a certain range of chemoattractant concentrations, indicating that they adapt poorly to altered stimulation. Due to these limitations, fibroblasts are often regarded as a stripped-down chemotactic system.2
However, fibroblasts possess unique regulatory mechanisms for migration and, perhaps, chemotaxis. Directional migration of fibroblasts requires synthesis of β-actin at the cell front.33 They can also locally sharpen PDGF gradients at the cell front through receptor-dependent endocytosis.34 The internalized receptors continue to signal in endosomes, leading to activation of Rac1-induced actin dynamics at the cell front.35 Given such behavior, adjacent fibroblasts move independently and randomly, yet persistently as long as the self-induced gradient of PDGF is maintained. Importantly, this intrinsic propensity of fibroblasts to develop and keep polarity, which is absent in amoeboid cells, contributes significantly to their ability to maintain direction and migrate persistently.36, 37 Because of this property, fibroblasts typically display an elongated appearance with a broad leading and narrow rear lamellae, even in the absence of stimulation.12, 14 Shallow gradients are not sufficient to counteract intrinsic polarity and repolarize fibroblasts.32 Only steep gradients support persistent migration along a polarity axis, otherwise, fibroblasts repolarize and change direction in response to adhesion to the extracellular matrix.
Thus, fibroblasts should not be considered as merely simplified amoeboid chemotactic systems. They have different functions and therefore different chemotactic behaviors. These differences are both morphological, based on distinct cytoskeletal organization, and functional due to specific differences in intracellular signaling. The following sections discuss the idea that signaling components play a leading role in these differences.
Cell motility
Morphologically, cell movement is commonly described by a four-step migration cycle, which has been extensively reviewed elsewhere.4, 26, 38, 39 Briefly, it involves leading protrusions, their attachment and adhesion formation, translocation of the cell body, and rear detachment associated with tail retraction. Remodeling of the actin cytoskeleton is the driving force for all types of motility, including that of mesenchymal and amoeboid cells. The tubulin (microtubule) cytoskeleton plays a lesser role in motility; however, it is involved in setting up polarized morphology.40
Protrusive activity at the cell front is determined by increased actin dynamics via formation of a branched actin filament meshwork.41, 42 Translocation of the cell body and tail retraction are predominantly mediated by the contractile activity of internal stress fibers composed of actin and myosin.43, 44 Substrate adhesions couple these two compartments and serve as mechanosensitive devices.45 When engaged with matrix proteins, these substrates bind actin and promote actomyosin assembly and contractility, which is then converted to traction. When disengaged, the actin meshwork grows in the leading lamella.46 Basically, cells coordinate protrusion and traction by alternating the two events. In turn, substrate adhesions are reinforced by contraction of the attached stress fibers.47 Thus, a positive feedback circuit is involved in adhesion-dependent motility: the more traction force that is generated by the stress fibers, the harder the grip of the substrate adhesions.37
This general model has been developed to describe single cell migration on flat 2D-surfaces. Soon thereafter, it was modified to fit within the 3D-context of physiological milieu48 and further refined.49 While a few specialized features have been added, such as integrin-independent migration, contact guidance, cellular strategies to overcome extracellular matrix resistance, and multicellular migration, the basic four-step concept holds for single cells. Several modes of migration have been defined, with the amoeboid and mesenchymal serving as the two basic types. However, a new paradigm has emerged that suggests a balance between protrusive, adhesive and contractile activities determines the mechanical mode of cell migration.49, 50
When protrusive activity dominates, single cells move effectively by extending leading protrusions and dragging the tail. Increased actin dynamics are sufficient to power protrusions and, in extreme circumstances, traction.43 When the contractile components predominate, cells use a ‘blebbing’ mode of motility.51 They first extrude cytoplasmic fluid in the form of membrane bulges (blebs), which are powered by hydrostatic pressure resulted from cortical actomyosin contraction. Subsequently, the blebs fill with the actin machinery and cytoplasm as a result of rear contraction. Because contractility and adhesion are intimately coupled,37, 45 blebbing motility can also be viewed as the adhesive mode of 2D-migration. However, in 3D-lattices, many cells move in an integrin-independent fashion purely by contraction-assisted blebbing motility.52, 53 In this case, compressed cells exert perpendicular forces to the surrounding matrix and squeeze themselves forward.51
According to the current multiparameter tuning model, each cell type uses a particular ‘default’ mode of migration but adapts to environmental cues by switching to other modes when most apt for their circumstances.49, 53 While amoeboid cells mostly rely on protrusive and blebbing modes, fibroblasts are adhesive and contractile. To move efficiently, they undergo a mesenchymal-to-amoeboid transition and then employ an amoeboid mode of motility.49 This indicates that the composition and function of the motile machinery is essentially the same for most amoeboid and fibroblasts cells; however, it manifests differently depending on the surrounding conditions.
Directional sensing
The current concept of the mechanism by which cells translate chemotactic signals into motile responses is that the external gradients first coax into internal gradients of signaling molecules, a process known as directional sensing.2, 5 These signaling molecules act as second messengers to activate downstream signaling pathways, thereby leading to protrusions, adhesions and contractions, which result in the asymmetric redistribution of the cytoskeleton and is known as polarization (Fig. 1).
Two quantitative parameters characterize spatial gradient sensing.16 The first is sensitivity to the relative steepness of the gradient or the degree of its slope. It is usually given as a percent difference in the concentration of chemoattractant across a cell. Expectedly, these values are much smaller for amoeboid cells than for fibroblasts due to cell size differences. Gradients are often called steep or shallow to qualitatively define the inclination. The second parameter is the average (midpoint) absolute concentration of a chemoattractant across a cell. This value depends on, and changes with, the distance of the cell from the chemoattractant source.
