Abstract
Y box binding protein-1 (YBX1) belongs to a DNA- and RNA-binding family of transcription factors, containing the highly conserved cold shock domain (CSD). YBX1 is involved in a number of cellular functions including transcription, translation, DNA damage repair etc., and it is upregulated during times of environmental stress. YBX1 is localized in both the cytoplasm and the nucleus. There, its nuclear translocation is observed in a number of cancers and is associated with poor prognosis and disease progression. Additionally, YBX1 expression is upregulated in a variety of cancers, pointing towards its role as a potential oncogene. Under certain circumstances, YBX1 also promotes the expression of multidrug resistance 1 (MDR1) gene, which is involved in the development of drug resistance. Thus, it is critical to understand the mechanism of YBX1 regulation and its downstream effects on promoting cancer development. A number of recent studies have highlighted the mechanisms of YBX1 regulation. Mass spectrometric analyses have reported several post-translational modifications that possibly play an important role in modulating YBX1 function. Phosphorylation is the most widely occurring post-translational modification in YBX1. In vivo analyses of sites like S102 and more recently, S165 illustrate the relationship of post-translational regulation of YBX1 in promoting cell proliferation and tumor growth. This review provides a comprehensive and up-to-date account of post-translational modifications identified in YBX1. This knowledge is a key in allowing us to better understand the mechanism of YBX1 regulation, which will aid in development of novel therapeutic strategies to target YBX1 in many types of cancer in the future.
Keywords: Cancer, Cold shock domain, Phosphorylation, Post-translational modification, Y box binding protein 1
Introduction
Y box binding protein 1 (YBX1) belongs to the YBX family of transcription factors that contain an ancient and evolutionarily conserved cold shock domain (CSD). The name “cold shock” originates from E. coli, which, when exposed to the stress of cold temperature, increases the expression of around 13 proteins containing the CSD by 2–10 folds. This helps the cell survive in low temperature.1 This observation found in bacteria is similar to the role of YBX1 in eukaryotic cell's response to stress, indicating the existence of not only structural, but also functional conservation over a wide evolutionary span in the YBX family.
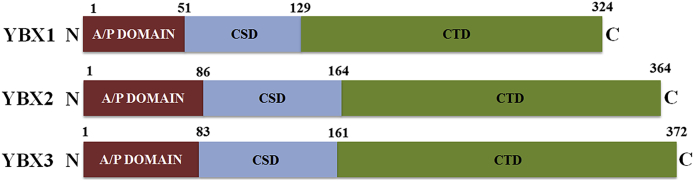
The name ‘YBX’ was coined due to the ability of the YBX protein family to bind the Y box sequence on DNA, defined as 5′-CTGATTGG-3′. The YBX family of proteins has high sequence homology across different species. There are three members of the YBX family: YBX1, YBX2 and YBX3. As shown in Fig. 1, all members have structural commonalities as follows: an N-terminal alanine (A)- and proline (P)-rich domain (A/P domain), a central CSD and a C-terminal domain (CTD) consisting of alternating base/acid amino acid repeats. The A/P domain is thought to be important for the transcriptional activity of YBX1 and has also been shown to interact with tumor suppressor p53 to mediate p53-dependent transcription.2
Figure 1.
Members of Y box binding protein family. All three family members, i.e. YBX1, YBX2 and YBX3 have common domains: the alanine/proline (A/P)-rich N terminal domain, the highly conserved cold shock domain (CSD), and the C-terminal domain (CTD).
The crystal structure of the human YBX1 CSD domain is known to be comprised of a five-stranded anti-parallel-barrel structure with a long flexible loop and has 40% homology with its bacterial counterpart.3 CSD has ribonucleoprotein particle 1 (RNP1) and RNP2 recognition sites, that mediate the site-specific interaction of YBX1 with DNA and RNA. On the other hand, CTD also mediates non-specific interactions with nucleic acids, especially with single-stranded DNA. Additionally, CTD consists of a nuclear localization signal and a cytoplasmic retention site. These two sites regulate YBX proteins shuttling between nucleus and cytoplasm.
