Abstract
Current estimates indicate that the hepatitis C (HCV) is the leading cause of mortality around the world, with infection rates steadily increasing in Egypt. The dual therapy for this silent epidemic with pegylated-interferon-α2b/ribavirin has markedly improved the success rates in genotype-4 patients. It was reported that apoptosis plays a vital mechanistic role in limiting viral replication. P53, a key regulator of apoptosis, induces CD95 gene expression and subsequently initiates apoptotic cascade to be activated. The current study examined the impact of P53 rs1042522 and CD95 rs1800682 polymorphisms on the treatment response. Three groups of 240 volunteers were enrolled in this study; 86 in sustained virological responders group, 74 in non-responders group, and 80 in control group. All patients had HCV genotype-4a and were interferon treatment naïve. Quantizations of HCV-RNA by qRT-PCR and histological scores were performed for all patients. In addition, genotyping of HCV-RNA, P53 rs1042522 Arg/Pro and CD95 rs1800682 A/G polymorphisms were investigated in all subjects. It was resulted that P53 Pro/Pro homozygous genotype has high significant increase, while CD95 A/A homozygous genotype has high significant decrease when comparing non-responders with responders. Finally, it was concluded that Pro variant of P53 rs1042522 may be used as a genetic predictor for non-responsiveness, while A/A variant of CD95 rs1800682 may be used as a sensitive biomarker for responsiveness to antiviral therapy of HCV genotype-4a infection. In addition, low prolactin, high total testosterone, and high GH levels may provide promising biomarkers for early prediction of the response when associated with these genetic polymorphisms.
Keywords: CD95 rs1800682, HCV gentotype-4a, P53 rs1042522, Response, SNPs
Introduction
Globally, the hepatitis C virus (HCV) is the main cause of mortality and it was identified 25 years ago.1 Since then, HCV infection has spread and its distribution varies broadly among various geographic areas. Africa and Asia have the highest reported rates of infections (more than 2.9%), while industrialized regions like America and Europe have lower rates of infections (less than 1%).2 Recent epidemiological data indicate that infection steadily increased from 2.3% to 2.8% between 1990 and 2005.3
Pre-clinical and clinical outputs in these industrial areas promise high response rates with some adverse effects following treatment with dual or triple direct-acting antiviral agents (DAA) therapy. However, such DAAs are designed with genotype 1 as the principle target and often have limited pan-genotypic efficacy or have yet to be approved for use against other genotypes. In 2015, it was reported that successful treatment for HCV is made difficult by the presence of various hosts, viruses, and treatment-related factors. The addition of telaprevir and boceprevir to the arsenal improved success rates in genotype 1 infected patients from roughly 40% in treatment-naïve patients with interferon-based dual therapy to 60.8% and 54.2%, respectively, but rapid development of resistance mechanisms, increases in adverse effects, and a low spectrum activity proved to be barriers to efficacious treatment.4
In late 2013, two new agents were approved – sofosbuvir and simeprevir – which have higher barriers to resistance and drastically improved success rates; however cost concerns could limit their utilization, especially in developing countries like Egypt. Sofosbuvir costs approximately $1000 per pill, because the wholesaler acquisition cost of a four week supply is $28,000. Depending on genotype, sofosbuvir costs $84,000 and $168,000 for 12 and 24 week-based treatment, without taking into considerations of costs of ribavirin and peginterferon. Not only does this add an additional burden to the health care system, but could also prevent patients from getting access.4, 5
There are 11 genetically distinct genotypes and more than 50 subtypes of the virus that have been identified.6 Genotype 1 and 4 are often considered more difficult to treat than genotype 2 and 3,7 leaving patients with genotype-4 with fewer treatment options, even though genotype-4 is common in Egypt and throughout the Middle East and Africa.8 Consequently, there is a continuous need for promising biomarkers for the interferon-based therapy.
IFN-l3, coded by the interleukin 28B (IL28B) gene, is a cytokine9 involved in the antiviral immune response and inhibits HCV replication in vitro.10 Recently, the impact of the IL28B gene SNPs on HCV treatment was discovered and it has been studied in several ethnicities and various viral genotypes.11, 12, 13 Major findings include the association of the minor alleles (rs12979860, rs8099917, and rs12980275) with non-responsiveness to interferon-based antiviral therapy. IL28B rs12980275 polymorphism was not found in HCV genotype 2 and 3 infected patients.14, 15 Although SNPs in the IL28B locus are probably the most well-known predictors of outcome of interferon therapy, the effect has been less widely examined in genotype-4a and there are unfavorable effects varied between ethnicities (European-Americans: OR 7.3, African-Americans: OR 6.1, Hispanics: OR 5.6), some interethnic variability in treatment response may be explained by this variant.11 Consequently, there is an unmet need for novel biomarkers for response to interferon-based therapy.
