Abstract
The intervertebral disc (IVD) comprises a gelatinous inner core (nucleus pulposus; NP) and concentric rings (annulus fibrosus; AF). The NP, an important structure for shock absorption in the vertebrate spinal motion segment, can be traced back to the notochord in ontogenetic lineage. In vertebrates, the notochord undergoes mucinoid changes, and had been considered vestigial until recently. However, observed correlations between IVD degeneration and back pain in humans have renewed interest in the IVD in biomedical fields.
Beyond its mechanical contribution to development, the notochord is also an essential signaling center, which coordinates formation of the neural tube and somites. The pertinent signaling molecules, particularly TGF-β and bone morphogenetic proteins (BMPs), continue to play roles in the adult tissues and have been utilized for tissue regeneration. Genetic factors are major determinants of who will develop IVD degeneration and related back pain, and seem to correlate better with disc degeneration and back pain than do external forces on the spine.
In summary, the spinal column is a landmark development in evolution. Genes directing the development of the IVD may also contribute to its maintenance, degeneration, and regeneration. Likewise, structural genes as well as genes responsible for maintenance of the structure are related to IVD degeneration. Finally, genes responsible for inflammation may play a dual role in exacerbating degeneration or facilitating repair responses depending on the context.
Keywords: Back pain, Development, Intervertebral disc (IVD), Notochord, Regeneration
“It's a Long Way from Amphioxus” was originally recorded by Sam Hilton and is the official theme song for the Biological Sciences Collegiate Division at The University of Chicago.
“A fish-like thing appeared among the annelids one day.
It hadn't any parapods nor setae to display.
It hadn't any eyes nor jaws, nor ventral nervous cord,
But it had a lot of gill slits and it had a notochord.
It wasn't much to look at and it scarce knew how to swim,
And Nereis was very sure it hadn't come from him.
The mollusks wouldn't own it and the arthropods got sore,
So the poor thing had to burrow in the sand along the shore.
He burrowed in the sand before a crab could nip his tail,
And he said “Gill slits and myotomes are all to no avail.
I've grown some metapleural folds and sport an oral hood,
But all these fine new characters don't do me any good.”
He sulked awhile down in the sand without a bit of pep,
Then he stiffened up his notochord and said, “I'll beat 'em yet!
Let 'em laugh and show their ignorance. I don't mind their jeers.
Just wait until they see me in a hundred million years.
My notochord shall turn into a chain of vertebrae
And as fins my metapleural folds will agitate the sea.
My tiny dorsal nervous cord will be a mighty brain,
And the vertebrates shall dominate the animal domain.”
Intervertebral disc development
Notochord development predates vertebrate development
The intervertebral disc (IVD) comprises a gelatinous inner core (nucleus pulposus; NP) and concentric rings (annulus fibrosus; AF). The NP, an important structure for shock absorption in the vertebrate spinal motion segment, can be traced back to the notochord in ontogenetic lineage.1 The notochord is believed to have first appeared in Amphioxus, a relict fish-like organism also known as a lancelet. The notochord in Amphioxus is a semi-flexible rod that stiffens the body and provides resistance to muscle contraction, which makes coordinated movements possible. Because of its mechanical properties, the notochord also serves to store energy for subsequent muscle contractions. In vertebrates, the notochord undergoes mucinoid changes, and had been considered vestigial until recently. However, recent studies on the biomechanical and biochemical function of the notochord have revealed that the IVD is an important structure,2 especially because IVD degeneration often correlates with back pain in humans.3
In vertebrates, the notochord is replaced by the vertebral column of the spine. The notochord transforms into the NP of the intervertebral disc, and in this new form continues to conserve energy during compression and, acting in concert with other disc substructures – the AF and the cartilaginous end plates – facilitates spinal motion. In addition to its biomechanical functions, the notochord serves as an important signaling center during development. Thus, in the first part of this review, we will discuss signaling functions of the notochord. This is of special relevance to current efforts to regenerate the intervertebral disc, since many related secreted factors have been utilized in attempts to direct various cells, including stem cells, towards an IVD cell-like phenotype.
