Abstract
The prevalence of diabetes and its complications is increasing at an alarming rate in both developed and deve1oping nations. The emerging evidences highlighted that both genetic and epigenetic mechanisms including histone modifications play a significant role in the pathogenesis of diabetic nephropathy (DN). Histone deacetylases (HDACs) and acetylation are involved in the regulation of autophagy as well as pathogenesis of DN. Both HDACs and histone acetyltransferases (HATs) play a key role in chromatin remodeling and affect the transcription of various genes involved in the cellular homeostasis, apoptosis, immunity and angiogenesis. Further, HDAC inhibitors are exert the renoprotective effects in DN and other diabetic complications. Thus, the cellular acetylation plays a crucial role in the regulation of autophagy and can be explored as a new therapeutic target for the treatment of DN. This review aimed to delineate the role of HDACs and associated molecular signaling/pathways in the regulation of autophagy with an emphasis on promising targets for the treatment of DN.
Keywords: Autophagy, Acetylation, Diabetic nephropathy and SIRTs, HDACs, HDAC inhibitors
Introduction
Diabetic nephropathy (DN), a micro-vascular complication, which leads to end-stage renal disease (ESRD).1 DN is characterized by the excessive deposition of extracellular matrix (ECM) with thickening of glomerular basement membranes and mesangial expansion in both glomerular and tubulo-interstitial compartments as well as persistent micro-albuminuria.2, 3 It is also associated with reduced renal function, podocyte damage and proteinuria. Approximately 30–40% of both type-1 and type-2 diabetic patients develop nephropathy, but evidences are higher in type-1 diabetic patients and ultimately lead to ESRD.4, 5 DN has several distinct phases of development. The structural and functional changes occur in the glomerulus and tubules such as glomerular hyper-filtration, hyper-perfusion, thickening of glomerular basement membrane and glomerular hypertrophy as well as mesangial expansion.6, 7 Excessive accumulation of ECM in glomerular mesangial and tubulo-interstitial compartments as well as, epithelial-to-mesenchymal transition (EMT) of renal tubular epithelial cells are the hallmark of DN.8 In DN, the renal hemodynamic changes involve the activation of various vaso-active systems such as rennin-angiotensin-aldosterone systems (RAAS), which results in the secretion of pro-fibrotic molecules such as transforming growth factor β1 (TGF-β1), advanced glycosylated end-products (AGEs), activation protein kinase C (PKC) and acceleration of the aldose reductase pathway increases systemic and intra-glomerular pressure, thereby hyper-perfusion, hyper-filtration and other functional changes, which lead to leakage of proteins from the glomerular capillaries.8, 9 Further, DN is also associated with podocytes damage; which are terminally differentiated highly specialized epithelial cells. It plays an important role in maintaining the integrity of glomerular filtration barrier and acts as a critical size and charge barrier to prevent proteinuria.10 Thus, DN is associated with compromised renal function, podocyte damage and subsequent proteinuria, which ultimately lead to ESRD.
Several mechanisms such as oxidative stress, inflammation, genetic and epigenetic alterations as well as autophagy are contributed in the pathogenesis of DN.11, 12, 13 However, the exact role of autophagy has not been studied in DN. Autophagy is a catabolic cellular process by which cells degrade and recycle the dysfunctional proteins and damaged organelles to maintain the cellular homeostasis under stress conditions such as starvation, hypoxia, endoplasmic reticulum (ER)-stress and hyperglycemia.14, 15 Several reports highlighted that the dysfunctional proteins and organelles are accumulated in DN due to reduce autophagy.16, 17 Further, deficiency of autophagy leads to chronic kidney injury, which is associated with ischemia-reperfusion, hypoxia and DN.18, 19, 20
Recent studies emphasized that the epigenetic mechanisms like histone modifications (acetylation), DNA methylation and microRNA play a significant role in the development and progression of DN.21, 22 Histone deacetylases (HDACs) are the enzymes, which remove the acetyl group from lysine residues of histone proteins, while histone acetyltransferases (HATs) add the acetyl group on the histone and ultimately regulate the gene expression.23, 24 HDACs are involved in several biochemical pathways and contribute in the pathogenesis and progression of DN.25, 26, 27 HDAC1 has been found to trigger fibroblast activation, proliferation and chemokine production in the interstitial renal fibroblasts and tubular epithelial cells.22 Recently, it has been reported that HDAC2/4/5 are up-regulated in both experimental and clinical DN and play a critical role in its progression by reducing autophagy.23, 28 Moreover, podocytes exposed to high glucose and transforming growth factor-β1 (TGF-β1) increased the expression of HDAC4, suggesting its contribution in DN.23, 29 On the other hand, HDAC inhibitors possess the renoprotective and anti-fibrotic activities against various pathophysiological insults in diabetes, which confirm that HDACs play a central role in DN.21, 30, 31 Further, valproic acid showed renoprotective effect in DN by decreasing the expression of HDAC4/5 and improving autophagy through HDAC inhibition and histone acetylation.32 The kidney of STZ-induced diabetic rats, db/db mice and TGF-β1-treated NRK52-E cells showed marked elevation of HDAC-2 activity, which increased the expression of fibronectin and SMA, while decreased the expression of E-cadherin.33 Moreover, HDAC inhibitors also exert beneficial effects in various nephrotic and non-nephrotic pathological conditions through chromatin-dependent and independent mechanisms.31, 34, 35, 36, 37 Considering the recent literature, it is clearly evident that HDACs and autophagy are the key contributors in the pathogenesis and progression of DN (Table 1). This review provides the novel insights on the role of HDACs, cellular acetylation and the associated molecular signaling/pathways, which modulate the autophagy and DN with an emphasis on promising therapeutic targets.
Table 1.