Teodoro and Branton have suggested that apoptosis, a biological phenomenon that plays a crucial role in immune regulation and tissue homeostasis, is an important machinery in limiting viral replication. In addition, apoptosis induction upon HCV infection may contribute to viral clearance, while its inhibition may result in HCV persistence and oncogenesis.16 BCL2 gene, a key regulator of apoptosis, has a single nucleotide polymorphism at codon 43 (127G/A) and it is recently reported that this polymorphic point can be used as a sensitive biological marker for early prediction of 56.1% response rate to interferon-based therapy.17
Over 25 years ago, a new P53 single nucleotide polymorphism located at codon 72 (rs1042522) and associated with arginine to proline (Arg/Pro) substitution has been identified.18 Thereafter, numerous observations examined the role of P53 rs1042522 Arg/Pro polymorphism on hepatocellular carcinoma (HCC) and HCV-associated HCC incidences. Scientists suggested that there was a frequent loss of P53 rs1042522 Pro allele in HCV-positive carriers.19 On the contrary, others found that there was no association between P53 rs1042522 genotypes and disease severity or hepatocarcinogenesis.20
In HCV infection, CD95 expression was found to be up-regulated within hepatocytes in accordance with the severity of liver inflammation.21 These CD95-expressing hepatocytes become susceptible to the apoptotic death signal and play a crucial role in liver cell injury caused by HCV infection.22 CD95-670A/G (rs1800682) polymorphism was associated potentially with fibrosis and cirrhosis in patients infected with HCV.23 In addition, authors have suggested that there was a higher frequency of CD95 rs1800682 A/A genotype in spontaneously recovered patients from HCV infection compared to patients with persistent HCV infection.24 On the contrary, other scientists have demonstrated that there was no significant correlation between CD95 rs1800682 polymorphism and fibrotic stages, while this polymorphism may account for some of the histopathological variations in HCV infections.25
Some hormones play a significant role in immune cells regulation such as B-cells, T-cells, and natural killer cells. This advantage supports their vital roles in chronic liver disease resistance and attack from early stages of infections.26, 27, 28, 29 It was suggested that HCV infected patients have high prolactin concentrations,30, 31 regardless of gender.32 Besides, prolactin concentrations decreased significantly after antiviral therapy in responders compared to non-responders33; regardless of gender.32 Meanwhile, there were high significant decreases of post-treatment total testosterone concentrations when comparing males of non-responders with responders.32 In addition, it was investigated that patients infected with HCV showed a significant decrease in human growth hormone levels compared to healthy subjects. Whereas, growth hormone concentrations increased after antiviral therapy in responders compared to non-responders.34, 35
Overall, these observations promoted us to study the impact of the P53 rs1042522 and CD95 rs1800682 polymorphisms on the response to pegylated-interferon-based antiviral therapy among HCV genotype-4a infected patients. In addition, the previous observations of hormonal profile alterations during antiviral therapy guided us to examine the association between these hormonal alterations and the genetic polymorphisms as well as their impact on the treatment response.
Materials and methods
Study patients
Two hundred and forty volunteers were enrolled in this prospective study and divided into 3 groups; 86 in sustained virological responders group, 74 in non-responders group, and 80 in healthy controls group. The 160 patients who were infected with HCV visited outpatient clinics of the Tropical Medicine and Hepatology Department, El-Kasr El-Aini Hospital, Cairo University, Egypt; where they were diagnosed with chronic HCV infection. All volunteers were enrolled in the current study after signing a consent form. The patients were treated with subcutaneous injections of pegylated-interferon-α 2b (Reiferon Retard®, Minapharm, Cairo, Egypt) (100 μg/week) combined with oral administration of ribavirin (Minapharm, Cairo, Egypt) (1000–1200 mg/day, body weight based) for 24 weeks. Patients were grouped according to treatment response. At week 24, 86 patients were tested negative with undetectable HCV-RNA and classified as responders, then continued treatment until week 48 to detect sustained virological response when the viral RNA remains undetected after six months of treatment cessation. Those who tested positive for HCV-RNA (n = 74) stopped treatment at week 24 and were classified as non-responders. The study protocol and informed consent were approved by the Ethics Committee of Cairo University.
Patients inclusion and exclusion criteria
Patients participated in the study were fulfilled the inclusion criteria included: elevated hepatic enzymes; ALT and AST (>37 IU/L); within 6 months prior to entry the study, hemoglobin (>11 g/dL), total leucocytes count (>3000/mm3), neutrophils (>1500/mm3), platelets (>100,000/mm3), prothrombin time within normal range (9.8–13.8 s, presence of antinuclear antibodies (ANA titre <1/160), positive HCV antibodies, detectable HCV-RNA, HCV genotype-4a, liver biopsy showing histological evidence of chronic hepatitis and they were never previously treated with interferon. On the other hand, patients presented with HCC, liver diseases other than hepatitis C such as hepatitis B surface antigen (HBsAg) seropositivity or autoimmune hepatitis or co-contamination with the human immunodeficiency virus (HIV) and active schistosomiasis were excluded from this study.
Biochemical and hormonal tests
Biochemical tests including liver enzymes (ALT, and AST), total bilirubin, direct bilirubin, albumin, and alpha fetoprotein (AFP) were performed on patients infected with HCV genotype-4a and treated with PEG-IFN-α/RBV antiviral therapy on Integra-400 (Roche, Mannheim, Germany). Human prolactin, total testosterone, and growth hormone were quantified in all patients infected with HCV using the enzyme-linked immunosorbent assay (ELISA). Enzyme immunoassay (EIA) was used for the quantitative determination of prolactin and total testosterone concentrations was measured using ELISA kits purchased from Immunospec Corporation (California, USA). Meanwhile, human growth hormone levels were measured using ELISA kits supplied by DRG International (Mountainside, USA). All these measurements were examined before, during and after the combination therapy.
Histological investigations
Patients infected with chronic HCV were subjected to abdominal ultrasound and liver biopsy was taken from each patient before the onset of therapy to estimate the grade of activity and fibrosis according to metavir scoring system.36 All biopsies were classified into 5 stages of fibrosis (F0: no fibrosis; F1: enlargement of portal tract without septa formation; F2: enlargement of portal tract with rare septa formation; F3: numerous septa without cirrhosis; F4: established cirrhosis) and 4 grades (inflammations) of histological activity (A0: none; A1: mild; A2: moderate; A3: severe) based on the intensity of necroinflammatory lesions.
HCV detection
The presence or absence of HCV antibodies was investigated by the third generation ELISA (DiaSorin, Torino, Italy). HCV-RNA in serum was measured before treatment and routinely at week 24 and 48 after treatment and graded into low (200,000–1,000,000 copies/mL), moderate (1,000,000–5,000,000 copies/mL), and high (5,000,000–25,000,000 copies/mL) viral levels.