The spinal column is the defining structure of the vertebrate body, and defects in this structure are either incompatible with life or result in serious disabilities (e.g., spina bifida). Even minor variations result in serious consequences. For example, genetic abnormalities leading to variations in cervical vertebra number can result in serious consequences, including miscarriage or childhood cancer.4 Therefore, in the second part of this review, we will discuss genetic polymorphisms of extracellular matrix genes in the IVD and their remodeling enzymes; many of these polymorphisms are related to increased disc degeneration, and a few of them are protective (resulting in less degeneration).
Vertebral column development
The anatomic position of the spine is specified early in development, shortly after gastrulation. HOX genes (also known as homeotic box genes) encode homeodomain proteins, which are highly conserved across vast evolutionary distances and are key transcription factors controlling anterior-posterior body patterning and vertebra number.5 For example, most vertebrates have 7 cervical vertebrae.4 Following establishment of the ectoderm, mesoderm, and endoderm, the notochord forms from the mesoderm at the cranial end of the embryo and extends towards the caudal end.6 The notochord defines the midline of the developing embryo as well as the dorsal–ventral and left–right axes.7 The notochord consists of epithelial-like outer sheath cells that produce a membrane composed primarily of type II collagen that isolates the notochord from the surrounding mesoderm, and an inner cell layer populated by large, vacuolated cells. The inner notochord cells absorb water, because of osmotic pressure exerted by vacuole glycosaminoglycans, and so increase in size. The mechanical interaction between the swollen inner notochord cells and the outer sheath-derived basement membrane results in the notochord's rigidity, which contributes to embryonic folding later in development.
Signaling functions of the notochord, and intervertebral disc (IVD) development
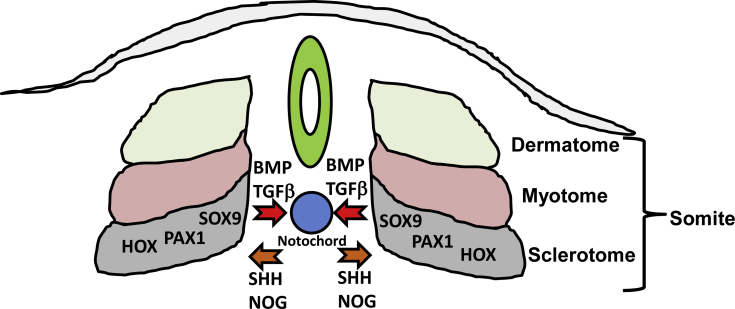
Beyond its mechanical contribution to development, the notochord is also an essential signaling center, which coordinates formation of the neural tube (precursor to the brain and spinal cord) and somites (musculoskeletal precursors; Fig.1). The notochord signals to surrounding embryonic tissue primarily via secreted signaling molecules. One primary molecule is Sonic Hedgehog (Shh), which acts on target cells through interaction with its receptor Patched (Ptch), to activate the Gli family of transcription factors. Shh specifies the floor plate of the neural tube and, critically for development of the axial skeleton, temporally regulates formation of the somites from the paraxial mesoderm.7, 8
Fig. 1.
Reciprocal communication between the sclerotome and notochord. During development, cells of the notochord release sonic hedgehog (SHH) and noggin (NOG) which facilitate segmentation of the somites into dermatome, myotome, and sclerotome. The sclerotome, in turn, governed by SOX9, PAX1, and HOX family transcription factors, releases bone morphogenetic proteins (BMPs) and transforming growth factor-β (TGF-β) which direct matrix development within the notochord.