Chemotactic receptors are evenly distributed on the cell surface of Dictyostelium54 and neutrophils,55 displaying little or no preference for a leading membrane. Although less data are available for fibroblasts,4 they apparently display a similar trend. Further studies have demonstrated that G-proteins, the immediate targets of chemotactic receptors, also have a distribution that matches the slope of external gradients.56, 57 These observations led to the conclusion that activation, but not clusterization, of receptors and their downstream targets set up internal gradients of signaling molecules. This is consistent with the chemotactic behavior of fibroblasts, which are not sensitive to shallow gradients. Rather, they respond to steep gradients in the narrow range of midpoint concentrations of chemoattractant.32 Low concentrations are insufficient to activate chemotactic signaling above a threshold level, whereas high concentrations activate signaling across the entire membrane. Thus, fibroblasts perceive information based on absolute concentrations of chemoattractant rather than on the degree of the gradient slope.
By contrast, amoeboid cells discern the steepness of gradients regardless of the midpoint value of the absolute concentration of chemoattractant. They are equally effective in navigating in steep and shallow gradients both close and far from their source. Accordingly, these cells must possess at least two specialized mechanisms that enable this type of sensing and behavior. One mechanism is ‘thresholding’, which is the ability to subtract the minimum level of signal induced in an exposed area by the lowest absolute concentration of chemoattractant. This can potentially be achieved via desensitization and downregulation of chemotactic receptors. The other mechanism is ‘amplification’, which addresses shallow gradient sensing. Small amoeboid cells experience little difference in the concentration of chemoattractant across the length of a cell. Thus, they must be amplified to achieve steep internal gradients. As discussed above, this amplification is achieved at the level of second messengers, downstream of receptors and their immediate targets (see Fig. 1).
Different types of receptors mediate chemotaxis of amoeboid and mesenchymal cells. Receptor tyrosine kinases (RTKs) are used to sense gradients of growth factors, which are the typical chemoattractants for fibroblasts.58 Among them, PDGF is the chief chemoattractant. Downstream signaling from PDGF receptors involves tyrosine phosphorylation, binding of modular adaptor proteins and activation of the monomeric G-proteins Ras, Rac, Rho and Cdc42 (see Ref. 4 and references therein). Ras GTPase functions as a primary hub to set| up chemotactic signaling at the membrane. Rac and Cdc42 act at the cell front to initiate membrane protrusions via regulated actin polymerization and dynamics.6 Rho GTPase functions in the rear to regulate myosin filament assembly and contractility. The reciprocity of Rac and Rho distribution and activation is essential for polarization and chemotaxis.14, 39
Amoeboid cells employ trimeric G-protein-coupled receptors (GPCRs) for chemotaxis. Although they differ in subunit composition between lower and higher eukaryotes,59 major chemotactic signaling occurs via the βγ-complex.57 Strikingly, both GPCRs and RTKs use the same set of signaling pathways to stimulate chemotaxis. These pathways similarly target small GTPases Ras, Rac, Rho and Cdc42, leading to polarization and activation of the cytoskeleton.39
Numerous studies have identified PI3-kinase signaling as the major chemotactic cascade in both amoeboid and mesenchymal cells (reviewed by Schneider and Haugh16 and Vorotnikov4). The pathway is activated directly by chemotactic receptors, with the participation of their immediate G-protein targets: the βγ-subunits of GPCR and Ras GTPase for RTK. Although different PI3-kinase isoforms are involved, they all generate a common PIP3 lipid that acts as a universal secondary messenger in chemotaxis. Locally elevated production of PIP3 at the site of the membrane facing the gradient is essential to establishing its internal gradient and cell polarity.
Live biosensor technology, especially the use of genetically modified fluorescent proteins,60 has been instrumental for imaging the dynamics of critical molecules inside cells. Among these, the activation dynamics of the Rho-family GTPases have been visualized to determine their function in directional migration (reviewed by Pertz61). Similarly, the intracellular dynamics of PIP3 have been widely studied by translocation-based biosensors composed of a fluorescent protein fused to isolated pleckstrin-homology (PH) domains specific for PIP3 binding (see Ref. 4 for references). These studies have unequivocally demonstrated that internal PIP3 gradients are significantly amplified in amoeboid cells compared to external chemotactic gradients. In contrast, this amplification has not been observed in fibroblasts.13 The final conclusion that it is absent has been made on the basis of chemotactic behavior modeling of fibroblasts.32 These results are consistent with experimental data obtained by imaging of PIP3 gradients in fibroblasts exposed to external gradients of varying steepness and magnitude.
Notably, this fundamental difference becomes clearly detectable at the second messenger level (see Fig. 1). Upstream signaling is well conserved and shared by GPCRs in amoeboid cells and RTKs in fibroblasts. Therefore, logic dictates that amplified levels of PIP3 are achieved by specific mechanisms that are present in amoeboid cells but absent in fibroblasts, either at or downstream from the second messenger level. These mechanisms are thought to involve specific redistribution of second messengers known as LEGI, and downstream feedback circuits, respectively.
The local excitation and global inhibition (LEGI) model has been suggested to explain directional sensing by small amoeboid cells in which shallow external gradients produce sharp excitation of internal gradients.2, 5 In addition, LEGI aims to explain the ‘thresholding’ phenomenon that renders amoeboid cells independent from the midpoint concentration of chemoattractants. It assumes that directed migration becomes persistent when protrusive activity is stimulated at the cell front and suppressed in the rear and lateral areas of the cell. According to this hypothesis, chemoattractants activate two types of signals. Strong activator molecules are produced locally by self-enhancing regional excitation. They diffuse slowly and accumulate in regions facing higher concentrations of chemoattractant. A lipid second messenger, such as PIP3, perfectly meets these criteria because it is confined to the membrane compartment and is boosted by escalating enzyme-catalyzed reactions in the proximity of activated chemotactic receptors. In contrast, the signal of second type is weak and inhibitory. It spreads out rapidly and acts at long range to cut off the activating signal everywhere in the cell except for the leading membrane. Soluble second messengers, such as cyclic nucleotides, are ideal for this purpose because they are produced in a receptor-dependent manner and quickly distribute throughout the cytoplasm. Thus, superposition of these two signals provides the required ‘thresholding’ of internal chemotactic signaling. Thus restricted to the cell front, the rest of the activating signal is subsequently amplified to yield a sharp internal gradient.