As described above, YBX1, 2, and 3 are the three family members that belong to the human YBX family. Different YBX family members are encoded by different genes and are localized on distinct chromosomes. For instance, human YBX1 is encoded by the ybx1 gene present on chromosome 1p34.2, while YBX2 is encoded by the ybx2 gene and is present on chromosome 17p13.1. On the other hand, YBX3 is encoded by the ybx3 gene, which is present on chromosome 12p13.1. Although YBX1, 2, and 3 share structural similarity, they do not share similar functions.4
Human YBX1 is generally expressed in adult somatic cells and is involved in the transcription of important genes that participate in tumor development.4 This will be discussed in the next section in detail. On the other hand, human YBX2 is expressed mainly in germ cells and is mainly involved in the maintenance of stability and/or translation of germ cell mRNAs. Human YBX3 is expressed during the embryonic stage of development, but is absent in adult cells. YBX3 has been shown to be a repressor of some growth factor promoters, such as the GM-CSF promoter.5 Overall, YBX1, 2, and 3 have important functions in almost all stages of the cell life cycle and have important DNA- and RNA-binding related functions.
In this review, we focus on the recent update regarding the role of YBX1 in cancer, especially with respect to its post-translational modifications. For broader knowledge regarding YBX1, please refer to a relevant review elsewhere.5
Role of YBX1 in cancer
YBX1 is one of the most evolutionarily conserved nucleic acid binding proteins. Initially, it was thought to bind only the conserved Y-box element in double stranded DNA (dsDNA). However, subsequent research has shown its ability to bind to single stranded DNA (ssDNA), damaged DNA, and RNA.5 In adult human cells, depending on its localization, the functions and protein–protein interactions of YBX1 differ. Nuclear localization of YBX1 promotes the transcription of genes containing the Y box consensus sequence in their promoter region.5 The target genes of YBX1 exert a broad scope of functions including transcriptional regulation, mRNA packaging, and DNA repair.6
YBX1 is also known to play important functions in cancer. Its levels are increased in various types of cancer, including cancers of the breast, colon, ovary, lung,7 prostate,8 stomach,9etc. Furthermore, nuclear localization of YBX1 is also associated with a more aggressive phenotype of the cancer and a poor survival rate.10, 11 Thus, both high expression levels and nuclear translocation of YBX1 in cancer make YBX1 an ideal diagnosis marker and a possible therapeutic target for cancer.12 To date, ample evidence has been accumulated regarding the role of YBX1 in cancers. YBX1 is widely studied in the field of breast cancer research. For instance, YBX1 is shown to be overexpressed in about 40% of breast cancers, but absent in normal breast tissue.13 This highlights the potential importance of YBX1 as an oncogene in breast cancer. Davies et al showed that YBX1 is capable of transforming normal human mammary epithelial cells to cancerous cells via p300-mediated chromatin remodeling, leading to the formation of basal-like breast cancer.14 Jung et al further reported that YBX1 is important for sustaining the stem-like nature and tumorigenic ability of a phenotypically distinct subset of breast cancer cells. YBX1 carries out this function mainly through the regulation of the sex determining region Y-box 2 gene (Sox2), leading to a relatively more aggressive cancer subtype.15 Besides the potential mechanisms discussed above, YBX1 also promotes breast cancer development by preventing cancer cells from being eliminated by apoptosis. For instance, YBX1 can interact with the tumor suppressor p53, which causes an increase in the nuclear localization of YBX1 and inhibits p53-dependent cell death, thus acting as a negative regulator of apoptosis in breast cancer.16 YBX1 has also been implicated in other types of cancers. In human non-small-cell lung cancers, YBX1 promotes the transcription of cyclin D1, an oncogenic cyclin that promotes cell growth.17 In human osteosarcoma cells, nuclear expression of YBX1 regulates cell cycle progression at the G1/S phase and not only promotes tumor growth, but is also associated with a poor prognosis in these cases.18 Interestingly, YBX1 also contributes to tumor metastasis and growth through the epithelial-to-mesenchymal transition (EMT) process. In prostate cancer, YBX1 promotes EMT, leading to a more invasive form of this disease.19 In patients with head and neck cancer, higher YBX1 expression is associated with a lower disease-specific survival rate in patients, supporting the association of YBX1 expression with poor prognosis.20
Besides its role in more common cancer types described above, YBX1 has also been shown to play a role in virus-induced cancers. Human papillomaviruses are implicated in cervical cancers and also in a subtype of head and neck cancers. Leiprecht et al found that YBX1 plays a critical role in papillomavirus-induced tumor formation by interacting with the viral regulatory protein E2, leading to increased expression of important oncogenes in rabbit keratinocyte cell lines.21 Collectively, the accumulated evidence supports the critical role of YBX1 in cancer development in both non-virus and virus-induced cancers.