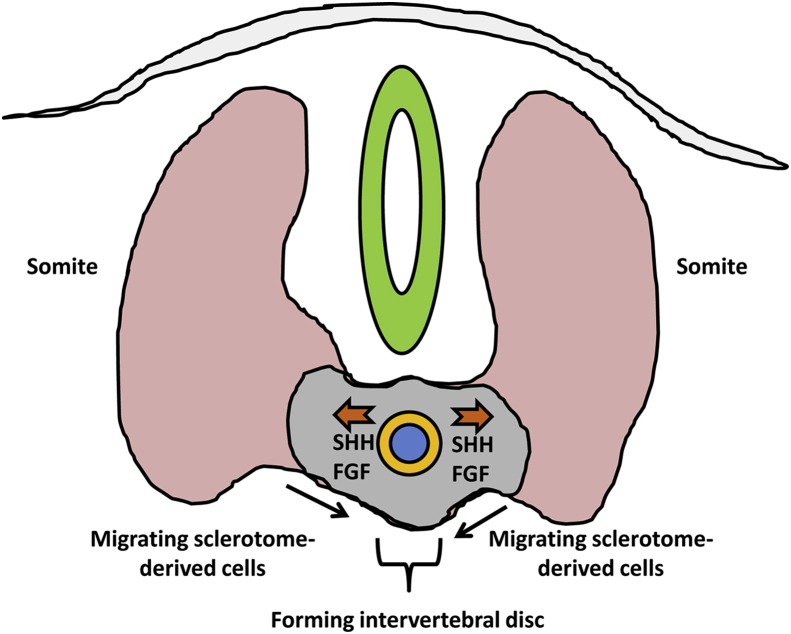
Specific regions of the somites themselves give rise to skeletal muscle (myotome), dermis (dermatome), and the AF with adjacent vertebral bodies (sclerotome; Fig.2). During formation of the vertebral bodies, the cranial half and caudal half of adjacent sclerotomes fuse, extend towards the midline, and mineralize via endochondral ossification. As discussed above, HOX genes determine the number and identity of the developing vertebral bodies.5 Two models offer explanations for inner notochord cell localization following vertebral body formation. According to the “pressure model,” as the sclerotome halves on either side of the notochord grow towards the midline, they exert pressure on the outer notochord sheath, which forces the notochord inner cell population out of the vertebral bodies and into the space between the vertebrae.9, 10 The “repulsion/attraction” model posits that chemoattractant and/or chemorepellent factors within the site of the presumptive intervertebral disc drive notochord cells into the intervertebral space; Eph/ephrin and Robo/Slit signaling pathways have been implicated in this mechanism.10 As the vertebral bodies form, other progenitors of the sclerotome migrate into the presumptive intervertebral space and form a dense matrix of fibronectin, which aligns the presumptive AF layers and fibroblast like cells which produce a matrix of collagens, including types I, II, III, and IV, in lamellar layers around the remnants of the notochord.11, 12 The proteoglycan-rich notochord remnants form the nucleus pulposus (NP).1 The development of the spine and intervertebral discs is coordinated by interplay between multiple signaling factors. Shh, described above, in addition to mediating direct effects, also alters cellular responses to Bone Morphogenetic Proteins (BMPs) to promote chondrogenesis of the nascent disc, a process supported further by Transforming Growth Factor-β (TGF-β) from the sclerotome and Noggin (Nog) from the notochord. Beyond specifying cell lineages, these molecules, particularly TGF-β and BMPs, continue to play roles in the adult and have been employed in tissue regeneration.13
Fig. 2.
Formation of the annulus fibrosus (AF; yellow) from sclerotome-derived cells and Nucleus Pulposus (NP; blue) from notochord-derived cells. As sclerotome-derived cells migrate around the notochord, cells of the notochord continue to secrete SHH and fibroblast growth factor (FGF) to drive endochondral ossification of the vertebral body. At the same time, matrix of the notochord outer sheath matures to form the intervertebral disc AF while inner notochord cells form NP.
Genes important for intervertebral disc (IVD) development
Genes important for IVD development are often required for survival of the organism, as illustrated by embryonic lethality following mutations of these genes (Table 1). Thus, neither animals nor humans with naturally occurring mutations are frequently observed. Because regeneration or repair processes often recapitulate development, genes and signaling molecules of development are often considered as a means to repair the IVD. These factors are also frequently used to stimulate cells in order to direct them into a more “IVD-like” phenotype, and/or to stimulate desirable extracellular matrix production.
Table 1.
Genes important for intervertebral disc development.