It is commonly accepted that PIP3 and cyclic nucleotides play a key role in chemotaxis of most eukaryotic cells. PI3-kinases that produce PIP3 are recruited to and activated by chemotactic receptors.62 PIP3 demonstrates sharp internal gradients in directionally migrating amoeboid cells55, 63 and fibroblasts.13 PIP3 acts as a local activator molecule that recruits to the membrane proteins that contain PIP3-binding PH-domains. They include most of guanine nucleotide exchange factors (GEFs) for Rac GTPase,64 which trigger local activation of Rac and actin dynamics. On the other hand, cyclic GMP is produced downstream of activated chemotactic receptors in Dictyostelium,65 and cyclic AMP is produced in neutrophils.66 The cyclic nucleotides seem to distribute diffusely throughout the cytoplasm and exert inhibitory functions.67 Cyclic GMP inhibits lateral protrusions in Dictyostelium,65 whereas cyclic AMP inhibits Rho-dependent activation of type II myosin and contractility in the cytoplasm.67 This is consistent with the LEGI mechanism, with PIP3 functioning as the activating signal and cyclic nucleotides as the inhibitory signal. However, other signaling molecules cannot be excluded.
Whereas the LEGI model accounts for most of the chemotactic behavior displayed by amoeboid cells, it has limitations. For example, it poorly explains internal gradient amplification and the stabilizing effect of cytoskeletal polarization as well as fibroblast chemotaxis. Thus, feedback loops and adaptive mechanisms need to be added to improve functionality. Some of these loops seem to be absent in fibroblasts, while others are less active. On the other hand, fibroblasts have much longer movement dynamics. To stabilize protrusions to maintain persistent migration, chemotactic signaling needs to have longer kinetics. Protrusions in fibroblasts appear and disappear stochastically unless stabilized by PIP3.68 This suggests that PIP3 levels must remain elevated longer in fibroblasts compared to amoeboid cells and that the LEGI mechanism may differ in fibroblasts to account for these temporal needs. Whether specific feedback mechanisms can fulfill this goal is discussed in the final section.
Chemotactic signaling
Signaling pathways that regulate chemotaxis have recently been extensively reviewed2, 4, 69 and will not be detailed here. In brief, those identified so far include the PI3-kinase/PIP3 module, phospholipase A2 (PLA2), guanylate/adenylate cyclases and the cyclic nucleotide module, mechanistic target of rapamycin complexes (mTORC1/2), mitogen-activated protein (MAP) kinases, and the non-receptor Src family of tyrosine kinases. Their relative importance for chemotaxis is compared below.
The common characteristics of chemotactic signaling are outlined in Fig. 1. Its major objective is to transmit external signals picked up by chemotactic receptors to the cytoskeleton to activate three major motility components (protrusion, adhesion, contraction) and initiate polarization. When comparing chemotactic signaling across different cell types, three specific aspects have to be emphasized.
First, chemotactic signaling is organized in two consecutive areas in all cells. The common part is composed of the cell surface receptors and associated G-proteins, either trimeric for GPCRs or monomeric Ras GTPase for RTKs. This enables transmembrane signal transduction and results in rapid accumulation of activating and inhibitory second messengers such as PIP3 lipids, cyclic nucleotides, Ca2+, diacylglycerol, and others. Thereafter, chemotactic signaling branches towards three major motility components (Fig. 1). These branches signal Rho-family GTPases, the master regulators of cytoskeletal dynamics,39 which become asymmetrically distributed and activated along the direction of the movement. Specifically, Cdc42 cooperates with microtubules at cell tips to establish polarity; when Cdc42 is defective, the directionality of migration is disrupted. Rac stimulates actin dynamics in the leading lamella, upregulating protrusive activity. Rho stimulates myosin II assembly and activity, upregulating adhesions and contractility.
Second, the same principal chemotactic pathways are shared by both types of cells. Inasmuch as the balance between protrusive, adhesive and contractile activities determines the mode of migration chosen by a certain cell type,48, 49, 50 chemotactic signaling aims to target a particular component of motility. Thus, amoeboid cells that move by protrusive motility would preferentially engage Rac-mediated actin dynamics at the cell front. For this, the PI3-kinase pathway is imperative. It provides local accumulation of PIP3, recruitment of Rac GEFs, and Rac activation.62, 64 The blebbing motility of amoeboid cells is mostly supported by contractility; therefore, signaling to actomyosin bundles in the cell body and rear would predominate. The guanylate cyclase/cyclic GMP system is responsible for regulation of myosin II in Dictyostelium,70 while Rho GTPase and Rho-activated kinase regulate myosin II in higher eukaryotes.39 Cyclic AMP is also involved in setting up cell polarity by acting via Epac proteins and Rap1 GTPase at the cell front and via Rho-mediated effects on myosin in the rear.66 Finally, adhesion component that is important for fibroblast migration is targeted by MAP-kinase pathways71 and Src-family tyrosine kinases, including focal adhesion kinase (FAK), Lyn, Fyn, Lck, and others.72
Third, another important aspect of chemotactic signaling is that all signaling pathways are highly redundant. This feature has been well illustrated with Dictyostelium as a chemotactic model (see Ref. 73 and references therein). Even the PI3-kinase/PIP3 module that is often regarded as critical for chemotaxis is dispensable in certain situations.74 Thus, switching off a few, but not all, chemotactic pathways at once may not fully disrupt chemotaxis.75 This indicates that chemotactic pathways can at least partially substitute for one another, which is consistent with the plasticity of cell migration.49, 50 Assuming that the motility components (protrusion, adhesion and contractility) are interconnected, disruption of signaling pathway(s) that activate one component is likely to cause a cell to alter its mode of motility and use another motility component to continue migration. Accordingly, the cell engages a redundant signaling pathway to activate the alternative component.