The multi-faceted effects of YBX1 in cancer are also reflected by its important role in cancer chemotherapy drug resistance.22, 23 Elevated levels of YBX1 have been associated with resistance to chemotherapy drugs in different types of cancers. This is a great challenge to clinicians, as it limits their ability to control disease progression and to improve patient survival. Various studies have highlighted novel factors that can be targeted to reduce YBX1-based drug resistance. For instance, YBX1 plays an important role in the chemotherapy drug taxane's resistance in ovarian cancer cells. It is reported that the focal adhesion kinase (FAK), a non-receptor tyrosine kinase that usually promotes cell growth, is involved in the regulation of YBX1-dependent drug resistance.24 Inhibition of FAK causes increased sensitivity to taxane by decreasing YBX1 phosphorylation and YBX1 nuclear accumulation in an Akt-dependent manner.23 In addition to taxane resistance, expression of YBX1 is also shown to confer resistance to cisplatin treatment in ovarian cancer cells.22
In addition to chemotherapy drug resistance, YBX1 also plays an important role in stress-induced drug resistance. One such important drug resistance gene regulated by YBX1 is the multi-drug resistance 1 (MDR1) gene. MDR1 has Y box in its promoter region. It was observed that during environmental stress, YBX1 caused the increased expression of MDR1, leading to environmental stress-dependent drug resistance.24 Basaki et al showed that Akt-mediated nuclear translocation of YBX1 is important for acquiring drug resistance through upregulation of MDR1 in human ovarian cancer cells.25 Knockdown of YBX1 also resulted in increased sensitization to DNA-damaging agents and ionizing radiation, thus suggesting it might exert protective effects against cytotoxic DNA-damaging agents in human colon cancer cells.26 Therefore, certain functions of YBX1, like managing the stress response, can also contribute to drug resistance in response to environmental factors.
Taken together, YBX1 plays an essential and unique role in cancer progression, chemotherapy drug resistance, and environmental factors related to its cytoprotective effects.
Post-translational modifications in YBX1
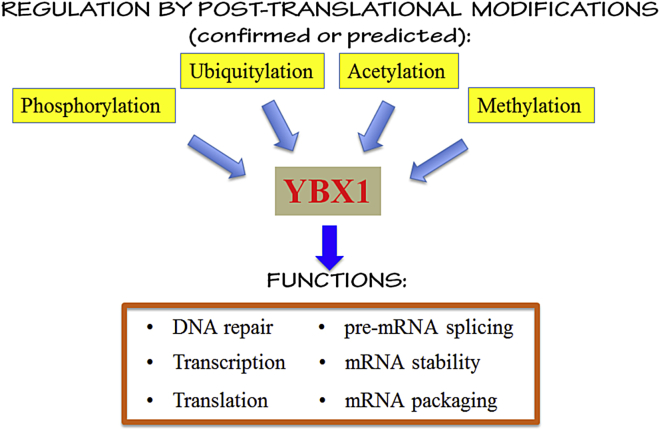
Like many other transcription factors, the activity and function of YBX1 is regulated at multiple levels. Recent emerging evidence has demonstrated that the functions of YBX1 can be regulated by post-translational modifications at various sites. As illustrated in Table 1 and Fig. 2, several post-translational modification sites with important biological functions have been identified and confirmed in vivo.
Table 1.
Known and confirmed post-translational modifications in human YBX1.
| Site on YBX1 | Location in YBX1 structure | Type of PTM | Known modulator |
|---|---|---|---|
| S10227 | CSD | Phosphorylation | Akt |
| Y16231, 33, 34 | CTD | Phosphorylation | FGFR1 |
| Y188, Y28130 | CTD | Phosphorylation | Unknown |
| S16529 | CSD | Phosphorylation | Unknown |
Akt: AKT8 virus oncogene cellular homolog; CSD: cold shock domain; CTD: C-terminal domain; FGFR1: fibroblast growth factor receptor 1.
Figure 2.
Regulation and functions of YBX1. YBX1 can be regulated by post-translational modifications like phosphorylation, ubiquitylation, acetylation and methylation. This affects the downstream functions of YBX1 that mediate a variety of cellular processes including DNA repair, transcription, translation, pre-mRNA splicing, mRNA packaging and mRNA stability.