| Protein (Gene) | Embryonic source tissue(s) | Contribution to disc development | References |
|---|---|---|---|
| Sonic Hedgehog (SHH) | Notochord | Notochord sheath formation, nucleus pulposus formation, chondrogenic commitment of sclerotome cells. | Choi et al and Murtaugh et al1, 14, 15, 16 |
| Homeodomain protein (HOX) | Somite | Patterning | Pearson et al5 |
| SRY-Box 9 (SOX) | Sclerotome | Regulates somite stem cell differentiation into chondrocytes | Zhoa et al and Sugimoto et al17, 18 |
| Forkhead Box A1/2 (FOXA1/2) | Notochord | Notochord sheath formation, nucleus pulposus formation | Maier et al19 |
| Paired Box 1 (PAX1) | Sclerotome | Chondrogenesis of sclerotome cells | Peters et al20 |
| Noggin (NOG) | Notochord | Antagonizes BMP signaling, promotes Shh signaling and Pax1 activation. | McMahon et al and Wijgerde et al21, 22 |
| Transforming Growth Factor-β (TGFB) | Sclerotome | Vertebral body formation | Baffi et al and Sohn et al23, 24 |
| Bone morphogenetic protein (BMP) family | Sclerotome | In the presence of Shh, promotes chondrogenesis of sclerotome-derived disc progenitors. | Murtaugh et al15 |
Disease-related genetic polymorphisms
Genes associated with human IVD degeneration
Genetic factors are major determinants of who will develop IVD degeneration and related back pain, and seem to correlate better with disc degeneration and back pain than do external forces on the spine.25 Genetic modulations associated with human disc degeneration or back pain were separated according to the functions of the genes (e.g., structural, enzymes cleaving extracellular matrix molecules, inflammatory mediators) as described by Tegeder and Lotsch26 and Yee and Chan27 (Table 2, Table 3). It is worth noting that most of the studies listed below in Table 2 have not been replicated or the study cohorts were small, and thus the studies were mostly under-powered. Furthermore, the effect sizes are relatively small, with odds ratios in the order of 1.3–1.8. Finally, most of the published studies were based on a candidate genes approach, and thus, it was not surprising that many extracellular matrix, inflammation and matrix degradation genes were studied. Therefore, the data should be interpreted carefully. There were only two unbiased studies, which identified carbohydrate sulfotransferase 3 (CHST3)43 and parkin RBR E3 ubiquitin protein ligase (PARK2).46 Given the principal role of the IVD (to withstand high biomechanical stress), defects in structural genes (types I, IX and XI collagen and aggrecan) unsurprisingly often lead to poor biomechanical properties and premature disc degeneration (Table 2).
Table 2.
Extracellular matrix and matrix-modifying gene polymorphisms associated with intervertebral disc degeneration.
| Protein (Gene) | Disc degeneration or pain | Increase in pathology, or protective | References |
|---|---|---|---|
| Collagen IX (COL9A2) | Disc degeneration and radicular pain | Increase | Annunen et al, Seki et al and Knoeringer et al28, 29, 30 |
| Collagen IX (COL9A3) | Disc degeneration | Increase | Paassilta et al and Toktas et al31, 32 |
| Collagen XI (COL11A1) | Disc degeneration | Increase | Mio et al33 |
| Collagen XI (COL11A2) | Disc degeneration | Protective | Noponen-Hietala et al34 |
| Collagen I (COL1A1) | Disc degeneration and osteoporosis | Increase | Toktas et al and Pluijm et al32, 35 |
| Aggrecan (ACAN) | Disc degeneration | Increase with short variable number of tandem repeats | Solovieva et al and Kawaguchi et al36, 37 |
| Cartilage intermediate layer protein (CILP) | Disc degeneration | Increase | Seki et al38 |
| Asporin (ASPN) | Disc degeneration | Increase | Song et al39 |
| Thrombospondin2 (THBS2) | Disc Herniation | Increase | Hirose et al40 |
| Vitamin D Receptor (VDR) | Disc degeneration and osteoporosis | Increase | Toktas et al, Videman et al and Kawaguchi et al32, 41, 42 |
| Carbohydrate sulfotransferase 3 (CHST3) | Disc degeneration | Increase | Song et al43 |
| Growth Differentiation Factor 5 (GDF5) | Disc degeneration | Increase | Williams et al and Mu et al44, 45 |
Table 3.
Polymorphism of genes encoding extracellular matrix-degrading enzymes and inflammatory mediators associated with intervertebral disc degeneration.