Finally, the mTORC pathway requires discussion as it has recently emerged as a critical regulator of cell migration (reviewed by Zhou and Huang76). mTOR is a ubiquitous kinase involved in the regulation of cell growth, survival and metabolism.77 It functions in two distinct signaling complexes, mTORC1 and mTORC2, which contain several partially overlapping accessory and regulatory components. Compromising the activity of any part of this complex seriously affects cell migration, and the underlying molecular mechanisms are just beginning to be discovered. In Dictyostelium78 and neutrophils,67 mTORC2 is required for directional migration; it functions via multiple pathways that are mediated by protein kinase B/Akt, cyclic AMP and Rho GTPase. Particularly, it seems to upregulate Rho activity by activating Rho GEFs76; therefore, mTORC2 is a potential regulator of cell contractility and adhesion. In fibroblasts, mTORC2 also upregulates matrix metalloproteinases and remodeling of the extracellular matrix,76 which is specifically required for interstitial migration of these cells.48
Thus, PI3-kinase/PIP3 is the major, but not unique, signaling pathway to regulate chemotaxis. Other signaling pathways exist and function in a redundant fashion. Overall, a great deal of similarity exists between amoeboid cells and fibroblasts. Whereas some signaling pathway specificity for a particular cell type may exist, it is unlikely to fully account for different chemotactic behavior in these cells. Rather, the activity of these pathways are coordinately balanced and translated into activities of the protrusive, adhesive and contractile components of motility as required for the effective migration of a given cell type.
Feedback and adaptation
Growing evidence suggests that fundamental differences in the chemotactic behavior of amoeboid cells and fibroblasts are due to feedback mechanisms that these cells employ to sharpen internal gradients, modulate signaling kinetics and adapt behavior to changes in external cues. These feedback circuits are designed to resolve major issues experienced while performing physiological functions. Amoeboid cells have to travel long distances, perceive shallow gradients independent of the midpoint concentration of chemoattractant, and respond effectively to changes in external cues. Their small size allows for the rapid delivery of second messengers to cell compartments but causes spatial problems in segregating the activating and inhibitory signals. This problem is addressed by sharpening internal gradients against external ones with the use of ‘threshold’ and ‘amplify’ functions at the second messenger level. While the LEGI system is thought to provide for ‘thresholding’ by an as yet unclear mechanism, two amplification feedback loops have been found in Dictyostelium and neutrophil cells (see Ref. 4 for details). The first involves the activation of Cdc42, while the second involves activation of Rac GTPase. They are responsible for a biphasic increase in actin polymerization and an actin-dependent boost of PIP3 production at the cell front, resulting in amplification of internal PIP3 gradients.
In contrast, fibroblasts move relatively short distances and are vaguely navigated by stable and steep gradients of chemoattractants that emanate from wounds. Due to their elongated shape, fibroblasts experience large differences in chemoattractant concentration over the length of the cell and have little problem segregating internal activating and inhibitory signals. For this reason, they lack the amplification feedback loops found in amoeboid cells. Instead, fibroblasts have temporal problems due to slow protrusion dynamics. To persistently move in one direction, they need to keep the growth of successive protrusions on one side of the cell.
Cells move by large protrusions (aka pseudopods); their formation obeys the principles of self-organizing systems.79 Studies on Dictyostelium have demonstrated that in the absence of chemotactic gradients, cells use correlated random walk (i.e., they grow protrusions that tend to arise close to previous ones).80 Pseudopods live through a certain ‘life’ cycle of extension, steadiness and retraction. Chemotactic gradients affect the spatial characteristics of pseudopods, but not their size, frequency, or duration of growth.24 Thus, external gradients act to ‘link’ successive pseudopods and bias their location toward the gradient. In Dictyostelium, the average growth time and pseudopod interval (period between the start times of two successive pseudopods) are as short as 13 and 15 s, respectively.80 This time frame requires that chemotactic signaling is adequately reactive and responsive to changes in receptor occupancy and gradient direction, which is necessary for effective adaptation responses. Most signaling cascades, including PI3-kinase, Src and MAP-kinase pathways, have a 2–4 min delay in peak activation as assessed by their target readout after in vitro stimulation. This suggests that the behavior of pseudopods is faster than signaling responses in amoeboid cells, which may help to explain the fast adaptive responses in these cells.
By contrast, fibroblasts display a slow behavior of protrusions that are responsible for cell body displacement. These intervals are as long as 40–60 min,14 which is more than two orders of magnitude longer than in amoeboid cells. This suggests that chemotactic signaling must be maintained long enough to ‘link’ two successive protrusions and to enable the correlated random walk. These temporal considerations become even more demanding for weakly stimulated fibroblasts migrating in shallow gradients.
Recent studies using live imaging of PIP3 in randomly moving fibroblasts demonstrated relatively short-lived stochastic hot spots of PI3-kinase activity that were enriched in regions of membrane protrusion and correlated with the direction and persistence of fibroblast movement.68 Furthermore, distant hot spots were found to be dynamically and stochastically coupled, which suggests that fibroblasts somehow convert rapid signaling events into long-term directional migration.81 Although the molecular details of this conversion are presently unknown, a few issues are worth noting. First, similar to amoeboid cells, a feedback or related circuit loop may maintain the activity of chemotactic signaling at the cell front. Second, this is likely to take place downstream of the second messenger level (Fig. 1) because it is not detected by PIP3 sensors.13, 68 Third, such a circuit may not result in signal amplification, but rather resolve the temporal problem of ‘linking’ successive protrusions. Finally, it has to be local and confined to regions of receptor activity, thus avoiding the need for a ‘thresholding’ system that is apparently absent in fibroblasts.