Phosphorylation
Confirmed phosphorylation sites on YBX1
The first phosphorylation site identified was serine 102 (S102) in the CSD region of YBX1 (Table 1).27 This is also the most extensively studied site in YBX1. Sutherland et al showed that mutating S102 of YBX1 to alanine (A) decreased tumor growth in breast cancer, strongly suggesting that S102 phosphorylation is critical for the oncogenic activity of YBX in breast cancer. Phosphorylation at S102 may promote the YBX1 oncogenic ability by facilitating nuclear translocation of YBX1 and enhancing its binding to DNA. Furthermore, it was reported that the level of phosphorylated S102 positively correlates with EGFR (epidermal growth factor receptor) and HER2 (human epidermal growth factor receptor 2) expression: both are upregulated in primary breast cancer specimens. On the other hand, S102A-YBX1 mutant prevented induction of both EGFR and HER2 and showed decreased tumor growth in breast cancer.28
In addition to phosphorylation of S102 in breast cancer, a recent study by our lab underlines the significance of the novel phosphorylation of S165 in the CSD region of YBX1 (Table 1).29 We provided the first evidence regarding the important connection between YBX1 and the nuclear factor kappa B (NF-κB). We suggest that YBX1 functions as a potential NF-κB activator. Using mass spectrometry (MS) analysis, we identified novel phosphorylation of S165 on YBX1. Overexpression of the S165A-YBX1 mutant in either 293 cells or colon cancer HT29 cells showed dramatically reduced NF-κB activating ability as compared to that of wt YBX1, confirming that S165 phosphorylation is critical for the activation of NF-κB by YBX1. We also showed that expression of the S165A-YBX1 mutant significantly decreased expression of NF-κB-inducible genes, reduced cell growth, and compromised tumorigenic ability as compared to wt YBX1. Taken together, we proved that YBX1 functions as a tumor promoter via NF-κB activation, and that phosphorylation of S165 of YBX1 is critical for this function. Therefore, our important discovery may lead to blocking S165 phosphorylation as a potential therapeutic strategy to treat colon cancer.
Other YBX1 phosphorylation sites have also been identified. Using deletion constructs of YBX1, von Roeyen et al identified two additional phosphorylation sites, tyrosine 188 (Y188) and Y281 in the CTD domain of YBX1 (Table 1). Using site-specific antibodies, they proved that phosphorylation of Y281, but not Y188 in YBX1 takes place and appears to correlate with nuclear protein shuttling of YBX1 in undifferentiated human monocytic THP-1 cells.30 Kasyapa et al reported that fibroblast growth factor receptor 2 (FGFR2), a receptor tyrosine kinase, can interact with YBX1, leading to the phosphorylation of tyrosine 162 (Y162) in the CTD of YBX1 in atypical myeloproliferative disorders (Table 1).31 This finding suggests that tyrosine phosphorylation of YBX1 may play an important role in the development of this devastating disease, in which FGFR2 has been widely implicated.
Predicted but unconfirmed phosphorylation sites on YBX1
As described above, very few phosphorylation sites on YBX1 have been confirmed in vivo. However, as shown in Table 2, multiple potential sites have been identified by MS analysis, but have yet to be confirmed by in vivo experiments.30, 32, 33, 34, 35, 36, 37, 38
Table 2.
Predicted but unconfirmed post-translational modifications in human YBX1.
| Site on YBX1 | Location in YBX1 structure | Type of PTM | Known modulator | Reference |
|---|---|---|---|---|
| Residues 262–32444 | CTD | Ubiquitylation | RBBP6 | Chibi et al 2008 |
| S174, S17632, 40 | CTD | Phosphorylation | EGF | Pan et al 2009, Sharma et al 2014 |
| K8145 | CSD | Acetylation | – | Choudhary et al 2009 |
| S16733, 40 | CTD | Phosphorylation | Ionizing Radiation | Bennetzen et al 2010, Sharma et al 2014 |
| T8039 | CSD | Phosphorylation | Chk1 | Blasius et al 2011 |
| S136, T27134 | CTD | Phosphorylation | – | Kettenbach et al 2011 |
| K64, K11843 | CSD | Ubiquitylation | – | Wagner, et al 2011 |
| K81, K92, K93, K13741, 43 | CSD | Ubiquitylation | – | Kim et al 2011, Wagner et al 2011 |
| K26441, 42 | CTD | Ubiquitylation | – | Shi et al 2011, Kim et al 2011 |
| T10837 | CSD | Phosphorylation | – | Stokes et al 2012 |
| Y15838 | CTD | Phosphorylation | – | Trost et al 2012 |
| Y196, S20835 | CTD | Phosphorylation | – | Bai et al 2012 |
| S313, S31428, 40 | CTD | Phosphorylation | RIP-3 | Wu et al 2012, Sharma et al 2014 |
| S2, S3, T7, S21, S32, S36, S4440 | A/P domain | Phosphorylation | – | Sharma et al 2014 |
| T8940 | CSD | Phosphorylation | – | Sharma et al 2014 |
| R199, R200, R23947 | CTD | Mono-methylation | – | Guo et al 2014 |
A/P domain: alanine- and proline-rich domain; Chk1: cell-cycle checkpoint kinase 1; CSD: cold shock domain; CTD: C-terminal domain; EGF: epidermal growth factor; RBBP6: retinoblastoma binding protein 6; RIP-3: receptor-interacting protein kinase 3.