Role of HDACs and their inhibitors in the development, progression and treatment of DN.
| S.No | Mechanism/Inference involved | Model/Species | References |
|---|---|---|---|
| 1 | HDAC4 contributes in the podocyte injury associated with compromise autophagy and exacerbates inflammation by HDAC4-STAT1 signaling in DN. | Rat and clinical DN | 23 |
| 2 | Sodium valproate ameliorates diabetes-induced fibrosis, proteinuria and renal damage by the inhibition of iNOS/NF-κB signaling and activation of autophagy through HDAC inhibition in diabetic rat. | Rat | 13, 32 |
| 3 | MS-275 inhibits renal fibroblast activation via TGF-β1 and EGFR signaling. | UUO-induced fibrosis in rat | 28 |
| 4 | Sodium butyrate decreases fibrosis, apoptosis and DNA damage in kidney by HDAC inhibition. | Juvenile diabetic rat | 11 |
| 5 | HDAC inhibition by vorinostat attenuates diabetes-associated kidney growth via reducing eNOS activity and oxidative stress as well as epigenetic modification of EGFR. | Proximal tubule cells culture and diabetic animals | 11, 65, 66 |
| 6 | Valproic acid and class I selective HDAC inhibitor SK-7041 prevent TGF-β1-induced ECM accumulation and EMT by the inhibition of HDAC2. | Rat and NRK52-E cells. | 33 |
| 7 | TSA prevents TGF-β1-induced apoptosis by inhibiting ERK activation in human renal proximal tubular epithelial cells. | In vitro | 29 |
Note: Sirtuins (SIRTs), particularly SIRT1 activators are also exerted the renoprotective effects in DN, but not listed here, because its activity is decreased in DN due to reduce level of NAD+, which is an essential co-factor for SIRTs activity.
Autophagy: an overview
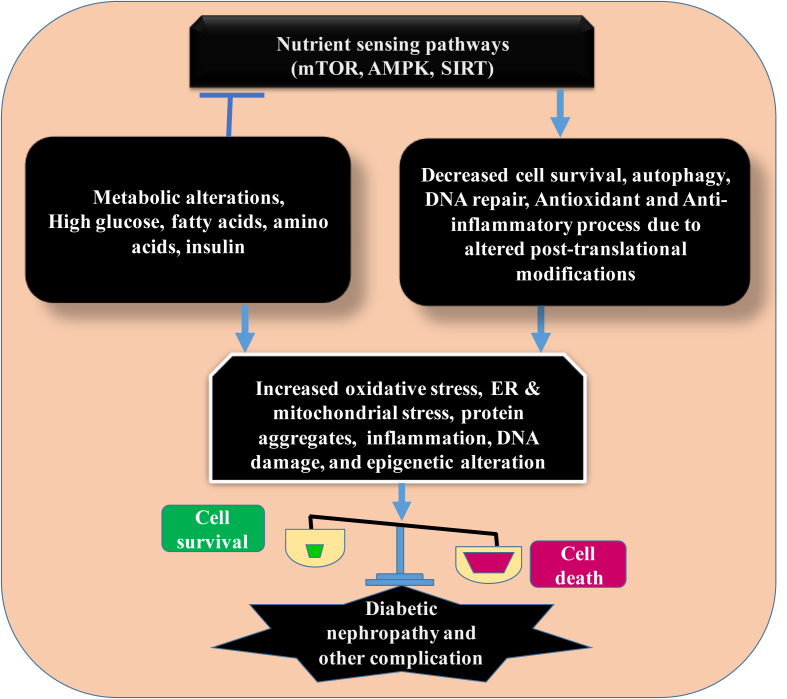
Autophagy is a cellular catabolic process, which degrades and recycles the unwanted proteins and organelles in cell to maintain cellular homeostasis under stress and pathological conditions.36, 38, 39 In the other words, autophagy recycles the intracellular energy resources in response to nutrient depletion, removes the cytotoxic proteins and organelles under physiological conditions (Fig. 1).14, 40 Every cell has a unique feature to sense the nutrients availability and produce a specific response via adenosine monophosphate (AMP)-activated protein kinase (AMPK), mammalian target of rapamycin (mTOR) and sirtuins (SIRTs) mediated nutrient sensing pathways for the cellular homeostasis.41, 42, 43 Apart from this, cellular energy level regulates autophagy through the nutrient-sensing signaling depending on cell requirement.19, 44 Depletion of energy level activates autophagy by the inhibition of mTOR or activation of AMPK and SIRT1, to degrade the unnecessary cellular components, thereby providing energy and other substrate to the cell.43, 44, 45, 46 The above pathways regulate the efficient nutrient utilization by autophagy for the cell growth and survival (Fig. 2). In general, these pathways are directly and indirectly involved in the pathogenesis of diabetes, cancer and obesity. Further, nutrient regulatory pathway modulates the post-translational modifications including HDAC-mediated de-acetylation of various target proteins of autophagy as well as many physiological processes.45, 47, 48
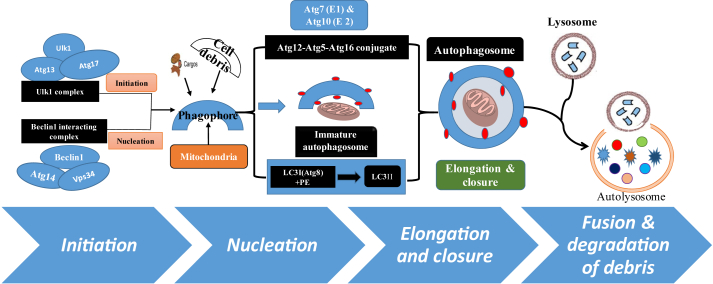
Figure 1.