| Protein (Gene) | Disc degeneration or pain | Increase in pathology, or protective | References |
|---|---|---|---|
| Matrix Metalloproteinase (MMP3) | Disc degeneration and Modic changes | Increase | Karppinen et al48 |
| Matrix Metalloproteinase (MMP9) | Disc herniation | Increase | Hirose et al40 |
| Interleukin-1 receptor antagonist (IL1RN) | Disc degeneration and back pain | Increase | Solovieva et al49, 50 |
| Interleukin-1α (IL-1A) | Disc degeneration and back pain | Increase | Solovieva et al49, 50 |
| Interleukin-1β (IL-1B) | Disc degeneration and back pain | Increase | Solovieva et al49, 50 |
| Interleukin-6 (IL-6) | Disc degeneration and radicular pain | Increase | Noponen-Hietala et al51 |
| Cyclooxygenase-2 (PTGS2) | Failure of NSAID analgesia | – | Skarke et al52 |
Genes known to modify extracellular matrix, such as Thrombospondin2 (THBS2), Vitamin D Receptor (VDR), carbohydrate sulfotransferase 3 (CHST3) and Growth Differentiation Factor (GDF)-5 are also included in this table (Table 2). Asporin (ASPN) and GDF5 are interesting from the perspective that mutations in these genes are also risk factors for osteoarthritis, as the disc resembles diarthrodial joints.47
Likewise, overactive matrix-degrading enzymes may also compromise integrity of the IVD and lead to premature disc degeneration through the breakdown of disc structural components (Table 3). For example, Matrix Metalloproteinase 3 (MMP3) gene polymorphism has been associated with IVD degeneration and Modic changes.48 Modic changes were defined by Modic et al as vertebral body changes seen on magnetic resonance (MR) images,53 likely in response to biomechanical changes with IVD degeneration.
Inflammatory cytokines such as interleukin (IL)-1β may inhibit extracellular matrix production and increase catabolism, and, thus, decrease the total amount of extracellular matrix.54 Furthermore, inflammatory cytokine/chemokine levels are found to be elevated in IVD tissues from patients with back pain.55 Similarly, polymorphisms in cytokines associated with disc degeneration and back pain may play significant roles in both loss of IVD tissue integrity and pain generation (Table 3).
The clinical relevance of the IVD tissue is the debilitating effects of back pain related to disc degeneration. Understanding the mechanism(s) of disc degeneration and related pain is critically important for developing strategies to repair the degenerating disc and thus to reduce pain. For example, the IVD cells are capable of producing multiple chemokines such as IL-8, IL-7 and IL-10,55 chemokine regulated on activation, normal T cell expressed and secreted (RANTES) and cytokines IL-1β56 and TNF-α,57 thus playing a role in pain generation. The outer one-third of the posterior AF is innervated, and nerve ingrowth occurs with degeneration, demonstrating an anatomical basis for discogenic pain.58 On a positive note, IVD stem cells persist into adulthood,59, 60 providing a potential mechanism for self-regeneration.
In summary, the spinal column is a landmark development in evolution. The intervertebral discs in total account for about 1/3 of the vertebral column length in humans, and permit spinal mobility. Genes directing the development of the IVD may also contribute to its maintenance, degeneration, and, potentially, regeneration. Likewise, structural genes as well as genes responsible for maintenance of the structure are related to IVD degeneration. Genes responsible for inflammation may also play a dual role in repair responses to injury, and thus may be crucial for the long-term health of the IVD, as well as linking disc structural deterioration to back pain.
Conflicts of interest
None of the authors have any conflicts to declare.