Several research groups carried out MS analyses to identify substrates of various kinases and other cellular factors in the proteomes of normal human as well as cancer cell lines. For instance, Blasius et al used MS analysis to identify substrates of cell-cycle checkpoint kinase 1 (Chk1), which plays an essential role in controlling cellular processes such as DNA replication, mitosis, and response to DNA-damage. One of the substrates Chk1 phosphorylates is threonine 80 (T80), which is present in the CSD domain of YBX1.39 Regulation of YBX1 by Chk1 seems plausible since the functions of both Chk2 and YBX1 are mostly similar and it is possible that Chk1 exerts some of its effects through post-translationally modifying YBX1. However, the function of T80 is yet to be confirmed in vivo.
The most recent advancement can be seen in the study by Sharma et al. In this study, a large-scale quantitative MS study of phosphoproteome in cervical cancer HeLa cells allowed for global identification of protein modifications.40 Among the various proteins that were modified, several sites were also found in YBX1 (Table 2). All the phosphorylation sites that were identified in this study were mostly in the A/P and CTD domains. The majority of these sites have never been defined before. Thus, this study opens an extensive area of research to explore the importance of these sites in contribution to cervical cancer.
It is worth noting that until these sites are confirmed by in vivo experiments, they are simply potential sites. Nevertheless, this information could guide any future endeavor to better understand the role of post-translational modifications in YBX1 regulation.
Other unconfirmed types of potential modifications
Data from Sharma et al and others suggest that YBX1 could be potentially modified at multiple amino acid residues. Besides the phosphorylation described above, other modifications on YBX1 include ubiquitylation, acetylation, caspase cleavage and methylation (Table 2).40
Ubiquitylation
Ubiquitylation is a type of post-translational modification that controls various cellular processes through proteasomal degradation of target proteins. As listed in Table 2, some potential ubiquitylation sites have been identified on YBX1. Notably, three studies employed genome wide mass spectrometric proteomics to scan for ubiquitylation in the genome and found several sites on YBX1.41, 42, 43 Chibi et al identified that retinoblastoma binding protein 6 (RBBP6), an E3 ubiquitin ligase, interacts with YBX1, leading to ubiquitylation and proteasomal degradation of YBX1.44 Thus, dysregulation of ubiquitylation may alter YBX1 function and further contribute to its oncogenic capability, and hence is worth exploring in future in vivo studies.
Acetylation
In 2009, Choudhary et al carried out a study using MS analysis to scan for global lysine acetylation sites in whole cell lysates from human acute myeloid leukemia cell lines. Among the 3600 lysine (K) acetylation sites found on 1750 proteins, one of them was K81 in the CSD domain of YBX1. Current evidence suggests that lysine acetylation is important for functions and interactions of various genes including p53.45 It is also known that p53 and YBX1 interact with each other and that nuclear localization of YBX1 can be regulated only by functional but not mutant p53.2, 46 Therefore, it would be worthwhile to confirm K81 acetylation in vivo, and to explore whether it affects the interaction between YBX1 and p53 or other proteins in the cell, and further, how the acetylation may modulate cell function in vivo.