RNA was extracted from sera using a Qiagen viral RNA kit (Qiagen, Hilden, Germany), then real time reverse transcription polymerase chain reaction (qRT-PCR) was performed using a LightCycler system (Roche, Mannheim, Germany). Amplification primers for HCV were 5′ primer K78F (CAAGCACCC TATCAGGCAGT) and 3′ primer K80R (AGCGTCTAGCCATGGCGT). Hybridization probes (FL) 5′-GCAGCCTCCAGGACCCCCC-3′ and (LC) 5′-CCCGGGAGAGCCATAGTGGTCTG-3′ were used to detect the product. Reaction mixtures included 7.5 μl of Lightcycler RNA Master HybProbe, 3.25 mM Mn(OAc)2, 0.5 μM concentration of each primer, 0.4 μM of hybridization probe mix and 1 μl of the RNA template in a total volume of 20 μl. HCV-RNA was first reverse-transcribed at 61 °C for 20 min. Following denaturation for 30 s at 95 °C, the LightCycler amplification was performed for 45 cycles, each cycle consisting of 5 s at 95 °C, annealing at 62 °C for 15 s and extension at 72 °C for 10 s. Fluorescence was monitored at 530/640 nm.
HCV-RNA genotyping
RNA was isolated from sera for HCV genotyping using a Qiagen viral RNA kit (Qiagen, Hilden, Germany). HCV-RNA genotyping was performed using Ohno method which depends on nested PCR amplification of the virus core gene using genotype specific primers mentioned previously.37
P53 rs1042522 and CD95 rs1800682 genotyping
The single nucleotide polymorphisms of P53 rs1042522 and CD95 rs1800682 were studied to find whether these polymorphisms have an impact on HCV response to interferon-based therapy. The DNA was extracted from whole blood of all individuals using QIAamp DNA blood Mini kit (Qiagen, Hilden, Germany). PCR was performed using the thermal cycler (Biometra®, Göttingen, Germany) for amplifying P53 and CD95 genes. PCR reagents were supplied by (Promega, Southampton, UK). Primer sequences were as follow: the forward primer (F): 5′- TTG CCG TCC CAA GCA ATG GAT GA-3′ and the reverse primer (R): 5′-TCT GGG AAG GGA CAG AAG ATG AC-3′ for P53 gene and the forward primer (F): 5′- CCT AAG AGC TAT CTA CCG TTC-3′ and the reversed primer (R): 5′-GGC TGT CCA TGT TGT GGC TGC-3′ for CD95 gene. The PCR products of P53 and CD95 gene amplification yield 199 bp and 233 amplicons respectively. The PCR conditions of P53 and CD95 were as follow: denaturation at 95 °C for 30 s, annealing at 60 °C and 58 °C respectively for 30 s, and extension at 72 °C for 30 s. These amplicons were enrolled in restriction fragment length polymorphism (RFLP) technique after purification.
The purified PCR products of P53 and CD95 genes were incubated at 37 °C overnight with Bsh1236I and BstNI restriction enzymes respectively to identify three different genotypes of each gene as described in numerous previous studies [19, 20, 23, 25, 38] and illustrated in Table 1. Restriction enzymes were purchased from (Fermentas, Ontario, Canada). Amplified DNA or digested PCR Products was loaded and electrophoresed on a 3% agarose gel prepared in 1 × TBE buffer using a gated mini-gel tank (BioRad, California, USA) and containing ethidium bromide (0.5 μg/ml) for 40 min at 100 V. DNA ladder (Fermentas, Ontario, Canada) was routinely used as a molecular weight standard as a reference in the agarose gel. The gel was visualized on a UV gel documentation computerized system. Images were manipulated by BioDocAnalyze software program (Biometra®, Göttingen, Germany).
Table 1.
Description of PCR and RFLP products of P53 and CD95 genes.
| Gene | SNPs | PCR amplicon | RFLP (R.E.) | Genotypes | Product sizes (bp) |
|---|---|---|---|---|---|
| P53 | +72 G/C (Arg72Pro) | 199 bp | BstUI (Bsh1236I) | +72 G/G | 113 + 86 |
| +72 C/C | 199 | ||||
| +72 G/C | 199 + 113 + 86 | ||||
| CD95 | −670 A/G | 233 bp | MvaI (BstNI) | −670 A/A | 233 + 99 |
| −670 G/G | 189 + 99 + 44 | ||||
| −670 A/G | 233 + 189 + 99 + 44 |
SNPs: single nucleotide polymorphisms.
RFLP: restriction fragment length polymorphism.
R.E.: restriction enzyme.
Biostatistics
The data have been expressed as mean ± standard error (S.E.) and the range was stated between parentheses. Comparison of variables between the study groups has been done using analysis of variance (ANOVA) test. Data were statistically described using statistical computer program: SPSS (Statistical Package for the Social Science, USA, version 17). For comparing categorical data, Chi-square (χ2) test was performed. A Chi-square (χ2) less than 3.84 was considered as a non-statistically significant value and (χ2) more than 3.84 was considered as a statistically significant value. A receiver operating characteristic (ROC) analysis was used to determine the area under ROC curve (AUC), sensitivity, and specificity for the studied prognostic markers. To rank the importance of predictor variables, logistic regressions of some promising variables were performed.
Results
Demographic and clinical data
Data for all HCV genotype-4a infected patients and treated with pegylated-interferon-α plus ribavirin (PEG-IFN-α/RBV) as well as control subjects were collected. There was no significant difference of age, gender, and body mass index (BMI) when comparing sustained virological responders (SVR) with either non-responders (NR) or controls (p > 0.05) as shown in Table 2. In addition, the hepatic function tests, including AST, ALT, total bilirubin, direct bilirubin, albumin, and AFP of 86 SVR against 74 NR patients (pre- and post-treatment with PEG-IFN-α/RBV) were analyzed; where there were high significant increases (p < 0.01) of post-treatment ALT, AST, direct bilirubin, and albumin as well as a significant increase (p < 0.05) of post-treatment total bilirubin when comparing responders with non-responders. On the contrary, there was no significant difference (p > 0.05) of pre-treatment AST, ALT, total bilirubin, direct bilirubin, albumin, and AFP as well as post-treatment AFP when comparing SVR with NR patients. Finally, there were high significant differences (p < 0.01) of pre-treatment ALT, AST, total bilirubin, direct bilirubin, and AFP levels when comparing either SVR or NR patients with controls.
Table 2.