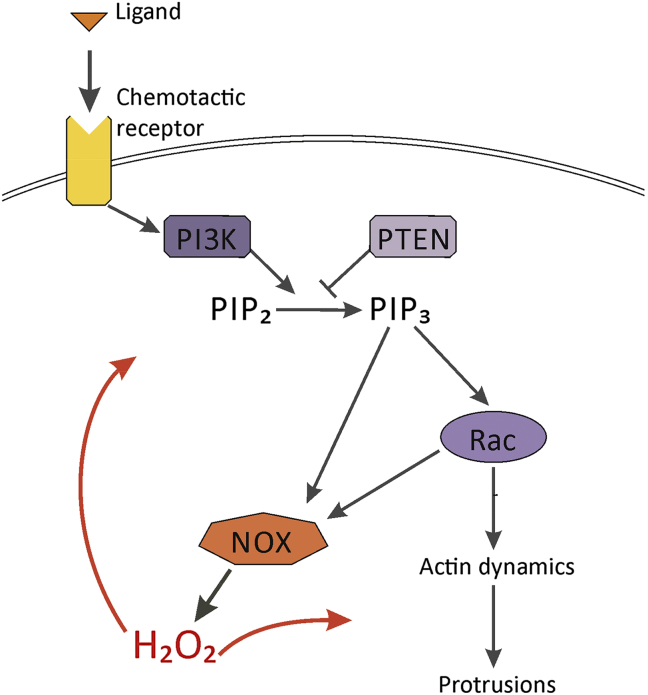
Growing evidence suggests that hydrogen peroxide (Н2О2) acts as a second messenger in signaling downstream of receptor tyrosine kinases.82, 83 It plays a critical role in cell proliferation,84, 85 differentiation,86 apoptosis,87 and, importantly, cell migration.82, 88, 89, 90 We hypothesize that Н2О2 may be a missing link in the feedback circuits of fibroblasts (Fig. 2).
Figure 2.
Proposed feedback loop mediated by hydrogen peroxide (H2O2). Shown is the fragment of PDGF signaling that is mediated by PI3-kinase and PTEN and leads to PIP3 production. In addition to conventional chemotactic pathway to Rac GTPase and actin dynamics, PIP3 activates NOX assembly on the plasma membrane both directly and via Rac. NOX produces a superoxide anion radical, which is further converted to H2O2 as the final metabolite. H2O2 mediates potential feedback loops to the PDGF receptor, PTEN and cytoskeletal proteins, leading to maintained PDGF signaling and sustained migration. See the text for details.
Using zebrafish as an animal model for chemotaxis, Niethammer et al demonstrated that Н2О2 forms tissue-scale gradients around acute wounds to recruit leukocytes.91 Blocking Н2О2 production significantly delayed leukocyte appearance in wounds. A follow-up study by Yoo et al identified the non-receptor tyrosine kinase Lyn as an intracellular target of Н2О2 in leukocytes.92 These studies demonstrate that Н2О2 is involved in the chemotaxis of amoeboid cells.
The intracellular mechanism of Н2О2 is thought to involve the reversible oxidation of cysteine residues in the active centers of a limited number of signaling enzymes.93 Н2О2 is produced in cells in response to physiological stimulation of RTKs (Fig. 2), which is in contrast to the non-physiological oxidative effects of high Н2О2 levels in inflammation and disease.94
Н2О2 accumulates in PDGF-stimulated fibroblasts.89, 95 In addition to the activation of conventional signaling pathways, PDGF triggers the assembly of NADPH-oxidase (NOX) complexes on the plasma membrane. Notably, this activation independently requires Rac GTPase, PIP3 and/or protein kinase C,87 all of which are critical for fibroblast chemotaxis.58 The major function of NOX is to produce superoxide anion radical О2•−. It is a short-lived precursor of other reactive oxygen species that all metabolize into relatively stable Н2О2.96
Tyrosine phosphatases are well known physiological targets of Н2О2 in cells because they contain an appropriate reactive cysteine residue in their active center.93, 97 Among them, tyrosine phosphatase РТР-1В is the best characterized.98 РТР-1В dephosphorylates and inactivates the PDGF receptor as its major substrate. When oxidized by Н2О2, РТР-1В is transiently inactivated and PDGF signaling is maintained. Lipid phosphatase PTEN, which dephosphorylates and inactivates PIP3 (Fig. 2), is another target of Н2О2.99 Similar to РТР-1В, it is transiently inactivated when oxidized by Н2О2. In this case, PIP3 levels are upregulated. Thus, Н2О2-mediated inactivation of PTP-1B and PTEN downstream of activated PDGF receptors may potentially maintain chemotactic signaling by PDGF and PIP3 in fibroblasts.
In addition to the inhibition of tyrosine phosphatases, Н2О2 has been shown to activate non-receptor tyrosine kinases. In addition to the Lyn kinase mentioned above, Н2О2 also activates Src.100 Given that Src is a critical regulator of focal adhesions and that fibroblasts actively use the adhesive component of motility, targeting Src by Н2О2 may specifically contribute to a mesenchymal vs. amoeboid type of migration.
Finally, evidence demonstrates that Н2О2 may act further downstream at the cytoskeletal level (Fig. 2). It has been suggested that cofilin, an early activator of actin dynamics, is activated via Н2О2-mediated release of Slingshot-1L phosphatase from its complex with the 14-3-3 scaffold protein.101 Two groups have also reported different mechanisms of redox regulation that affect cysteine oxidation and subsequent actin assembly.102, 103
These observations lead us to hypothesize that Н2О2 may act as an unrecognized activating secondary messenger in the LEGI model. According to the LEGI concept, the activating signal must be highly localized and limited in diffusion.2, 5 Consistent with these requirements, we have recently shown that Н2О2 is produced in the cytoplasm and restricted to the endocytic compartment.95 This controlled and localized production potentially allows Н2О2 to escape degradation and toxic intracellular effects. Whether intracellular Н2О2 contributes to LEGI and chemotaxis remains to be studied.
Conclusions and future directions
The directional migration of mesenchymal cells is a potential target in a variety of diseases. It is involved in both physiological and pathological angiogenesis during cancer progression. Cancer metastasis and dissemination involves the epithelial-to-mesenchymal transition of cancer cells and their intra- and extravasation by directional migration. Wound healing undoubtedly requires fibroblast chemotaxis, but hyperactivity of fibroblasts impairs regeneration and results in scar formation. Regeneration of damaged areas also depends on the effective recruitment of mesenchymal precursor cells, which occurs via their directional migration. Thus, an approach is needed to selectively target mesenchymal vs. amoeboid cell chemotaxis.