Schematic diagram showing the overview of basic and molecular processes involved in autophagy. The process of autophagy involves a series of dynamic membrane rearrangements regulated by a set of ATG proteins, which can be modulated by the post-translational modifications (re-review in ref. 52, 53 and 70). In brief, it involves the following key steps i) control of phagophore formation by beclin-1/VPS34 at the ER and other membranes in response to stress such as starvation or diabetes; ii) Atg5–Atg12 conjugation, interaction with Atg16L and polymerization at the immature phagophore; iii) the LC3 processing and insertion into the extending phagophore membrane; iv) then engulfing of random or selective targets for degradation, completion of the autophagosome and recycling of some LC3-II/ATG8 v) and finally fusion of the autophagosome with the lysosome and proteolytic degradation by lysosomal proteases of engulfed molecules or cellular debris.
Figure 2.
Schematic diagram showing the influences on cell survival and death in diabetes. The metabolic changes in diabetes induce various cellular stresses, which inhibit the regulation of protective mechanisms by nutrient/energy sensing pathways. The equilibrium between the two opposing processes is altered during the progression of DN, which modulate the cell survival pathways.
The process of autophagy involves a series of dynamic membrane rearrangements regulated by a set of ATG proteins.49 This comprise of four protein complexes, which constitute core molecular machinery i.e., the kinase complex ATG1–ATG13 (initiator), phosphatidylinositol-3 kinase (PI3K) complex I (BECN1, ATG14, PIK3C3/VPS34 and PIK3R4/VPS15) as well as two ubiquitin-like protein conjugation complexes i.e, ATG12–ATG5-ATG16L1 and LC3–PE.24, 50 In brief, the process of autophagy is accomplished in the following steps.
Initiation
Autophagy is triggered by Ulk1 (unc-51-like kinase), a Ser–Thr protein kinase/ATG1 mediated phosphorylation of ATG13 and FIP 200 or RB1-inducible coiled-coil protein 1 (RB1CC1 or ATG 17), which are necessary for phagophore formation.51
Nucleation
Formation of the phagophore (pre-autophagosomal) by beclin-1 complex, composed of beclin-1/ATG6, B-cell lymphoma 2 (BCL-2) family proteins and large macro-molecule complex also known as class III PI3K vacuolar protein sorting 34 and ATG14 as well as Vps15.52
Elongation and closure
Autophagosomal maturation involves two ubiquitin-like conjugation systems; ATG12, which form the ATG12–ATG5–ATG16 complex and microtubule-associated protein light chain 3 (LC3/ATG8) system.53 The LC3-phosphatidylethanolamine (PE)-conjugation occurs by a pair of consecutive ubiquitination-like reactions catalyzed by E1-like enzyme ATG7 and E2-like enzyme ATG3, which forms LC3-phosphatidylethanolamine conjugate (LC3-II).53, 54 Thus, LC3-II formation is recognized as a marker for the presence of autophagosomes in cell culture and animal experiments.55, 56
Fusion and breakdown of contents
After formation the autophagosomes, it merges with the lysosomal compartment to form autolysosomes for acid hydrolase-mediated hydrolysis/degradation of cellular unwanted and toxic materials.53
Based on the mechanism and function, autophagy is divided into three types; macroautophagy, microautophagy and chaperone-mediated autophagy, which are controlled by the evolutionarily conserved autophagy related genes (ATG). The activity of core ATG, transcription factors and cytoskeleton proteins are tightly regulated under normal physiological condition to maintain the basal level of autophagy.24, 50
Role of autophagy in the progression and treatment of DN
Several studies reported the compromised autophagy in DN, obesity, aging and neurodegenerative disorders.20, 57, 58 The kidney cells seem to be more susceptible to the metabolic alterations due to high level of basal autophagy.15, 19 Thus, deficiency of autophagy might play a critical role in the progression of diabetes-associated renal damages.15, 59 In diabetes, the altered nutrient levels (glucose and others) triggered the specific response in the kidney cell and modulated the post-translational modifications like acetylation of target proteins and ultimately autophagy.45, 47 In experimental models of type-1 and type-2 diabetes, the expression and activity of SIRT1 have been reduced in the kidney.60, 61, 62 In addition, SIRT1 plays essential role in metabolic alterations and protects against the high-fat diet (HFD)-induced insulin-resistance and hyperglycemia due to decrease activity of the SIRT1.63 Moreover, selenium nanoparticles and rhein reduced the oxidative stress and renal injury in type-1 and type-2 diabetes-associated DN by the increased action of SIRT1, respectively.46, 61 Further, accelerated renal fibrosis and apoptosis are observed in unilateral ureteral obstruction (UUO) model due to decrease activity of SIRT1.64 Thus, SIRT1 might affect the energy balance of cell and/or acetylation of various proteins, which ultimately modulates the autophagy, thereby contributes in the DN.
Autophagy influences glomerular disease susceptibility and also serves as an essential physiological process for the maintenance of post-mitotic cells like podocyte.19 Deletion of podocyte specific ATG5 leads to glomerulopathy and proteinuria in aging mice due to limited capacity of regeneration in the podocyte.15 Valproic acid, a HDAC inhibitor exerts reno-protective effects and prevents the podocyte effacement by inactivation of iNOS/NF-κB signaling as well as facilitates the autophagy through HDAC inhibition.13, 32 Another report highlighted that HDACs and histone acetylation control the nitric oxide (NO) production through eNOS/iNOS activity in DN and HDAC inhibition ameliorated the TGF-β1-induced fibrogenesis, apoptosis and DNA damage.11, 65, 66 It can be postulated that kidney damage in diabetes might be triggered by hyperglycemia, which subsequently leads to perturbations in autophagy and associated signaling. Thus, it can be concluded that autophagy is an important survival process, which, helps to overcome stress-induced pathological perturbations and might be a promising strategy for the prevention and treatment of DN.