Methylation
Methylation of proteins occurring at lysine and arginine (R) is also one of the most critical post-translational modifications seen in nature. Methylation of proteins has been implicated in a number of cellular processes like gene transcription, DNA damage and repair, protein translocation etc. As shown in Table 2, YBX1 is methylated at residues R199, 200 and 239 using immunoaffinity purification of methylated peptides followed by MS analysis.47 Interestingly, previous work from our laboratory also showed that the transcription factor NF-κB is methylated on K218/221 and R30 of its p65 subunit.48, 49, 50 Since we found that YBX1 is a direct activator of NF-κB,29 it would be interesting to further examine whether methylation of YBX1 plays an important role in regulating the interaction between YBX1 and NF-κB, contributing to dysfunction of these two proteins in cancer. This possibility further justifies the necessity of confirming the methylation sites of YBX1 in vivo.
Perspective
Post-translational modification is an important way to regulate protein function. Different sites on a single protein allow for triggering varied functions of the same protein, depending on the cellular requirements and/or environmental stimuli. This allows for fine-tuning of the protein function. YBX1 has emerged as one of the proteins substantially post-translationally modified, as identified by advanced mass spectrometric proteomics (Table 1, Table 2). This vast amount of information makes post-translational modification of YBX1 an exciting research area for further exploration.
Based on the fact that many post-translational modification sites have been identified on YBX1, while few were confirmed in vivo, an obvious and immediate task that scientists are facing is to confirm most if not all of the identified sites in vivo. If confirmed, the important biological role of each modified site can be further examined in cancer or other YBX1 related diseases. Another daunting task once the modification site is confirmed is that efforts need to be put into the discovery of the enzymes that may catalyze these specific modifications. Often, enzymes are better targets in the development of drugs to treat cancer. Furthermore, as cutting edge technologies emerge at an amazing pace, it should not come as a major surprise that more post-translational modification sites will be discovered on YBX1 in the near future. Moreover, post-translational modification of proteins is often cellular-context dependent. It is not difficult to imagine that quite different modifications of YBX1 could be identified in different types of cancer and disease states. These unique post-translational modifications of YBX1 could represent a disease-specific signature which may help with diagnosis and treatment of that specific disease.
To conclude, understanding the fascinating state of post-translational modification of YBX1 will not only lead to a better understanding of the mechanisms underlying biological processes regulated by YBX1, but will also help to develop effective pathway-specific therapeutic strategies against YBX1 in different types of cancer and YBX1-related diseases.
Conflicts of interest
All authors have none to declare.
Acknowledgment
We sincerely thank Ms. Lisa King at the Department of Pharmacology and Toxicology at Indiana University for her professional help with editing this review. This research is supported by grants 4186265 (American Cancer Society) and 23-862-07 and 036433730102 (Indiana University) to TL.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Obokata J., Ohme M., Hayashida N. Nucleotide sequence of a cDNA clone encoding a putative glycine-rich protein of 19.7 kDa in Nicotiana sylvestris. Plant Mol Biol. 1991;17:951–955. doi: 10.1007/BF00037080. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto T., Izumi H., Imamura T. Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene. 2000;19:6194–6202. doi: 10.1038/sj.onc.1204029. [DOI] [PubMed] [Google Scholar]
- 3.Kloks C.P., Spronk C.A., Lasonder E. The solution structure and DNA-binding properties of the cold-shock domain of the human Y-box protein YB-1. J Mol Biol. 2002;316:317–326. doi: 10.1006/jmbi.2001.5334. [DOI] [PubMed] [Google Scholar]
- 4.Kent J., Sugnet C., Terrence S. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eliseeva I.A., Kim E.R., Guryanov S.G., Ovchinnikov L.P., Lyabin D.N. Y-box-binding protein 1 (YB-1) and its functions. Biochem (Mosc) 2011;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 6.Lyabin D., Eliseeva I., Ovchinnikov L. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2013;5:95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- 7.Shibahara K., Sugio K., Osaki T. Nuclear expression of the Y-box binding protein, YB-1, as a novel marker of disease progression in non-small cell lung cancer. Clin Cancer Res. 2001;7:3151–3155. [PubMed] [Google Scholar]