Acknowledgments
Jason W. Ashley, PhD is supported by the University of Pennsylvania Postdoctoral Opportunities in Research and Teaching (PENN-PORT) fellowship funded by the National Institute of General Medical Sciences Institutional Research and Career Development Award (IRACDA; 5 K12 GM081259-09). Yejia Zhang, MD, PhD has been supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, 1K08 HD049598). This work is supported, in part, by research grants from the Department of Veterans Affairs (VA RR&D I01 RX001321 and VA1I21RX001896), and the Research Grants Committee of Hong Kong (T-12-708/12N).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Choi K.S., Cohn M.J., Harfe B.D. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237:3953–3958. doi: 10.1002/dvdy.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urban J.P., Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120–130. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luoma K., Riihimaki H., Luukkonen R., Raininko R., Viikari-Juntura E., Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine (Phila Pa 1976) 2000;25:487–492. doi: 10.1097/00007632-200002150-00016. [DOI] [PubMed] [Google Scholar]
- 4.Galis F. Why do almost all mammals have seven cervical vertebrae? developmental constraints, hox genes, and cancer. J Exp Zool. 1999;285:19–26. [PubMed] [Google Scholar]
- 5.Pearson J.C., Lemons D., McGinnis W. Modulating hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 6.Stemple D.L. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–2512. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- 7.Corallo D., Trapani V., Bonaldo P. The notochord: structure and functions. Cell Mol Life Sci. 2015;72:2989–3008. doi: 10.1007/s00018-015-1897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Resende T.P., Ferreira M., Teillet M.A., Tavares A.T., Andrade R.P., Palmeirim I. Sonic hedgehog in temporal control of somite formation. Proc Natl Acad Sci U S A. 2010;107:12907–12912. doi: 10.1073/pnas.1000979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scaal M. Early development of the vertebral column. Semin Cell Dev Biol. 2016;49:83–91. doi: 10.1016/j.semcdb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Lawson L., Harfe B.D. Notochord to nucleus pulposus transition. Curr Osteoporos Rep. 2015;13:336–341. doi: 10.1007/s11914-015-0284-x. [DOI] [PubMed] [Google Scholar]
- 11.Hayes A.J., Benjamin M., Ralphs J.R. Extracellular matrix in development of the intervertebral disc. Matrix Biol. 2001;20:107–121. doi: 10.1016/s0945-053x(01)00125-1. [DOI] [PubMed] [Google Scholar]
- 12.Hayes A.J., Benjamin M., Ralphs J.R. Role of actin stress fibres in the development of the intervertebral disc: cytoskeletal control of extracellular matrix assembly. Dev Dyn. 1999;215:179–189. doi: 10.1002/(SICI)1097-0177(199907)215:3<179::AID-AJA1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 13.Smith L.J., Nerurkar N.L., Choi K.S., Harfe B.D., Elliott D.M. Degeneration and regeneration of the intervertebral disc: lessons from development. Dis Model Mech. 2011;4:31–41. doi: 10.1242/dmm.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi K.S., Lee C., Harfe B.D. Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech Dev. 2012;129:255–262. doi: 10.1016/j.mod.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murtaugh L.C., Chyung J.H., Lassar A.B. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13:225–237. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi K.S., Harfe B.D. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proc Natl Acad Sci U S A. 2011;108:9484–9489. doi: 10.1073/pnas.1007566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q., Eberspaecher H., Lefebvre V., De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 18.Sugimoto Y., Takimoto A., Akiyama H. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development. 2013;140:2280–2288. doi: 10.1242/dev.096354. [DOI] [PubMed] [Google Scholar]
- 19.Maier J.A., Lo Y., Harfe B.D. Foxa1 and Foxa2 are required for formation of the intervertebral discs. PLoS One. 2013;8:e55528. doi: 10.1371/journal.pone.0055528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters H., Wilm B., Sakai N., Imai K., Maas R., Balling R. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126:5399–5408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- 21.McMahon J.A., Takada S., Zimmerman L.B., Fan C.M., Harland R.M., McMahon A.P. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wijgerde M., Karp S., McMahon J., McMahon A.P. Noggin antagonism of BMP4 signaling controls development of the axial skeleton in the mouse. Dev Biol. 2005;286:149–157. doi: 10.1016/j.ydbio.