Demographic and clinical data of all subjects.
| Characteristics (Mean ± S.E.) | Controls (n = 80) | SVR (n = 86/160, 53.7%) | NR (n = 74/160, 46.3%) |
|---|---|---|---|
| Age, (years) Range, (min–max) |
39.85 ± 6.61 (19.00–53.00) |
41.17 ± 6.91d (20.00–53.00) |
39.08 ± 6.94d (20.00–54.00) |
| Gender | |||
| Females, n (%) | 36 (45%) | 44 (51.2%)d | 38 (51.4%)d |
| Males, n (%) | 44 (55%) | 42 (48.8%)d | 36 (48.6%)d |
| BMI, (kg/m2) | 24.30 ± 6.26 | 25.20 ± 5.24d | 26.04 ± 6.53d |
| ALT, (IU/L), A | 30.40 ± 0.94 | 60.00 ± 2.54a∗ | 54.00 ± 3.47b∗ |
| ALT, (IU/L), B | – | 33.73 ± 2.83 | 41.88 ± 4.17c∗ |
| AST, (IU/L), A | 32.12 ± 1.19 | 62.13 ± 2.61a∗ | 60.35 ± 3.85b∗,d |
| AST, (IU/L), B | – | 34.26 ± 1.19 | 65.11 ± 4.39c∗ |
| Total Bilirubin, (mg/dL), A | 0.78 ± 0.03 | 1.05 ± 0.08a∗ | 1.15 ± 0.04b∗,d |
| Total Bilirubin, (mg/dL), B | – | 0.78 ± 0.01 | 1.22 ± 0.1c |
| Direct Bilirubin, (mg/dL), A | 0.15 ± 0.01 | 0.24 ± 0.02a∗ | 0.26 ± 0.03b∗,d |
| Direct Bilirubin, (mg/dL), B | – | 0.14 ± 0.01 | 0.31 ± 0.04c∗ |
| Albumin, (g/dL), A | 4.34 ± 0.13 | 3.86 ± 0.04a | 3.77 ± 0.1b∗,d |
| Albumin, (g/dL), B | – | 3.28 ± 0.02 | 3.90 ± 0.07c∗ |
| AFP, (U/L), A | 4.11 ± 0.19 | 5.77 ± 0.37a | 6.02 ± 0.41b∗,b |
| AFP, (U/L), B | – | 4.64 ± 0.29 | 5.34 ± 0.46b |
S.E.: standard error.
n: size of samples.
A: before treatment.
B: after treatment.
a/a*: a significant/a high significant correlation (χ2 > 3.84) when comparing responders with controls.
b/b*: a significant/a high significant correlation (χ2 > 3.84) when comparing non-responders with controls.
c/c*: a significant/a high significant correlation (χ2 > 3.84) when comparing non-responders with responders.
d: a non-significant correlation (χ2 < 3.84) either when comparing non-responders or responders with controls.
Virological and histological data
The metavir scoring system including hepatic activity scores (A0/1 and A2/3) and fibrotic stages (F0/1, F2, and F3/4) as well as the low, moderate, and high viral loads were collected from 86 SVR and 74 NR patients (pre- and post-treatment with PEG-IFN-α/RBV); where there were high frequency and significant differences of the low, moderate, and high viral loads when comparing post-treated SVR with NR patients (χ2 = 48.31, 66.69, 6.38; respectively). On the contrary, there was no significant difference (p > 0.05) of the low, moderate, and high viral loads when comparing pre-treated SVR with NR patients (χ2 = 3.183, 0.247, 3.678; respectively) as illustrated in Table 3.
Table 3.
Baseline virological and histological features of HCV sustained virological responders (SVR) against non-responders (NR).
| Virological and histological features | SVR (n = 86) |
NR (n = 74) |
χ2 |
P |
||
|---|---|---|---|---|---|---|
| Size (n) | Frequency (%) | Size (n) | Frequency (%) | |||
| HCV-RNA Level, before therapy | ||||||
| Low viral load | 43 | 50.0 | 27 | 36.5 | 3.1 | NS |
| Moderate viral load | 39 | 45.3 | 37 | 50.0 | 0.2 | NS |
| High viral load | 4 | 4.7 | 10 | 13.5 | 3.6 | NS |
| HCV-RNA Level, after therapy | ||||||
| Low viral load | 0 | 0 | 30 | 40.5 | 48.3 | S* |
| Moderate viral load | 0 | 0 | 38 | 51.4 | 66.6 | S* |
| High viral load | 0 | 0 | 6 | 8.1 | 6.3 | S |
| Metavir Scoring System, before therapy | ||||||
| Activity Score | ||||||
| A0/1 | 72 | 83.7 | 52 | 70.3 | 4.3 | S |
| A2/3 | 14 | 16.3 | 22 | 29.7 | 4.3 | S |
| Fibrosis Score | ||||||
| F0/1 | 54 | 62.8 | 18 | 24.3 | 28.6 | S* |
| F2 | 20 | 23.3 | 30 | 40.5 | 6.04 | S |
| F3/4 | 12 | 13.9 | 26 | 35.2 | 11.1 | S |
| Metavir Scoring System, after therapy | ||||||
| Activity Score | ||||||
| A0/1 | 76 | 88.4 | 44 | 59.5 | 20.2 | S* |
| A2/3 | 10 | 11.6 | 30 | 40.5 | 20.2 | S* |
| Fibrosis Score | ||||||
| F0/1 | 56 | 65.1 | 30 | 40.5 | 11.1 | S |
| F2 | 24 | 27.9 | 12 | 16.2 | 3.3 | NS |
| F3/4 | 6 | 7 | 32 | 43.3 | 33.0 | S* |
n: size of samples.
NS: represents no significant difference.
S: represents significant difference with Chi-square value (χ2 > 3.84).
S*: high significant difference with Chi-square value (χ2 > 3.84).