Role of protein acetylation in autophagy regulation and DN
Autophagy is regulated by the post-translational modifications of ATG proteins such as acetylation and phosphorylation in response to nutrients status or growth factor levels as well as various stress and pathological conditions. Starvation activates the SIRT1, which deacetylates the ATG7, ATG12, ATG5 and LC-3 (ATG8).19, 67 Further, starvation also facilitates the acetylation of ATG3 by KAT5, which regulates its interaction with microtubule-associated proteins 1A/1B light chain 3B (LC3B) as well as lipidation of LC3B.24 Nutrients excess (diabetes) increase the HDACs activity and reduce the acetylation of ATG3 and autophagy in skeletal muscle homeostasis in mice.24, 31 Forkhead box protein O (FOXO), an evolutionarily conserved transcription factor, regulates many biological processes like autophagy, cell proliferation, cell death, DNA repair and metabolism.68, 69 The acetylation status of FOXO1 and FOXO3 controls its transcription activation and modulates the various genes required for autophagosome formation.70, 71 Moreover, the de-acetylated FOXO also facilitates the expression of Ras-related protein Rab-7 (RAB7), which is responsible for the fusion of autophagosome and lysosome.24, 69 Similarly, HDAC6 regulates the autophagy by recruiting the actin and tubulin networks as well as cortactin to ubiquitinated protein aggregates by de-acetylation.72 Thus, it can be considered that HDAC6-mediated partial LC3B-II de-acetylation might be involved in autophagy during serum starvation.2 Additionally, it has also been reported that acetylated microtubules are necessary for the fusion of autophagosomes with lysosomes to form autolysosomes.24, 69 In contrast, acetylation of ATG5, ATG7, ATG8 and ATG12 by p300, an acetyltransferase inhibits the process of elongation and maturation of the autophagosome membranes in in-vitro experiments.24, 73 on the other hand, different HDACs can deacetylate the lysine residue of histone H3 (H3K56) and modulate the transcription of particular ATG genes, thereby autophagy.74, 75 Considering the current literature, it can be postulated that the protein acetylation plays a critical role in the regulation of autophagy. However, the overall role of protein acetylation on autophagy is still controversial, because both acetylation and de-acetylation of protein at specific targets can modulate the process of autophagy.70, 71
Recently, HDAC inhibition by VPA ameliorates the podocyte and renal injuries by the activation of autophagy and the inactivation of NF-kB/iNOS signaling in kidney of diabetic rat.32 Further, HDAC4 deacetylates the STAT1, which facilitates its phosphorylation and the subsequent nuclear trans-location and ultimately leads to inflammation and apoptosis as well as reduce the expression of ATGs (autophagy).23, 76 In contrast, it has been reported that the acetylation of NF-κB (p65) and STAT3 are increased in both mouse and human diabetic kidney with the progression of DN due to decrease activity of SIRT1, most probably because of reduced level of NAD+, which is an essential co-factor for SIRTs activity.77 Moreover, a growing number of non-histone proteins, such as p53, FOXO, NF-κB, STAT3 and LC3II-B are identified as substrates of HDACs or SITRs and de-acetylation of these proteins can modulate the autophagy.78 On the other hand, TGF-β1 and other genes expression including HDACs have been up-regulated during the fibrogenesis and renal damage in diabetic kidney of rats.13, 48 However, it has been reported that exposure of TGF-β1 to human renal proximal tubular epithelial cells up-regulated the autophagy-related genes such as ATG5, ATG7 and beclin-1. Despite the critical role of acetylation/de-acetylation in autophagy, the exact molecular signaling of autophagy should be explored to answer how HDACs and SIRTs suppress the autophagy in DN despite their opposite level exist in diabetes.20, 41, 76 Taken together, HDACs with other molecular signaling can compromise the autophagy and significantly contributed in the development and progression of DN (Fig. 3). Further, there is urgent need to investigate the exact role of different HDAC isoforms not only in the regulation of autophagy, but also in the other physiological conditions to understand their exact contribution in physiology and development of DN (Table 2).
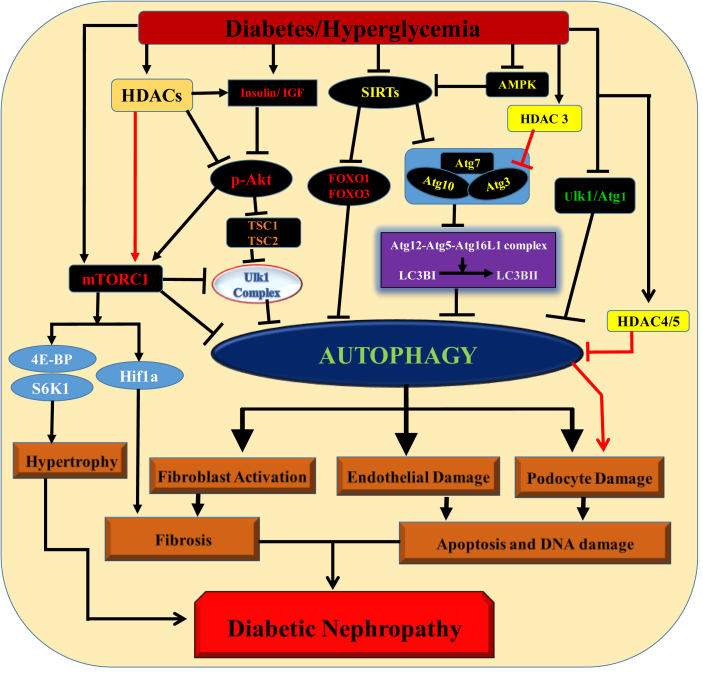
Figure 3.