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Baffi M.O., Moran M.A., Serra R. Tgfbr2 regulates the maintenance of boundaries in the axial skeleton. Dev Biol. 2006;296:363–374. doi: 10.1016/j.ydbio.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohn P., Cox M., Chen D., Serra R. Molecular profiling of the developing mouse axial skeleton: a role for Tgfbr2 in the development of the intervertebral disc. BMC Dev Biol. 2010;10 doi: 10.1186/1471-213X-10-29. 29–213X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Battie M.C., Videman T., Kaprio J. The twin spine study: contributions to a changing view of disc degeneration. Spine J. 2009;9:47–59. doi: 10.1016/j.spinee.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Tegeder I., Lotsch J. Current evidence for a modulation of low back pain by human genetic variants. J Cell Mol Med. 2009;13:1605–1619. doi: 10.1111/j.1582-4934.2009.00703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yee A., Chan D. Genetic basis of intervertebral disc degeneration. In: Shapiro I.M., Risbud M.V., editors. The Intervertebral Disc: Molecular and Structural Studies of the Disc in Health and Disease. Springer; 2014. pp. 157–176. [Google Scholar]
- 28.Annunen S., Paassilta P., Lohiniva J. An allele of COL9A2 associated with intervertebral disc disease. Science. 1999;285:409–412. doi: 10.1126/science.285.5426.409. [DOI] [PubMed] [Google Scholar]
- 29.Seki S., Kawaguchi Y., Mori M. Association study of COL9A2 with lumbar disc disease in the Japanese population. J Hum Genet. 2006;51:1063–1067. doi: 10.1007/s10038-006-0062-9. [DOI] [PubMed] [Google Scholar]
- 30.Knoeringer M., Reinke A., Trappe A.E., Schlegel J. Absence of the mutated Trp2 allele but a common polymorphism of the COL9A2 collagen gene is associated with early recurrence after lumbar discectomy in a German population. Eur Spine J. 2008;17:463–467. doi: 10.1007/s00586-007-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paassilta P., Lohiniva J., Goring H.H. Identification of a novel common genetic risk factor for lumbar disk disease. JAMA. 2001;285:1843–1849. doi: 10.1001/jama.285.14.1843. [DOI] [PubMed] [Google Scholar]
- 32.Toktas Z.O., Eksi M.S., Yilmaz B. Association of collagen I, IX and vitamin D receptor gene polymorphisms with radiological severity of intervertebral disc degeneration in southern European ancestor. Eur Spine J. 2015;24:2432–2441. doi: 10.1007/s00586-015-4206-5. [DOI] [PubMed] [Google Scholar]
- 33.Mio F., Chiba K., Hirose Y. A functional polymorphism in COL11A1, which encodes the alpha 1 chain of type XI collagen, is associated with susceptibility to lumbar disc herniation. Am J Hum Genet. 2007;81:1271–1277. doi: 10.1086/522377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noponen-Hietala N., Kyllonen E., Mannikko M. Sequence variations in the collagen IX and XI genes are associated with degenerative lumbar spinal stenosis. Ann Rheum Dis. 2003;62:1208–1214. doi: 10.1136/ard.2003.008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pluijm S.M., van Essen H.W., Bravenboer N. Collagen type I alpha1 Sp1 polymorphism, osteoporosis, and intervertebral disc degeneration in older men and women. Ann Rheum Dis. 2004;63:71–77. doi: 10.1136/ard.2002.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solovieva S., Noponen N., Mannikko M. Association between the aggrecan gene variable number of tandem repeats polymorphism and intervertebral disc degeneration. Spine (Phila Pa 1976) 2007;32:1700–1705. doi: 10.1097/BRS.0b013e3180b9ed51. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi Y., Osada R., Kanamori M. Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine (Phila Pa 1976) 1999;24:2456–2460. doi: 10.1097/00007632-199912010-00006. [DOI] [PubMed] [Google Scholar]
- 38.Seki S., Kawaguchi Y., Chiba K. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar disc disease. Nat Genet. 2005;37:607–612. doi: 10.1038/ng1557. [DOI] [PubMed] [Google Scholar]
- 39.Song Y.Q., Cheung K.M., Ho D.W. Association of the asporin D14 allele with lumbar-disc degeneration in Asians. Am J Hum Genet. 2008;82:744–747. doi: 10.1016/j.ajhg.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirose Y., Chiba K., Karasugi T. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar-disc herniation. Am J Hum Genet. 2008;82:1122–1129. doi: 10.1016/j.ajhg.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Videman T., Gibbons L.E., Battie M.C. The relative roles of intragenic polymorphisms of the vitamin d receptor gene in lumbar spine degeneration and bone density. Spine (Phila Pa 1976) 2001;26:E7–E12. doi: 10.1097/00007632-200102010-00003. [DOI] [PubMed] [Google Scholar]