Regarding the metavir scoring system, there were high frequency and significant differences of pre-treatment hepatic activity scores (A0/1 and A2/3) and liver fibrotic stages (F0/1, F2, and F3/4) when comparing SVR with NR patients (χ2 = 4.34, 4.34, 28.6, 6.04, 11.12; respectively). On the other hand, there were high frequency and significant differences of post-treatment hepatic activity scores (A0/1 and A2/3) and liver fibrotic scores (F0/1 and F3/4) when comparing SVR with NR patients (χ2 = 20.20, 20.20, 11.17, 33.09; respectively). On the contrary, there was no significant difference (p > 0.05) of pre-and post-treatment hepatic fibrotic score (F2) when comparing SVR with NR patients (χ2 = 0.01, 3.33; respectively) as shown in Table 3.
Identification of P53 rs1042522 and CD95 rs1800682 genetic variants
In all patients infected with HCV and healthy individuals, Bsh1236I restriction enzyme was used to distinguish between P53 rs1042522 Arg/Arg, Pro/Pro, and Arg/Pro. Bsh1236I digestion of the PCR amplicon (199 bp) was performed to identify the three different genotypes of P53: the homozygous Arg72Arg (+72 G/G) genotype at (113 bp+86 bp), the homozygous Pro72Pro (+72 C/C) genotype at (199 bp), and the heterozygous Arg72Pro (+72 G/C) genotype at (199 bp+113 bp+86 bp) as illustrated in Fig. 1.
Figure 1.
Detection of the P53 polymorphism by PCR-BstUI digestion. BstUI restriction enzyme cuts the PCR products of the homozygous Arg72Arg and the heterozygous Arg72Pro but does not cut the homozygous Pro72Pro PCR fragment. Lane M represents DNA Marker (100 bp), lane 1 and 2 represent the homozygous Pro72Pro (+72 C/C) genotype at (199 bp), lane 3 and 5 represent the heterozygous Arg72Pro (+72 G/C) genotype at (199 bp +113 bp + 86 bp), and lane 4 represents the homozygous Arg72Arg (+72 G/G) genotype at (113 bp +86 bp).
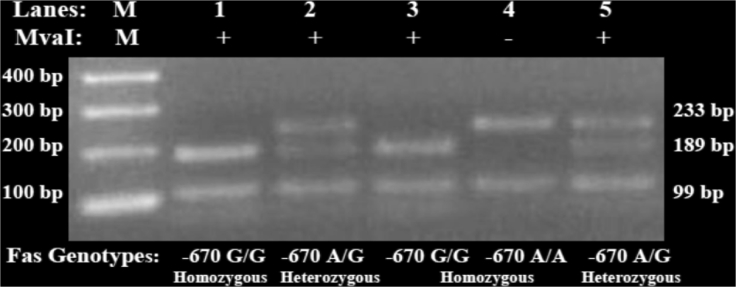
Additionally, the BstNI digestion of the PCR amplicon (233 bp) was used to discriminate CD95 rs1800682 A/A, G/G, and A/G among HCV infected patients and healthy controls. BstNI/RFLP analysis was performed to identify the different three types of CD95 genotypes; the homozygous −670 A/A genotype at (233 bp+99 bp), the homozygous −670 G/G genotype at (189 bp+99 bp+44 bp), and the heterozygous −670 A/G genotype at (233 bp+189 bp+99 bp+44 bp) as illustrated in Fig. 2.
Figure 2.
Detection of the CD95 polymorphism by PCR-MvaI digestion. MvaI restriction enzyme cuts the PCR products of the homozygous −670 G/G and the heterozygous −670 A/G but does not cut the homozygous −670 A/A PCR fragment. Lane M represents DNA Marker (100 bp), lane 1 and 3 represent the homozygous −670 G/G genotype at (189 bp+99 bp+44 bp), lane 2 and 5 represent the heterozygous −670 A/G genotype at (233 bp+189 bp+99 bp+44 bp), and lane 4 represent the homozygous −670 A/A genotype at (233 bp+99 bp).
Frequencies of P53 rs1042522 and CD95 rs1800682 polymorphisms
Table 4 shows the frequencies of P53 rs1042522 and CD95 rs1800682 genetic polymorphisms at +72 Arg/Pro and −670 A/G respectively in patients treated with PEG-IFN/RBV and healthy controls. Non-responders have high frequencies and significant increases of P53 rs1042522 Pro/Pro genotype and allele (n = 24, 32.43% vs. n = 4, 4.6%; n = 74, 50% vs. n = 52, 30.23%, respectively), while have a low frequency and significant decrease of P53 rs1042522 Arg/Pro genotype (n = 26, 35.14% vs. n = 44, 51.2%, respectively) compared to responders. On the other hand, responders have a high frequency and significant increases of CD95 rs1800682 A/A genotype and allele (n = 28, 32.6% vs. n = 8, 10.8%; n = 98, 57% vs. n = 61, 41.2%, respectively), while there was no significant difference of P53 rs1042522 Arg/Arg genotype as well as CD95 rs1800682 A/G and G/G genotypes when comparing NR with SVR patients.
Table 4.
Frequencies of P53 and CD95 polymorphic spots at +72 G/C and −670 A/G respectively in all subjects.
| Genotypes/Alleles/models | Controls (n = 80) |
SVR (n = 86) |
NR (n = 74) |
|||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| P53 General genotypes | ||||||
| 72 G/G (Arg/Arg) | 42 | 52.5 | 38 | 44.2c | 24 | 32.4a |
| 72 G/C (Arg/Pro) | 33 | 41.3 | 44 | 51.2c | 26 | 35.2c |
| 72 C/C (Pro/Pro) | 5 | 6.2 | 4 | 4.6c | 24 | 32.4a∗,b∗ |
| P53 Alleles frequency | ||||||
| 72 G (Arg) | 117 | 73.1 | 120 | 69.8c | 76 | 50a |
| 72 C (Pro) | 43 | 26.9 | 52 | 30.2c | 74 | 50a |
| P53 Dominant model | ||||||
| Arg/Arg | 42 | 52.5 | 38 | 44.2c | 24 | 32.4a |
| Arg/Pro + Pro/Pro | 38 | 47.5 | 48 | 55.8c | 50 | 67.6a |
| P53 Recessive model | ||||||
| Arg/Arg + Arg/Pro | 75 | 93.7 | 82 | 95.4c | 50 | 67.6a∗,b∗ |
| Pro/Pro | 5 | 6.3 | 4 | 4.6c | 24 | 32.4a∗,b∗ |
| CD95 Genotypes | ||||||
| −670 A/A | 29 | 36.3 | 28 | 32.6c | 8 | 10.8a,b |
| −670 A/G | 34 | 42.5 | 42 | 48.8c | 45 | 60.8a |
| −670 G/G | 17 | 21.2 | 16 | 18.6c | 21 | 28.4c |
| CD95 Alleles frequency | ||||||
| −670 A | 92 | 57.5 | 98 | 57c | 61 | 41.2c |
| −670 G | 68 | 42.5 | 74 | 43c | 87 | 58.8c |
| CD95 Dominant model | ||||||
| A/A | 29 | 36.3 | 28 | 32.6c | 8 | 10.8a,b |
| A/G + G/G | 51 | 63.7 | 58 | 67.4c | 66 | 89.2a,b |
| CD95 Recessive model | ||||||
| A/A + A/G | 63 | 78.7 | 70 | 81.4c | 53 | 71.6c |
| G/G | 17 | 21.3 | 16 | 18.6c | 21 | 28.4c |
n: size of samples.