Role of HDACs and associated pathways involved in the processes of autophagy during the progression of DN. High glucose level under diabetic condition inhibits PI3K/AKT, AMPK and SIRT1 signaling as well as activates mTORC1 pathway, which ultimately lead to the development and progression of DN. Reduced autophagy activates fibroblast, endothelial and podocyte damage leading to fibrosis, apoptosis and DNA damage, which ultimately contributes in the progression of DN.
Table 2.
Role of different HDACs or SIRTs isoforms and protein acetylation status in the regulation of autophagy in DN.
| HDAC classes | HDAC isoforms | Specific role and involvement in DN | References |
|---|---|---|---|
| Class I | HDAC 1 | Did not contribute in DN as well as autophagy. | 23 |
| HDAC 2 | Up-regulated and contributed in DN and may contribute in autophagy. | 23, 33 | |
| HDAC 3 | Did not contribute in DN as well as autophagy. | 23 | |
| HDAC 8 | Did not contribute in DN as well as autophagy. | 23 | |
| Class IIa | HDAC 4& 5 | Up-regulated in DN and played a major role in the pathogenesis of DN by the inhibition of autophagy. | 23, 32 |
| HDAC 7 | Up-regulated and contributed in the diabetic kidney damages, but its effect on autophagy is still unexplored. | 13 | |
| HDAC 9 | Did not contribute in DN as well as autophagy. | 23 | |
| Class IIb | HDAC 6 | Did not contribute in DN as well as autophagy. | 23 |
| HDAC 10 | Did not contribute in DN as well as autophagy. | 23 | |
| Class III (Sirtuins or SIRTs) | SIRT1 | Reduced activity and subsequently compromised the autophagy in DN and aging. Reduced activity of SIRT1 can acetylate the various proteins such as FOXO, NF-kB, STAT3, p53 and LC3II-B, which directly or indirectly suppressed autophagy in DN. | 19, 41, 46 |
| SIRT2-7 | No report available on their role in the pathogenesis of DN and autophagy so far. | – | |
| Class IV | HDAC 11 | Did not contribute in DN as well as autophagy. | 23 |
Note: The above reports presented here are summarizing the current status of HDACs and SIRTs and their contribution in DN and autophagy. Since different isoform of HDACs and SIRTs are expressed in a tissue specific manner under physiological and pathological conditions. Therefore, the role of different HDACs and SIRTs may vary in different pathological conditions, even in the different tissues of the same pathological condition. Therefore, the acetylation/de-acetylation of different autophagic proteins by HDACs and SIRTs, particularly in DN might play a very complex role in autophagy, which cannot be explained in general.
The process of de-acetylation differs markedly between sirtuins (SIRTs) and HDACs. While class I, II, and IV HDACs transfer the final acetyl group and are sensitive to the inhibitor trichostatin A (TSA), while sirtuins (SIRTs) require NAD+ as an enzymatic co-factor, transfer the acetyl group from the substrate to an ADP-ribose molecule, and are insensitive to TSA or other HDAC inhibitors. So the mechanisms and activities as well as the de-acetylation by SIRTs and HDACs are greatly modulated by the pathological conditions, particularly in diabetes.79 Further, class I, II and IV HDACs are up-regulated in DN (are pathogenic in nature) and their inhibitors exert beneficial effects in diabetic patients and rodents. In contrast, SIRTs (III HDACs) are suppressed in diabetes due to decreased NAD+ level and activation of SIRTs by resveratrol or calorie restriction promotes energy metabolism, thereby exerts beneficial effects in diabetes/DN.47, 80 Additionally, both type of enzymes also have non-histone targets and directly or indirectly modulate the cellular acetylation status and subsequently affect the activities of various autophagy proteins.76, 81 Thus, HDACs and SIRTs may regulate the conformation, localization, molecular interaction, and function of various autophagy proteins by controlling their acetylation status.
Although, there is an opposite levels of SIRTs and HDACs in DN, but overall, the global de-acetylation by the SIRTs and HDACs in DN suppresses the autophagy through regulation of the conformation, localization, molecular interaction and function of various target proteins and transcription factors, which directly and/or indirectly promote the autophagy. However, de-acetylation on specific molecules may have a very critical role on autophagy. Therefore, targeting de-acetylation/acetylation on specific target molecule will provide the exact role of SIRTs and HDACs on the regulation of autophagy in DN. Further, both type of enzymes have non-histone targets and directly or indirectly modulate the cellular acetylation status and subsequently affect the activities of various autophagy proteins.24, 80, 82 Moreover, SIRTs (SIRT1) activators exert an anti-apoptotic and pro-autophagic responses in cells under stress conditions as well as diabetes by directly de-acetylating essential ATGs such as ATG5, ATG7 and ATG8 as well as by de-acetylation of various transcription factors such as FOXO3 and ultimately increased the expression of autophagy proteins.62 Similarly, HDAC inhibitors also promote the autophagy through transcriptional activation of FOXO1 and up-regulating ATGs as well as suppression of mTOR signaling by phosphorylation of AKT.20, 71 Considering the current literature, protein acetylation, an important form of post-translation modification control by HATs and HDACS, is involved in the regulation of autophagy. Intriguingly, it is still controversial how protein acetylation modulates autophagy. In diabetes, over expression of HDACs may de-acetylated the histones and non-histone target proteins at specific sites and modulate the molecular signaling of autophagy, thereby ultimately suppress the autophagy.23, 62 In contrast, reduced activity of SIRTs can acetylate the various proteins such as FOXO, NF-kB, STAT3, p53 and LC3II-B, which directly or indirectly suppress autophagy in DN. Therefore, acetylation/de-acetylation of different autophagy proteins by HDACs and SIRTs, particularly in DN might have very complex role in autophagy, which cannot be explained in general. The future studies will elucidate that how different HDACs or SIRTs and subsequent protein acetylation are involved in autophagy and eventually in DN.