Despite being clearly different from amoeboid cells, mesenchymal cell chemotaxis seems to use similar strategies and common principles of directional sensing, chemotactic signaling and cell motility. Therefore, parallels can be drawn to identify key participants as potential therapeutic targets. We further reason that notable differences appear at the level of feedback mechanisms, which are specifically suited for the biology and physiological performance of a given cell type. Identifying molecular players in these mechanisms is a prerequisite for the future development of means to selectively target mesenchymal cell chemotaxis in disease.
Conflicts of interest
All authors have none to declare.
Acknowledgments
This work was supported, in part, by the Russian Foundation for Basic Research grant 14-04-01746 (feedback, adaptation, and directional sensing) and the Russian Scientific Foundation grant 14-15-00439 (other sections).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.King J.S., Insall R.H. Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol. Oct 2009;19:523–530. doi: 10.1016/j.tcb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Swaney K.F., Huang C.H., Devreotes P.N. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. Jun 9 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Philipsborn A., Bastmeyer M. Mechanisms of gradient detection: a comparison of axon pathfinding with eukaryotic cell migration. Int Rev Cytol. 2007;263:1–62. doi: 10.1016/S0074-7696(07)63001-0. [DOI] [PubMed] [Google Scholar]
- 4.Vorotnikov A.V. Chemotaxis: movement, direction, control. Biochemistry (Mosc) Dec 2011;76:1528–1555. doi: 10.1134/S0006297911130104. [DOI] [PubMed] [Google Scholar]
- 5.Xiong Y., Huang C.H., Iglesias P.A., Devreotes P.N. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci U S A. Oct 5 2010;107:17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pankov R., Endo Y., Even-Ram S. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol. Aug 29 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X., Bruzewicz D.A., Wong A.P., Piel M., Whitesides G.M. Directing cell migration with asymmetric micropatterns. Proc Natl Acad Sci U S A. Jan 25 2005;102:975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Annesley S.J., Fisher P.R. Dictyostelium discoideum – a model for many reasons. Mol Cell Biochem. Sep 2009;329:73–91. doi: 10.1007/s11010-009-0111-8. [DOI] [PubMed] [Google Scholar]
- 9.Drayer A.L., van Haastert P.J. Transmembrane signalling in eukaryotes: a comparison between higher and lower eukaryotes. Plant Mol Biol. Dec 1994;26:1239–1270. doi: 10.1007/BF00016473. [DOI] [PubMed] [Google Scholar]
- 10.Shaw T.J., Martin P. Wound repair at a glance. J Cell Sci. Sep 15 2009;122:3209–3213. doi: 10.1242/jcs.031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irie H.Y., Pearline R.V., Grueneberg D. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. Dec 19 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abercrombie M. The crawling movement of metazoan cells. The Croonian lecture 1978. Proc R Soc Lond B Biol Sci. 1980;207:129–147. [Google Scholar]
- 13.Haugh J.M., Codazzi F., Teruel M., Meyer T. Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J Cell Biol. Dec 11 2000;151:1269–1280. doi: 10.1083/jcb.151.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Small J.V., Anderson K., Rottner K. Actin and the coordination of protrusion, attachment and retraction in cell crawling. Biosci Rep. Oct 1996;16:351–368. doi: 10.1007/BF01207261. [DOI] [PubMed] [Google Scholar]
- 15.Devreotes P.N., Zigmond S.H. Chemotaxis in eukaryotic cells: a focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 16.Schneider I.C., Haugh J.M. Mechanisms of gradient sensing and chemotaxis: conserved pathways, diverse regulation. Cell Cycle. Jun 2006;5:1130–1134. doi: 10.4161/cc.5.11.2770. [DOI] [PubMed] [Google Scholar]
- 17.De Bruyn P.P. The amoeboid movement of the mammalian leukocyte in tissue culture. Anat Rec. Jun 1946;95:177–191. doi: 10.1002/ar.1090950209. [DOI] [PubMed] [Google Scholar]
- 18.Weijer C.J. Dictyostelium morphogenesis. Curr Opin Genet Dev. Aug 2004;14:392–398. doi: 10.1016/j.gde.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Chisholm R.L., Gaudet P., Just E.M. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. Jan 1 2006;34:D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niggli V. Signaling to migration in neutrophils: importance of localized pathways. Int J Biochem Cell Biol. Dec 2003;35:1619–1638. doi: 10.1016/s1357-2725(03)00144-4. [DOI] [PubMed] [Google Scholar]
- 21.Foxman E.F., Kunkel E.J., Butcher E.C. Integrating conflicting chemotactic signals. The role of memory in leukocyte navigation. J Cell Biol. Nov 1 1999;147:577–588. doi: 10.1083/jcb.147.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zigmond S.H. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. Nov 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrew N., Insall R.H. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Biol. Feb 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 24.Bosgraaf L., Van Haastert P.J. Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PLoS One. 2009;4:e6842. doi: 10.1371/journal.pone.0006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puck T.T., Cieciura S.J., Fisher H.W. Clonal growth in vitro of human cells with fibroblastic morphology; comparison of growth and genetic characteristics of single epithelioid and fibroblast-like cells from a variety of human organs. J Exp Med. Jul 1 1957;106:145–158. doi: 10.1084/jem.106.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauffenburger D.A., Horwitz A.F. Cell migration: a physically integrated molecular process. Cell. Feb 9 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 27.Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr Opin Cell Biol. Feb 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Deuel T.F., Kawahara R.S., Mustoe T.A., Pierce A.F. Growth factors and wound healing: platelet-derived growth factor as a model cytokine. Annu Rev Med. 1991;42:567–584. doi: 10.1146/annurev.me.42.020191.003031. [DOI] [PubMed] [Google Scholar]
- 29.De Donatis A., Comito G., Buricchi F. Proliferation versus migration in platelet-derived growth factor signaling: the key role of endocytosis. J Biol Chem. Jul 18 2008;283:19948–19956. doi: 10.1074/jbc.M709428200. [DOI] [PubMed] [Google Scholar]