- 8.Gimenez-Bonafe P., Fedoruk M.N., Whitmore T.G. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate. 2004;59:337–349. doi: 10.1002/pros.20023. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y., Wang K.Y., Li Z. Y-box binding protein 1 expression in gastric cancer subtypes and association with cancer neovasculature. Clin Transl Oncol. 2015;17:152–159. doi: 10.1007/s12094-014-1208-4. [DOI] [PubMed] [Google Scholar]
- 10.Fujii T., Kawahara A., Basaki Y. Expression of HER2 and estrogen receptor alpha depends upon nuclear localization of Y-box binding protein-1 in human breast cancers. Cancer Res. 2008;68:1504–1512. doi: 10.1158/0008-5472.CAN-07-2362. [DOI] [PubMed] [Google Scholar]
- 11.Kashihara M., Azuma K., Kawahara A. Nuclear Y-box binding protein-1, a predictive marker of prognosis, is correlated with expression of HER2/ErbB2 and HER3/ErbB3 in non-small cell lung cancer. J Thorac Oncol. 2009;4:1066–1074. doi: 10.1097/JTO.0b013e3181ae2828. [DOI] [PubMed] [Google Scholar]
- 12.Kosnopfel C., Sinnberg T., Schittek B. Y-box binding protein 1-a prognostic marker and target in tumour therapy. Eur J Cell Biol. 2014;93:61–70. doi: 10.1016/j.ejcb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Habibi G., Leung S., Law J.H. Redefining prognostic factors for breast cancer: YB-1 is a stronger predictor of relapse and disease-specific survival than estrogen receptor or HER-2 across all tumor subtypes. Breast Cancer Res. 2008;10:R86. doi: 10.1186/bcr2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies A.H., Reipas K.M., Pambid M.R. YB-1 transforms human mammary epithelial cells through chromatin remodeling leading to the development of basal-like breast cancer. Stem Cells. 2014;32:1437–1450. doi: 10.1002/stem.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung K., Wu F., Wang P., Ye X., Abdulkarim B.S., Lai R. YB-1 regulates Sox2 to coordinately sustain stemness and tumorigenic properties in a phenotypically distinct subset of breast cancer cells. BMC Cancer. 2014;14:328. doi: 10.1186/1471-2407-14-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homer C., Knight D.A., Hananeia L. Y-box factor YB1 controls p53 apoptotic function. Oncogene. 2005;24:8314–8325. doi: 10.1038/sj.onc.1208998. [DOI] [PubMed] [Google Scholar]
- 17.Harada M., Kotake Y., Ohhata T. YB-1 promotes transcription of cyclin D1 in human non-small-cell lung cancers. Genes Cells. 2014;19:504–516. doi: 10.1111/gtc.12150. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara-Okada Y., Matsumoto Y., Fukushi J. Y-box binding protein-1 regulates cell proliferation and is associated with clinical outcomes of osteosarcoma. Br J Cancer. 2013;108:836–847. doi: 10.1038/bjc.2012.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan M.I., Adhami V.M., Lall R.K. YB-1 expression promotes epithelial-to-mesenchymal transition in prostate cancer that is inhibited by a small molecule fisetin. Oncotarget. 2014;5:2462–2474. doi: 10.18632/oncotarget.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolk A., Jubitz N., Mengele K. Expression of Y-box-binding protein YB-1 allows stratification into long- and short-term survivors of head and neck cancer patients. Br J Cancer. 2011;105:1864–1873. doi: 10.1038/bjc.2011.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leiprecht N., Notz E., Schuetz J., Haedicke J., Stubenrauch F., Iftner T. A novel recombinant papillomavirus genome enabling in vivo RNA interference reveals that YB-1, which interacts with the viral regulatory protein E2, is required for CRPV-induced tumor formation in vivo. Am J Cancer Res. 2014;4:222–233. [PMC free article] [PubMed] [Google Scholar]
- 22.Guay D., Evoy A., Paquet E. The strand separation and nuclease activities associated with YB-1 are dispensable for cisplatin resistance but overexpression of YB-1 in MCF7 and MDA-MB-231 breast tumor cells generates several chemoresistance signatures. Int J Biochem Cell B. 2008;40:2492–2507. doi: 10.1016/j.biocel.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Kang Y., Hu W., Ivan C. Role of focal adhesion kinase in regulating YB-1-mediated paclitaxel resistance in ovarian cancer. J Natl Cancer Inst. 2013;105:1485–1495. doi: 10.1093/jnci/djt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chattopadhyay R., Das S., Maiti A.K. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basaki Y., Hosoi F., Oda Y. Akt-dependent nuclear localization of Y-box-binding protein 1 in acquisition of malignant characteristics by human ovarian cancer cells. Oncogene. 2006;26:2736–2746. doi: 10.1038/sj.onc.1210084. [DOI] [PubMed] [Google Scholar]