Conclusion and future perspective
Several molecular signaling are involved in the pathogenesis and progression of DN, which can be modulated by both genetic and epigenetic mechanisms. The emerging evidences highlighted that epigenetic mechanisms including histone modifications play a critical role in the pathogenesis of DN. The altered cellular acetylation and subsequent modulation of genes and transcription factors affect the autophagy in DN. Modulation of acetylation status of ATG proteins may be one of the best possible targets to promote the autophagy and tissue repair to attenuate kidney damage in DN. Thus, modulation of cellular acetylation by multiple ways can emerge as a new therapeutic approach for the prevention of DN. The different HDACs serve very distinct physiological functions and currently available HDAC inhibitors are mostly nonselective (pan-inhibitors) in nature and inhibit multiple HDACs. In addition, the use of pan-HDAC inhibitors might be associated with adverse side effects due to unknown physiological functions.83, 84 Therefore, it is very important to delineate the exact biological role of individual HDACs in the normal physiology and pathological conditions, particularly in DN. Thus, targeting the most relevant HDAC isoform in a particular signaling during the progression of DN may greatly improve the efficacy of novel HDAC inhibitors with minimal chance of toxicities.
In conclusion, here we present the role of cellular acetylation in the regulation of autophagy and its possible usefulness for the treatment of DN. Thus, we conclude that the regulation of autophagy could be an attractive therapeutic strategy for the prevention and treatment of DN. The detail understanding of the biological and pathological roles of HDACs in tissue specific manner will provide pinpoint targets for the DN, which can promote the designing and synthesis of more selective inhibitors for promising drug molecules for the treatment of DN. On the other hand, advancement of the synthetic technology and drug designing can allow the medicinal chemists to design and synthesize more specific HDAC inhibitors, which can provide a new dimension to drug discovery and development for the treatment of DN.
Conflicts of interest
The authors declare that there is no conflict of interest.
Acknowledgments
This work has been funded by National Institute of Pharmaceutical Education and Research, SAS Nagar, Punjab, India.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Yamahara K., Yasuda M., Kume S. The role of autophagy in the pathogenesis of diabetic nephropathy. J Diabetes Res. 2013;2013:193757. doi: 10.1155/2013/193757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoi H., Mukoyama M., Mori K. Overexpression of connective tissue growth factor in podocytes worsens diabetic nephropathy in mice. Kidney Int. 2008;73:446–455. doi: 10.1038/sj.ki.5002722. [DOI] [PubMed] [Google Scholar]
- 3.Riser B.L., Najmabadi F., Perbal B. CCN3/CCN2 regulation and the fibrosis of diabetic renal disease. J Cell Commun Signal. 2010;4:39–50. doi: 10.1007/s12079-010-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozieh M.N., Dismuke C.E., Lynch C.P. Medical care expenditures associated with chronic kidney disease in adults with diabetes: United States 2011. Diabetes Res Clin Pract. 2015;109:185–190. doi: 10.1016/j.diabres.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schena F.P., Gesualdo L. Pathogenetic mechanisms of diabetic nephropathy. J Am Soc Nephrol. 2005;16(suppl 1):S30–S33. doi: 10.1681/asn.2004110970. [DOI] [PubMed] [Google Scholar]
- 6.Osterby R., Gundersen H.J. Glomerular size and structure in diabetes mellitus. I. Early abnormalities. Diabetologia. 1975;11:225–229. doi: 10.1007/BF00422326. [DOI] [PubMed] [Google Scholar]
- 7.Vallon V., Richter K., Blantz R.C. Glomerular hyperfiltration in experimental diabetes mellitus: potential role of tubular reabsorption. J Am Soc Nephrol. 1999;10:2569–2576. doi: 10.1681/ASN.V10122569. [DOI] [PubMed] [Google Scholar]
- 8.Raptis A.E., Viberti G. Pathogenesis of diabetic nephropathy. Exp Clin Endocrinol Diabetes. 2001;109(suppl 2):S424–S437. doi: 10.1055/s-2001-18600. [DOI] [PubMed] [Google Scholar]
- 9.Soetikno V., Arozal W., Louisa M. New insight into the molecular drug target of diabetic nephropathy. Int J Endocrinol. 2014;2014 doi: 10.1155/2014/968681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankland S.J. The podocyte's response to injury: role in proteinuria and glomerulosclerosis. Kidney Int. 2006;69:2131–2147. doi: 10.1038/sj.ki.5000410. [DOI] [PubMed] [Google Scholar]
- 11.Khan S., Jena G. Sodium butyrate, a HDAC inhibitor ameliorates eNOS, iNOS and TGF-beta1-induced fibrogenesis, apoptosis and DNA damage in the kidney of juvenile diabetic rats. Food Chem Toxicol. 2014;73:127–139. doi: 10.1016/j.fct.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Ding Y., Choi M.E. Regulation of autophagy by TGF-beta: emerging role in kidney fibrosis. Semin Nephrol. 2014;34:62–71. doi: 10.1016/j.semnephrol.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S., Jena G., Tikoo K. Sodium valproate ameliorates diabetes-induced fibrosis and renal damage by the inhibition of histone deacetylases in diabetic rat. Exp Mol Pathol. 2015;98:230–239. doi: 10.1016/j.yexmp.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartleben B., Godel M., Meyer-Schwesinger C. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins G.C., Coughlan M.T. Mitochondrial dysfunction and mitophagy: the beginning and end to diabetic nephropathy? Br J Pharmacol. 2014;171:1917–1942. doi: 10.1111/bph.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kume S., Yamahara K., Yasuda M. Autophagy: emerging therapeutic target for diabetic nephropathy. Semin Nephrol. 2014;34:9–16. doi: 10.1016/j.semnephrol.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Kimura T., Takabatake Y., Takahashi A. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol. 2011;22:902–913. doi: 10.1681/ASN.2010070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kume S., Uzu T., Horiike K. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Y., Choi M.E. Autophagy in diabetic nephropathy. J Endocrinol. 2015;224:R15–R30. doi: 10.1530/JOE-14-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert R.E., Huang Q., Thai K. Histone deacetylase inhibition attenuates diabetes-associated kidney growth: potential role for epigenetic modification of the epidermal growth factor receptor. Kidney Int. 2011;79:1312–1321. doi: 10.1038/ki.2011.39. [DOI] [PubMed] [Google Scholar]