a/a*: a significant/a high significant correlation when comparing non-responders with controls.
b/b*: a significant/a high significant correlation when comparing non-responders with responders.
c: a non-significant correlation either when comparing non-responders or responders with controls.
In addition, Table 4 shows that there was no significant difference of all P53 and CD95 genetic variants (χ2> 3.84) when comparing responders with controls, while there were high significant differences of P53 rs1042522 Pro/Pro genotype and recessive model (Pro/Pro vs Arg/Arg plus Arg/Pro) as well as significant differences (χ2< 3.84) of P53 rs1042522 Arg/Arg genotype and dominant model (Arg/Arg vs Pro/Pro plus Arg/Pro) when comparing non-responders with controls. On the other side, non-responders have significant differences (χ2< 3.84) of CD95 rs1800682 A/A and A/G genotypes as well as the dominant model (A/A vs. G/G plus A/G) compared to controls.
Response rates of P53 rs1042522 and CD95 rs1800682 polymorphisms
Table 5 shows the treatment response frequencies of P53 rs1042522 Arg/Pro and CD95 rs 1800682 A/G genetic polymorphisms in patients treated with PEG-IFN/RBV in order to study the contribution rates of their genotypes, alleles, dominant, and recessive models in SVR and NR patients as well as to detect which variant has the potentiality to be used for early response prediction. It was observed that P53 rs1042522 Pro/Pro genotype and allele have high frequencies and significant increases when comparing NR with SVR (n = 24, 85.7% vs. n = 4, 14.3%; n = 74, 58.7% vs. n = 52, 41.3%, respectively). Additionally, P53 rs1042522 Arg/Arg genotype and allele, Arg/Pro genotype, and P53 recessive model (Arg/Arg + Arg/Pro) have high frequencies and significant increases when comparing SVR with NR patients (n = 38, 61.3% vs. n = 24, 38.7%; n = 44, 62.9% vs. n = 26, 37.1%; n = 82, 62.1% vs. n = 50, 37.9%; n = 120, 61.2% vs. n = 76, 38.8%, respectively). On the other hand, CD95 rs1800682 A/A genotype and allele have high frequencies and significant increases when comparing SVR with NR patients (n = 28, 77.8% vs. n = 8, 22.2%; n = 98, 61.6% vs. n = 61, 38.4%, respectively).
Table 5.
Response rates of P53 and CD95 genotypes and alleles as well as dominant and recessive models at +72 G/C and −670 A/G polymorphic points.
| Genotypes/Alleles/models | SVR |
NR |
χ2 | P | ||
|---|---|---|---|---|---|---|
| Size (n) | Frequency (%) | Size (n) | Frequency (%) | |||
| P53 General genotypes | ||||||
| 72 G/G (Arg/Arg) | 38 | 61.3 | 24 | 38.7 | 9.3 | S |
| 72 G/C (Arg/Pro) | 44 | 62.9 | 26 | 37.1 | 12.3 | S |
| 72 C/C (Pro/Pro) | 4 | 14.3 | 24 | 85.7 | 99.1 | S* |
| P53 Alleles frequency | ||||||
| 72 G (Arg) | 120 | 61.2 | 76 | 38.8 | 9.1 | S |
| 72 C (Pro) | 52 | 41.3 | 74 | 58.7 | 5.3 | S |
| P53 Dominant model | ||||||
| Arg/Arg | 38 | 61.3 | 24 | 38.7 | 9.3 | S |
| Arg/Pro + Pro/Pro | 48 | 49 | 50 | 51 | 0.0 | NS |
| P53 Recessive model | ||||||
| Arg/Arg + Arg/Pro | 82 | 62.1 | 50 | 37.9 | 10.7 | S |
| Pro/Pro | 4 | 14.3 | 24 | 85.7 | 99.1 | S* |
| CD95 Genotypes | ||||||
| −670 A/A | 28 | 77.8 | 8 | 22.2 | 59.6 | S* |
| −670 A/G | 42 | 48.3 | 45 | 51.7 | 0.1 | NS |
| −670 G/G | 16 | 43.2 | 21 | 56.8 | 3.1 | NS |
| CD95 Alleles frequency | ||||||
| −670 A | 98 | 61.6 | 61 | 38.4 | 9.8 | S |
| −670 G | 74 | 46 | 87 | 54 | 0.9 | NS |
| CD95 Dominant model | ||||||
| A/A | 28 | 77.8 | 8 | 22.2 | 59.6 | S* |
| A/G + G/G | 58 | 46.8 | 66 | 53.2 | 0.5 | NS |
| CD95 Recessive model | ||||||
| A/A + A/G | 70 | 56.9 | 53 | 43.1 | 3.2 | NS |
| G/G | 16 | 43.2 | 21 | 56.8 | 3.1 | NS |
n: size of samples.
χ2-value; represent Chi-square calculated value.
NS: no significant difference with Chi-square value (χ2 < 3.84).