- 42.Kawaguchi Y., Kanamori M., Ishihara H., Ohmori K., Matsui H., Kimura T. The association of lumbar disc disease with vitamin-D receptor gene polymorphism. J Bone Jt Surg Am. 2002;84-A:2022–2028. doi: 10.2106/00004623-200211000-00018. [DOI] [PubMed] [Google Scholar]
- 43.Song Y.Q., Karasugi T., Cheung K.M. Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Invest. 2013;123:4909–4917. doi: 10.1172/JCI69277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams F.M., Popham M., Hart D.J. GDF5 single-nucleotide polymorphism rs143383 is associated with lumbar disc degeneration in northern European women. Arthritis Rheum. 2011;63:708–712. doi: 10.1002/art.30169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mu J., Ge W., Zuo X., Chen Y., Huang C. A SNP in the 5'UTR of GDF5 is associated with susceptibility to symptomatic lumbar disc herniation in the Chinese Han population. Eur Spine J. 2014;23:498–503. doi: 10.1007/s00586-013-3059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams F.M., Bansal A.T., van Meurs J.B. Novel genetic variants associated with lumbar disc degeneration in northern Europeans: a meta-analysis of 4600 subjects. Ann Rheum Dis. 2013;72:1141–1148. doi: 10.1136/annrheumdis-2012-201551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro I.M., Vresilovic E.J., Risbud M.V. Is the spinal motion segment a diarthrodial polyaxial joint: what a nice nucleus like you doing in a joint like this? Bone. 2012;50:771–776. doi: 10.1016/j.bone.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karppinen J., Daavittila I., Solovieva S. Genetic factors are associated with modic changes in endplates of lumbar vertebral bodies. Spine (Phila Pa 1976) 2008;33:1236–1241. doi: 10.1097/BRS.0b013e318170fd0e. [DOI] [PubMed] [Google Scholar]
- 49.Solovieva S., Kouhia S., Leino-Arjas P. Interleukin 1 polymorphisms and intervertebral disc degeneration. Epidemiology. 2004;15:626–633. doi: 10.1097/01.ede.0000135179.04563.35. [DOI] [PubMed] [Google Scholar]
- 50.Solovieva S., Leino-Arjas P., Saarela J., Luoma K., Raininko R., Riihimaki H. Possible association of interleukin 1 gene locus polymorphisms with low back pain. Pain. 2004;109:8–19. doi: 10.1016/j.pain.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 51.Noponen-Hietala N., Virtanen I., Karttunen R. Genetic variations in IL6 associate with intervertebral disc disease characterized by sciatica. Pain. 2005;114:186–194. doi: 10.1016/j.pain.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 52.Skarke C., Reus M., Schmidt R. The cyclooxygenase 2 genetic variant -765G>C does not modulate the effects of celecoxib on prostaglandin E2 production. Clin Pharmacol Ther. 2006;80:621–632. doi: 10.1016/j.clpt.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 53.Modic M.T., Steinberg P.M., Ross J.S., Masaryk T.J., Carter J.R. Degenerative disk disease: assessment of changes in vertebral body marrow with MR imaging. Radiology. 1988;166:193–199. doi: 10.1148/radiology.166.1.3336678. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y., An H.S., Toofanfard M., Li Z., Andersson G.B., Thonar E.J. Low-dose interleukin-1 partially counteracts osteogenic protein-1-induced proteoglycan synthesis by adult bovine intervertebral disk cells. Am J Phys Med Rehabil. 2005;84:322–329. doi: 10.1097/01.phm.0000159972.85053.7e. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y., Chee A., Shi P. Intervertebral disc cells produce interleukins found in patients with back pain. Am J Phys Med Rehabil. 2015 [Epub ahead of print] doi: 10.1097/PHM.0000000000000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kepler C.K., Markova D.Z., Dibra F. Expression and relationship of proinflammatory chemokine RANTES/CCL5 and cytokine IL-1beta in painful human intervertebral discs. Spine (Phila Pa 1976) 2013;38:873–880. doi: 10.1097/BRS.0b013e318285ae08. [DOI] [PubMed] [Google Scholar]
- 57.Le Maitre C.L., Hoyland J.A., Freemont A.J. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res Ther. 2007;9:R77. doi: 10.1186/ar2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melrose J., Roberts S., Smith S., Menage J., Ghosh P. Increased nerve and blood vessel ingrowth associated with proteoglycan depletion in an ovine anular lesion model of experimental disc degeneration. Spine (Phila Pa 1976) 2002;27:1278–1285. doi: 10.1097/00007632-200206150-00007. [DOI] [PubMed] [Google Scholar]
- 59.Sivakamasundari V., Lufkin T. Stemming the degeneration: IVD stem cells and stem cell regenerative therapy for degenerative disc disease. Adv Stem Cells. 2013;2013:724547. doi: 10.5171/2013.724547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakai D., Nakamura Y., Nakai T. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. doi: 10.1038/ncomms2226. [DOI] [PMC free article] [PubMed] [Google Scholar]