- 22.Pang M., Ma L., Liu N. Histone deacetylase 1/2 mediates proliferation of renal interstitial fibroblasts and expression of cell cycle proteins. J Cell Biochem. 2011;112:2138–2148. doi: 10.1002/jcb.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Liu J., Zhen J. Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int. 2014;86:712–725. doi: 10.1038/ki.2014.111. [DOI] [PubMed] [Google Scholar]
- 24.Banreti A., Sass M., Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013;9:819–829. doi: 10.4161/auto.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H.B., Noh H., Seo J.Y. Histone deacetylase inhibitors: a novel class of therapeutic agents in diabetic nephropathy. Kidney Int Suppl. 2007:S61–S66. doi: 10.1038/sj.ki.5002388. [DOI] [PubMed] [Google Scholar]
- 26.Villeneuve L.M., Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 2010;299:F14–F25. doi: 10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan S., Kumar S., Jena G. Valproic acid reduces insulin-resistance, fat deposition and FOXO1-mediated gluconeogenesis in type-2 diabetic rat. Biochimie. 2016;125:42–52. doi: 10.1016/j.biochi.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Liu N., He S., Ma L. Blocking the class I histone deacetylase ameliorates renal fibrosis and inhibits renal fibroblast activation via modulating TGF-beta and EGFR signaling. PLoS One. 2013;8:e54001. doi: 10.1371/journal.pone.0054001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshikawa M., Hishikawa K., Idei M. Trichostatin a prevents TGF-beta1-induced apoptosis by inhibiting ERK activation in human renal tubular epithelial cells. Eur J Pharmacol. 2010;642:28–36. doi: 10.1016/j.ejphar.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 30.Marumo T., Hishikawa K., Yoshikawa M. Histone deacetylase modulates the proinflammatory and -fibrotic changes in tubulointerstitial injury. Am J Physiol Renal Physiol. 2010;298:F133–F141. doi: 10.1152/ajprenal.00400.2009. [DOI] [PubMed] [Google Scholar]
- 31.Moresi V., Carrer M., Grueter C.E. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc Natl Acad Sci U S A. 2012;109:1649–1654. doi: 10.1073/pnas.1121159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan S., Jena G., Tikoo K. Valproate attenuates the proteinuria, podocyte and renal injury by facilitating autophagy and inactivation of NF-kappaB/iNOS signaling in diabetic rat. Biochimie. 2015;110:1–16. doi: 10.1016/j.biochi.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Noh H., Oh E.Y., Seo J.Y. Histone deacetylase-2 is a key regulator of diabetes- and transforming growth factor-beta1-induced renal injury. Am J Physiol Renal Physiol. 2009;297:F729–F739. doi: 10.1152/ajprenal.00086.2009. [DOI] [PubMed] [Google Scholar]
- 34.Khan S., Jena G.B. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: study in juvenile diabetic rat. Chem Biol Interact. 2014;213:1–12. doi: 10.1016/j.cbi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Aher J.S., Khan S., Jain S. Valproate ameliorates thioacetamide-induced fibrosis by hepatic stellate cell inactivation. Hum Exp Toxicol. 2015;34:44–55. doi: 10.1177/0960327114531992. [DOI] [PubMed] [Google Scholar]
- 36.Wilson C.M., Magnaudeix A., Yardin C. Autophagy dysfunction and its link to Alzheimer's disease and type II diabetes mellitus. CNS Neurol Disord Drug Targets. 2014;13:226–246. doi: 10.2174/18715273113126660146. [DOI] [PubMed] [Google Scholar]
- 37.Kanika G., Khan S., Jena G. Sodium butyrate ameliorates l-arginine-induced pancreatitis and associated fibrosis in wistar rat: role of inflammation and nitrosative stress. J Biochem Mol Toxicol. 2015;29:349–359. doi: 10.1002/jbt.21698. [DOI] [PubMed] [Google Scholar]
- 38.Hill B.G., Benavides G.A., Lancaster J.R., Jr. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murrow L., Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu M., Waguri S., Ueno T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kume S., Thomas M.C., Koya D. Nutrient sensing, autophagy, and diabetic nephropathy. Diabetes. 2012;61:23–29. doi: 10.2337/db11-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallows K.R., Mount P.F., Pastor-Soler N.M. Role of the energy sensor AMP-activated protein kinase in renal physiology and disease. Am J Physiol Renal Physiol. 2010;298:F1067–F1077. doi: 10.1152/ajprenal.00005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J., Kundu M., Viollet B. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee I.H., Cao L., Mostoslavsky R. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imai S., Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar G.S., Kulkarni A., Khurana A. Selenium nanoparticles involve HSP-70 and SIRT1 in preventing the progression of type 1 diabetic nephropathy. Chem Biol Interact. 2014;223C:125–133. doi: 10.1016/j.cbi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 47.Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowes A.J., Khan M.I., Shi Y. Valproate attenuates accelerated atherosclerosis in hyperglycemic apoE-deficient mice: evidence in support of a role for endoplasmic reticulum stress and glycogen synthase kinase-3 in lesion development and hepatic steatosis. Am J Pathol. 