S: significant difference with Chi-square value (χ2 > 3.84).
S*: high significant difference with Chi-square value (χ2 > 20).
Association between hormonal profile and genotypes of P53 and CD95 in term of treatment response
The association between the hormonal profile, including prolactin, total testosterone, and growth hormone levels, as well as the genetic polymorphisms of P53 rs1042522 and CD95 rs1800682 was summarized in Table 6 among SVR and NR patients. This table showed that responders have high significant decreases of prolactin levels in case of P53 rs1042522 C/C genotype and CD95 rs1800682 A/A genotype (p < 0.01), high significant increases of total testosterone concentrations in case of P53 rs1042522 C/C genotype (p < 0.01), and significant increases of growth hormone levels in case of P53 rs1042522 G/G genotype (p < 0.01) and CD95 rs1800682 A/A genotype (p < 0.05) compared to non-responders.
Table 6.
Correlation between hormonal profile and genotypes of P53 and CD95 at +72 G/C and −670 A/G polymorphic points.
| Hormonal profile (mean ± SE) | Gene | Genotypes | SVR (n = 86) | NR (n = 74) | P |
|---|---|---|---|---|---|
| Prolactin, (ng/mL) | P53 | 72 G/G | 12.91 ± 0.69 | 11.29 ± 0.71 | >0.05 |
| 72 G/C | 16.80 ± 0.99 | 16.25 ± 0.87 | >0.05 | ||
| 72 C/C | 9.32 ± 0.28 | 12.39 ± 0.20 | <0.01 | ||
| CD95 | −670 A/A | 10.24 ± 0.51 | 13.73 ± 0.55 | <0.01 | |
| −670 A/G | 14.02 ± 0.79 | 13.12 ± 0.57 | >0.05 | ||
| −670 G/G | 14.77 ± 0.65 | 13.09 ± 0.65 | >0.05 | ||
| Total Testosterone, (ng/mL) | P53 | 72 G/G | 1.74 ± 0.21 | 1.83 ± 0.36 | >0.05 |
| 72 G/C | 1.87 ± 0.22 | 2.09 ± 0.26 | >0.05 | ||
| 72 C/C | 2.84 ± 0.23 | 1.55 ± 0.22 | <0.01 | ||
| CD95 | −670 A/A | 2.09 ± 0.19 | 1.75 ± 0.30 | >0.05 | |
| −670 A/G | 2.26 ± 0.22 | 2.01 ± 0.28 | >0.05 | ||
| −670 G/G | 2.12 ± 0.25 | 1.80 ± 0.26 | >0.05 | ||
| GH, (ng/ml) | P53 | 72 G/G | 0.98 ± 0.02 | 0.41 ± 0.13 | <0.01 |
| 72 G/C | 0.75 ± 0.10 | 0.66 ± 0.36 | >0.05 | ||
| 72 C/C | 0.82 ± 0.12 | 0.83 ± 0.16 | >0.05 | ||
| CD95 | −670 A/A | 0.84 ± 0.06 | 0.56 ± 0.12 | <0.05 | |
| −670 A/G | 0.85 ± 0.08 | 0.57 ± 0.33 | >0.05 | ||
| −670 G/G | 0.86 ± 0.11 | 0.76 ± 0.20 | >0.05 |
S.E.: standard error.
n: size of samples.
<0.01: represents high significant difference (P < 0.01).
<0.05: represents significant difference (P < 0.05).
>0.05: represents no significant difference (P > 0.05).
Logistic regression analyses
Table 7 summarizes the different logistic regressions of the human pre- and post-treatment prolactin and testosterone against different independent variables of hormonal and biochemical measured parameters in HCV responders and non-responders. Regression analysis for pre-treatment prolactin showed a significant positive correlation with post-treatment prolactin and a significant negative correlation with post-treatment testosterone in HCV responders and non-responders (p < 0.01). Meanwhile, pre-treatment prolactin showed a significant positive correlation with pre-treatment total bilirubin in responders (p < 0.005). On the other side, post-treatment testosterone had a significant positive correlation with pre-treatment testosterone and vice versa in HCV responders and non-responders (p < 0.01).
Table 7.
Logistic regression analyses of hormonal and biochemical variables in responders (SVR) and non-responders (NR).
| Dependent Variable (Log-transformed) | Independent Variables (Log-transformed) | SVR |
NR |
||||||
|---|---|---|---|---|---|---|---|---|---|
| B-regression coefficient | SE of regression coefficient | t | P | B-regression coefficient | SE of regression coefficient | t | P | ||
| Prolactin (A) | Intercept | 0.326 | 0.439 | 0.743 | >0.05 | 0.257 | 0.489 | 0.525 | >0.05 |
| BMI | 0.035 | 0.042 | 0.826 | >0.05 | 0.047 | 0.049 | 0.966 | >0.05 | |
| Testosterone (A) | 0.032 | 0.047 | 0.680 | >0.05 | 0.038 | 0.053 | 0.716 | >0.05 | |
| Prolactin (B) | 0.538 | 0.078 | 6.897 | <0.01 | 0.522 | 0.087 | 5.968 | <0.01 | |
| Testosterone (B) | −0.176 | 0.051 | −3.473 | <0.01 | −0.192 | 0.057 | −3.343 | <0.01 | |
| ALT | 0.094 | 0.048 | 1.960 | >0.05 | 0.094 | 0.058 | 1.607 | >0.05 | |
| AST | −0.010 | 0.049 | −0.199 | >0.05 | 0.006 | 0.058 | 0.100 | >0.05 | |
| T. Bilirubin | 0.177 | 0.079 | 2.227 | <0.05 | 0.175 | 0.089 | 1.956 | >0.05 | |