- 30.Wiesner S., Legate K.R., Fassler R. Integrin-actin interactions. Cell Mol Life Sci. May 2005;62:1081–1099. doi: 10.1007/s00018-005-4522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodham E.F., Machesky L.M. Polarised cell migration: intrinsic and extrinsic drivers. Curr Opin Cell Biol. Jun 19 2014;30C:25–32. doi: 10.1016/j.ceb.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Schneider I.C., Haugh J.M. Quantitative elucidation of a distinct spatial gradient-sensing mechanism in fibroblasts. J Cell Biol. Dec 5 2005;171:883–892. doi: 10.1083/jcb.200509028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shestakova E.A., Singer R.H., Condeelis J. The physiological significance of beta-actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci U S A. Jun 19 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haugh J.M. Deterministic model of dermal wound invasion incorporating receptor-mediated signal transduction and spatial gradient sensing. Biophys J. Apr 1 2006;90:2297–2308. doi: 10.1529/biophysj.105.077610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawada K., Upadhyay G., Ferandon S. Cell migration is regulated by platelet-derived growth factor receptor endocytosis. Mol Cell Biol. Aug 2009;29:4508–4518. doi: 10.1128/MCB.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasiliev J.M. Cytoskeletal mechanisms responsible for invasive migration of neoplastic cells. Int J Dev Biol. 2004;48:425–439. doi: 10.1387/ijdb.041806jv. [DOI] [PubMed] [Google Scholar]
- 37.Bershadsky A., Kozlov M., Geiger B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr Opin Cell Biol. Oct 2006;18:472–481. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Mitchison T.J., Cramer L.P. Actin-based cell motility and cell locomotion. Cell. Feb 9 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- 39.Ridley A.J., Schwartz M.A., Burridge K. Cell migration: integrating signals from front to back. Science. Dec 5 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 40.Small J.V., Geiger B., Kaverina I., Bershadsky A. How do microtubules guide migrating cells? Nat Rev Mol Cell Biol. Dec 2002;3:957–964. doi: 10.1038/nrm971. [DOI] [PubMed] [Google Scholar]
- 41.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. Feb 21 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 42.Carlier M.F., Pantaloni D. Control of actin assembly dynamics in cell motility. J Biol Chem. Aug 10 2007;282:23005–23009. doi: 10.1074/jbc.R700020200. [DOI] [PubMed] [Google Scholar]
- 43.Cramer L.P. Mechanism of cell rear retraction in migrating cells. Curr Opin Cell Biol. Oct 2013;25:591–599. doi: 10.1016/j.ceb.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Fournier M.F., Sauser R., Ambrosi D., Meister J.J., Verkhovsky A.B. Force transmission in migrating cells. J Cell Biol. Jan 25 2010;188:287–297. doi: 10.1083/jcb.200906139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bershadsky A.D., Balaban N.Q., Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 46.Gardel M.L., Schneider I.C., Aratyn-Schaus Y., Waterman C.M. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. Nov 10 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chrzanowska-Wodnicka M., Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. Jun 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedl P., Brocker E.B. The biology of cell locomotion within three-dimensional extracellular matrix. Cell Mol Life Sci. Jan 20 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedl P., Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. Jan 11 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lammermann T., Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr Opin Cell Biol. Oct 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Charras G., Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. Sep 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 52.Lammermann T., Bader B.L., Monkley S.J. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. May 1 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 53.Renkawitz J., Schumann K., Weber M. Adaptive force transmission in amoeboid cell migration. Nat Cell Biol. Dec 2009;11:1438–1443. doi: 10.1038/ncb1992. [DOI] [PubMed] [Google Scholar]
- 54.Xiao Z., Zhang N., Murphy D.B., Devreotes P.N. Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J Cell Biol. Oct 20 1997;139:365–374. doi: 10.1083/jcb.139.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Servant G., Weiner O.D., Herzmark P., Balla T., Sedat J.W., Bourne H.R. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. Feb 11 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janetopoulos C., Jin T., Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. Mar 23 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 57.Jin T., Zhang N., Long Y., Parent C.A., Devreotes P.N. Localization of the G protein betagamma complex in living cells during chemotaxis. Science. Feb 11 2000;287:1034–1036. doi: 10.1126/science.287.5455.1034. [DOI] [PubMed] [Google Scholar]
- 58.Anand-Apte B., Zetter B. Signaling mechanisms in growth factor-stimulated cell motility. Stem Cells. 1997;15:259–267. doi: 10.1002/stem.150259. [DOI] [PubMed] [Google Scholar]
- 59.Xu J., Wang F., Van Keymeulen A. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. Jul 25 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 60.Chudakov D.M., Matz M.V., Lukyanov S., Lukyanov K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. Jul 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 61.Pertz O. Spatio-temporal Rho GTPase signaling – where are we now? J Cell Sci. Jun 1 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 62.Hawkins P.T., Anderson K.E., Davidson K., Stephens L.R. Signalling through class I PI3Ks in mammalian cells. Biochem Soc Trans. Nov 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 63.Parent C.A., Blacklock B.J., Froehlich W.M., Murphy D.B., Devreotes P.N. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. Oct 2 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 64.Welch H.C., Coadwell W.J., Stephens L.R., Hawkins P.T. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. Jul 3 2003;546:93–97. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- 65.Bosgraaf L., Russcher H., Smith J.L., Wessels D., Soll D.R., Van Haastert P.J. A novel cGMP signalling pathway mediating myosin phosphorylation and chemotaxis in Dictyostelium. Embo J. Sep 2 2002;21:4560–4570. doi: 10.1093/emboj/cdf438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grandoch M., Roscioni S.S., Schmidt M. The role of Epac proteins, novel cAMP mediators, in the regulation of immune, lung and neuronal function. Br J Pharmacol. Jan 1 2010;159:265–284. doi: 10.1111/j.1476-5381.2009.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu L., Das S., Losert W., Parent C.A. mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell. Dec 14 2010;19:845–857. doi: 10.1016/j.devcel.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiger M.C., Wang C.C., Krajcovic M., Melvin A.T., Rhoden J.J., Haugh J.M. Spontaneous phosphoinositide 3-kinase signaling dynamics drive spreading and random migration of fibroblasts. J Cell Sci. Feb 1 2009;122:313–323. doi: 10.1242/jcs.037564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Haastert P.J., Devreotes P.N. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. Aug 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 70.Bosgraaf L., Van Haastert P.J. A model for cGMP signal transduction in Dictyostelium in perspective of 25 years of cGMP research. J Muscle Res Cell Motil. 2002;23:781–791. doi: 10.1023/a:1024431813040. [DOI] [PubMed] [Google Scholar]