2009;174:330–342. doi: 10.2353/ajpath.2009.080385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z., Klionsky D.J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganley I.G., Lam du H., Wang J. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pattingre S., Espert L., Biard-Piechaczyk M. Regulation of macroautophagy by mTOR and beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 53.Glick D., Barth S., Macleod K.F. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravikumar B., Sarkar S., Davies J.E. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 55.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klionsky D.J., Abeliovich H., Agostinis P. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frake R.A., Ricketts T., Menzies F.M. Autophagy and neurodegeneration. J Clin Invest. 2015;125:65–74. doi: 10.1172/JCI73944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui J., Shi S., Sun X. Mitochondrial autophagy involving renal injury and aging is modulated by caloric intake in aged rat kidneys. PLoS One. 2013;8:e69720. doi: 10.1371/journal.pone.0069720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang L., Zhou Y., Cao H. Autophagy attenuates diabetic glomerular damage through protection of hyperglycemia-induced podocyte injury. PLoS One. 2013;8:e60546. doi: 10.1371/journal.pone.0060546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tikoo K., Singh K., Kabra D. Change in histone H3 phosphorylation, MAP kinase p38, SIR 2 and p53 expression by resveratrol in preventing streptozotocin induced type I diabetic nephropathy. Free Radic Res. 2008;42:397–404. doi: 10.1080/10715760801998646. [DOI] [PubMed] [Google Scholar]
- 61.Chen W., Chang B., Zhang Y. Rhein promotes the expression of SIRT1 in kidney tissues of type 2 diabetic rat. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2015;31:615–619. [PubMed] [Google Scholar]
- 62.Yacoub R., Lee K., He J.C. The role of SIRT1 in diabetic kidney disease. Front Endocrinol (Lausanne) 2014;5:166. doi: 10.3389/fendo.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfluger P.T., Herranz D., Velasco-Miguel S. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He W., Wang Y., Zhang M.Z. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest. 2010;120:1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Advani A., Huang Q., Thai K. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. Am J Pathol. 2011;178:2205–2214. doi: 10.1016/j.ajpath.2011.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu Z., Zhang W., Kone B.C. Histone deacetylases augment cytokine induction of the iNOS gene. J Am Soc Nephrol. 2002;13:2009–2017. doi: 10.1097/01.asn.0000024253.59665.f1. [DOI] [PubMed] [Google Scholar]
- 67.Kume S., Koya D., Uzu T. Role of nutrient-sensing signals in the pathogenesis of diabetic nephropathy. Biomed Res Int. 2014;2014:315494. doi: 10.1155/2014/315494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brunet A., Sweeney L.B., Sturgill J.F. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 69.Hariharan N., Maejima Y., Nakae J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wani W.Y., Boyer-Guittaut M., Dodson M. Regulation of autophagy by protein post-translational modification. Lab Invest. 2015;95:14–25. doi: 10.1038/labinvest.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J., Ng S., Wang J. Histone deacetylase inhibitors induce autophagy through FOXO1-dependent pathways. Autophagy. 2015;11:629–642. doi: 10.1080/15548627.2015.1023981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iwata A., Riley B.E., Johnston J.A. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 73.Pietrocola F., Lachkar S., Enot D.P. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 2015;22:509–516. doi: 10.1038/cdd.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaquero A., Sternglanz R., Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 75.Oh M., Choi I.K., Kwon H.J. Inhibition of histone deacetylase1 induces autophagy. Biochem Biophys Res Commun. 2008;369:1179–1183. doi: 10.1016/j.bbrc.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 76.Wei Q., Dong Z. HDAC4 blocks autophagy to trigger podocyte injury: non-epigenetic action in diabetic nephropathy. Kidney Int. 2014;86:666–668. doi: 10.1038/ki.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu R., Zhong Y., Li X. Role of transcription factor acetylation in diabetic kidney disease. Diabetes. 2014;63:2440–2453. doi: 10.2337/db13-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Periyasamy-Thandavan S., Jiang M., Wei Q. Autophagy is cytoprotective during cisplatin injury of renal proximal tubular cells. Kidney Int. 2008;74:631–640. doi: 10.1038/ki.2008.214. [DOI] [PubMed] [Google Scholar]
- 79.Martinez-Redondo P., Vaquero A. The diversity of histone versus nonhistone sirtuin substrates. Genes Cancer. 2013;4:148–163. doi: 10.1177/1947601913483767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ye J. Improving insulin sensitivity with HDAC inhibitor. Diabetes. 2013;62:685–687. doi: 10.2337/db12-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koya D., Kitada M., Kume S. Interventions against nutrient-sensing pathways represent an emerging new therapeutic approach for diabetic nephropathy. Clin Exp Nephrol. 2014;18:210–213. doi: 10.1007/s10157-013-0908-3. [DOI] [PubMed] [Google Scholar]
- 82.Zhao Y., Yang J., Liao W. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 83.Khan S., Ahmad T., Parekh C.V. Investigation on sodium valproate induced germ cell damage, oxidative stress and genotoxicity in male Swiss mice. Reprod Toxicol. 2011;32:385–394. doi: 10.1016/j.reprotox.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 84.Khan S., Jena G.B. Effect of sodium valproate on the toxicity of cyclophosphamide in the testes of mice: influence of pre- and post-treatment schedule. Toxicol Int. 2013;20:68–76. doi: 10.4103/0971-6580.111562. [DOI] [PMC free article] [PubMed] [Google Scholar]