- 26.Ohga T., Koike K., Ono M. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56:4224–4228. [PubMed] [Google Scholar]
- 27.Sutherland B.W., Kucab J., Wu J. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 28.Wu J., Lee C., Yokom D. Disruption of the Y-Box binding protein-1 results in suppression of the Epidermal Growth Factor Receptor and HER-2. Cancer Res. 2006;66:4872–4879. doi: 10.1158/0008-5472.CAN-05-3561. [DOI] [PubMed] [Google Scholar]
- 29.Prabhu L., Mundade R., Wang B. Critical role of phosphorylation of serine 165 of YBX1 on the activation of NF-κB in colon cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.5120. [in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Roeyen C.R., Scurt F.G., Brandt S. Cold shock Y-box protein-1 proteolysis autoregulates its transcriptional activities. Cell Commun Signal. 2013;11:63. doi: 10.1186/1478-811X-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasyapa C., Gu T.L., Nagarajan L., Polakiewicz R., Cowell J.K. Phosphorylation of the SSBP2 and ABL proteins by the ZNF198-FGFR1 fusion kinase seen in atypical myeloproliferative disorders as revealed by phosphopeptide-specific MS. Proteomics. 2009;9:3979–3988. doi: 10.1002/pmic.200800852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan C., Olsen J.V., Daub H., Mann M. Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol Cell Proteomics. 2009;8:2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennetzen M.V., Larsen D.H., Bunkenborg J., Bartek J., Lukas J., Andersen J.S. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics. 2010;9:1314–1323. doi: 10.1074/mcp.M900616-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kettenbach A.N., Schweppe D.K., Faherty B.K., Pechenick D., Pletnev A.A., Gerber S.A. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai Y., Li J., Fang B. Phosphoproteomics identifies driver tyrosine kinases in sarcoma cell lines and tumors. Cancer Res. 2012;72:2501–2511. doi: 10.1158/0008-5472.CAN-11-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robitaille A.M., Christen S., Shimobayashi M. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 37.Stokes M.P., Farnsworth C.L., Moritz A. PTMScan direct: identification and quantification of peptides from critical signaling proteins by immunoaffinity enrichment coupled with LC-MS/MS. Mol Cell Proteomics. 2012;11:187–201. doi: 10.1074/mcp.M111.015883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trost M., Sauvageau M., Hérault O. Posttranslational regulation of self-renewal capacity: insights from proteome and phosphoproteome analyses of stem cell leukemia. Blood. 2012;12:17–27. doi: 10.1182/blood-2011-12-397844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blasius M., Forment J.V., Thakkar N., Wagner S.A., Choudhary C., Jackson S.P. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma K., D'Souza R.C.J., Tyanova S. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Kim W., Bennett E.J., Huttlin E.L. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y., Chan D.W., Jung S.Y., Malovannaya A., Wang Y., Qin J. A data set of human endogenous protein ubiquitination sites. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002089. M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner S.A., Beli P., Weinert B.T. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics. 2012;11:1578–1585. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chibi M., Meyer M., Skepu A., Rees D.J.G., Moolman-Smook J.C., Pugh D.J.R. RBBP6 interacts with multifunctional protein YB-1 through its RING Finger domain, leading to ubiquitination and proteosomal degradation of YB-1. J Mol Biol. 2008;384:908–916. doi: 10.1016/j.jmb.2008.09.060. [DOI] [PubMed] [Google Scholar]
- 45.Choudhary C., Kumar C., Gnad F. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y.F., Homer C., Edwards S.J. Nuclear localization of Y-box factor YB1 requires wild-type p53. Oncogene. 2003;22:2782–2794. doi: 10.1038/sj.onc.1206357. [DOI] [PubMed] [Google Scholar]
- 47.Guo A., Gu H., Zhou J. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics. 2014;13:372–387. doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu T., Jackson M.W., Wang B. Regulation of NF-B by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu T., Yang M., Huang D., Ghosh G., Stark G.R. Role of lysine methylation of NF-κB in differential gene regulation. Proc Natl Acad Sci U S A. 2013;110:13510–13515. doi: 10.1073/pnas.1311770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei H., Wang B., She Y. PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc Natl Acad Sci U S A. 2013;110:13516–13521. doi: 10.1073/pnas.1311784110. [DOI] [PMC free article] [PubMed] [Google Scholar]