- 71.Fincham V.J., James M., Frame M.C., Winder S.J. Active ERK/MAP kinase is targeted to newly forming cell-matrix adhesions by integrin engagement and v-Src. Embo J. Jun 15 2000;19:2911–2923. doi: 10.1093/emboj/19.12.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geiger B., Yamada K.M. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol. 2011;3(5) doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rericha E.C., Parent C.A. Steering in quadruplet: the complex signaling pathways directing chemotaxis. Sci Signal. 2008;1:pe26. doi: 10.1126/scisignal.122pe26. [DOI] [PubMed] [Google Scholar]
- 74.Hoeller O., Kay R.R. Chemotaxis in the absence of PIP3 gradients. Curr Biol. May 1 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 75.Chen L., Iijima M., Tang M. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev Cell. Apr 2007;12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou H., Huang S. Role of mTOR signaling in tumor cell motility, invasion and metastasis. Curr Protein Pept Sci. Feb 2011;12:30–42. doi: 10.2174/138920311795659407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. Apr 13 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai H., Das S., Kamimura Y., Long Y., Parent C.A., Devreotes P.N. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol. Jul 26 2010;190:233–245. doi: 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karsenti E. Self-organization in cell biology: a brief history. Nat Rev Mol Cell Biol. Mar 2008;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- 80.Bosgraaf L., Van Haastert P.J. The ordered extension of pseudopodia by amoeboid cells in the absence of external cues. PLoS One. 2009;4:e5253. doi: 10.1371/journal.pone.0005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weiger M.C., Ahmed S., Welf E.S., Haugh J.M. Directional persistence of cell migration coincides with stability of asymmetric intracellular signaling. Biophys J. Jan 6 2010;98:67–75. doi: 10.1016/j.bpj.2009.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hurd T.R., Degennaro M., Lehmann R. Redox regulation of cell migration and adhesion. Trends Cell Biol. Feb 2012;22:107–115. doi: 10.1016/j.tcb.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stone J.R., Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal. Mar–Apr 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 84.Geiszt M., Leto T.L. The Nox family of NAD(P)H oxidases: host defense and beyond. J Biol Chem. Dec 10 2004;279:51715–51718. doi: 10.1074/jbc.R400024200. [DOI] [PubMed] [Google Scholar]
- 85.Tyurin-Kuzmin P.A., Agaronian K.M., Morozov IaI., Mishina N.M., Belousov V.V., Vorotnikov A.V. NADPH oxidase controls EGF-induced proliferation via the ERK1/2-independent mechanism. Biofizika. Nov–Dec 2010;55:959–965. [PubMed] [Google Scholar]
- 86.Rocic P., Lucchesi P.A. NAD(P)H oxidases and TGF-beta-induced cardiac fibroblast differentiation: Nox-4 gets Smad. Circ Res. Oct 28 2005;97:850–852. doi: 10.1161/01.RES.0000190403.87462.bf. [DOI] [PubMed] [Google Scholar]
- 87.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. Jan 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 88.Ushio-Fukai M. Redox signaling in angiogenesis: role of NADPH oxidase. Cardiovasc Res. Jul 15 2006;71:226–235. doi: 10.1016/j.cardiores.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 89.Sundaresan M., Yu Z.X., Ferrans V.J., Irani K., Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. Oct 13 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 90.San Martin A., Griendling K.K. Redox control of vascular smooth muscle migration. Antioxid Redox Signal. Mar 1 2010;12:625–640. doi: 10.1089/ars.2009.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niethammer P., Grabher C., Look A.T., Mitchison T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. Jun 18 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brzostowski J.A., Sawai S., Rozov O. Phosphorylation of chemoattractant receptors regulates chemotaxis, actin reorganization and signal relay. J Cell Sci. Oct 15 2013;126:4614–4626. doi: 10.1242/jcs.122952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rhee S.G., Bae Y.S., Lee S.R., Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. Oct 10 2000;2000:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 94.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. Apr 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 95.Mishina N.M., Tyurin-Kuzmin P.A., Markvicheva K.N. Does cellular hydrogen peroxide diffuse or act locally? Antioxid Redox Signal. Jan 1 2011;14:1–7. doi: 10.1089/ars.2010.3539. [DOI] [PubMed] [Google Scholar]
- 96.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. Jan 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 97.Tonks N.K. Redox redux: revisiting PTPs and the control of cell signaling. Cell. Jun 3 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 98.den Hertog J., Groen A., van der Wijk T. Redox regulation of protein-tyrosine phosphatases. Arch Biochem Biophys. Feb 1 2005;434:11–15. doi: 10.1016/j.abb.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 99.Kwon J., Lee S.R., Yang K.S. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci U S A. Nov 23 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McCubrey J.A., Steelman L.S., Chappell W.H. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascade inhibitors: how mutations can result in therapy resistance and how to overcome resistance. Oncotarget. Oct 2012;3:1068–1111. doi: 10.18632/oncotarget.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim J.S., Huang T.Y., Bokoch G.M. Reactive oxygen species regulate a slingshot-cofilin activation pathway. Mol Biol Cell. Jun 2009;20:2650–2660. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lassing I., Schmitzberger F., Bjornstedt M. Molecular and structural basis for redox regulation of beta-actin. J Mol Biol. Jul 6 2007;370:331–348. doi: 10.1016/j.jmb.2007.04.056. [DOI] [PubMed] [Google Scholar]
- 103.Hung R.J., Pak C.W., Terman J.R. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. Dec 23 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]