| D. Bilirubin | −0.056 | 0.057 | −0.993 | >0.05 | −0.060 | 0.063 | −0.954 | >0.05 | |
| Albumin | 0.291 | 0.184 | 1.582 | >0.05 | 0.292 | 0.203 | 1.437 | >0.05 | |
| Testosterone (A) | Intercept | 1.494 | 1.079 | 1.384 | >0.05 | 1.469 | 1.144 | 1.284 | >0.05 |
| BMI | 0.012 | 0.104 | 0.116 | >0.05 | −0.028 | 0.116 | −0.244 | >0.05 | |
| Prolactin (A) | 0.195 | 0.287 | 0.680 | >0.05 | 0.214 | 0.299 | 0.716 | >0.05 | |
| Prolactin (B) | −0.441 | 0.242 | −1.819 | >0.05 | −0.470 | 0.253 | −1.860 | >0.05 | |
| Testosterone (B) | 0.924 | 0.083 | 11.166 | <0.01 | 0.925 | 0.089 | 10.360 | <0.01 | |
| ALT | −0.032 | 0.122 | −0.264 | >0.05 | 0.013 | 0.141 | 0.090 | >0.05 | |
| AST | −0.009 | 0.122 | −0.074 | >0.05 | −0.027 | 0.137 | −0.200 | >0.05 | |
| T. Bilirubin | −0.212 | 0.202 | −1.053 | >0.05 | −0.201 | 0.216 | −0.929 | >0.05 | |
| D. Bilirubin | 0.057 | 0.142 | 0.402 | >0.05 | 0.054 | 0.149 | 0.365 | >0.05 | |
| Albumin | −0.633 | 0.458 | −1.383 | >0.05 | −0.570 | 0.483 | −1.182 | >0.05 | |
| Prolactin (B) | Intercept | 1.542 | 0.480 | 3.209 | <0.01 | 1.589 | 0.530 | 2.999 | <0.01 |
| BMI | −0.016 | 0.049 | −0.323 | >0.05 | −0.028 | 0.057 | −0.495 | >0.05 | |
| Prolactin (A) | 0.728 | 0.106 | 6.897 | <0.01 | 0.699 | 0.117 | 5.968 | <0.01 | |
| Testosterone (A) | −0.097 | 0.053 | −1.819 | >0.05 | −0.112 | 0.060 | −1.860 | >0.05 | |
| Testosterone (B) | −0.010 | 0.064 | −0.152 | >0.05 | 0.001 | 0.072 | 0.010 | >0.05 | |
| ALT | −0.024 | 0.057 | −0.421 | >0.05 | −0.035 | 0.069 | −0.510 | >0.05 | |
| AST | −0.083 | 0.056 | −1.477 | >0.05 | −0.070 | 0.066 | −1.056 | >0.05 | |
| T. Bilirubin | −0.132 | 0.094 | −1.400 | >0.05 | −0.134 | 0.105 | −1.273 | >0.05 | |
| D. Bilirubin | 0.089 | 0.066 | 1.361 | >0.05 | 0.094 | 0.072 | 1.305 | >0.05 | |
| Albumin | −0.247 | 0.216 | −1.145 | >0.05 | −0.206 | 0.237 | −0.868 | >0.05 | |
| Testosterone (B) | Intercept | 1.278 | 0.925 | 1.381 | >0.05 | 1.030 | 0.989 | 1.042 | >0.05 |
| BMI | 0.031 | 0.089 | 0.344 | >0.05 | 0.063 | 0.100 | 0.632 | >0.05 | |
| Prolactin (A) | −0.796 | 0.229 | −3.473 | <0.01 | −0.796 | 0.238 | −3.343 | <0.01 | |
| Testosterone (A) | 0.679 | 0.061 | 11.166 | <0.01 | 0.685 | 0.066 | 10.360 | <0.01 | |
| Prolactin (B) | −0.032 | 0.212 | −0.152 | >0.05 | 0.002 | 0.223 | 0.010 | >0.05 | |
| ALT | 0.099 | 0.104 | 0.950 | >0.05 | 0.054 | 0.121 | 0.449 | >0.05 | |
| AST | −0.079 | 0.104 | −0.760 | >0.05 | −0.014 | 0.118 | −0.119 | >0.05 | |
| T. Bilirubin | 0.279 | 0.171 | 1.631 | >0.05 | 0.246 | 0.185 | 1.331 | >0.05 | |
| D. Bilirubin | −0.025 | 0.121 | −0.209 | >0.05 | −0.025 | 0.128 | −0.192 | >0.05 | |
| Albumin | 0.570 | 0.392 | 1.454 | >0.05 | 0.538 | 0.415 | 1.298 | >0.05 | |
A: pre-treatment.
B: post-treatment.
S.E.: standard error.
<0.01: represents high significant difference (P < 0.01).
<0.05: represents significant difference (P < 0.05).
>0.05: represents no significant difference (P > 0.05).
Response rates of P53 rs1042522, CD95 rs1800682 versus IL28B rs12979860 and BCL2 rs1800477 polymorphisms
The treatment response rates of IL28B rs1297986039 versus P53 rs1042522, CD95 rs1800682, and BCL2 rs180047717 polymorphisms in HCV genotype-4 infected patients were illustrated in Fig. 3. IL28B rs12979860 C/C, P53 rs1042522 Arg/Arg (G/G), CD95 rs1800682 A/A, and BCL2 rs1800477 A/A genotypes have high treatment response rates compared to the remaining genotypes of the same polymorphic points. There was no significant difference of sustained virological response rates when comparing IL28B rs12979860 C/C variant with CD95 rs1800682 A/A variant and IL28B rs12979860 C/T variant with P53 rs1042522 Arg/Pro (G/C) variant. On the other side, there were significant increases of sustained virological response rates when comparing IL28B rs12979860 C/C variant with either P53 rs1042522 G/G variant or BCL2 rs1800477 G/G variant (p > 0.01), IL28B rs12979860 C/T variant with either CD95 rs1800682 A/G (p < 0.05) or BCL2 rs1800477 G/A variant (p < 0.01), and CD95 rs1800682 G/G variant with IL28B rs12979860 T/T variant (p < 0.05) or P53 rs1042522 Pro/Pro (C/C) variant (p < 0.01) or BCL2 rs1800477 A/A variant (p < 0.01).
Figure 3.
Response rates of IL28B rs12979860 versus P53 rs1042522, CD95 rs1800682, and BCL-2 rs1800477 polymorphisms according to sustained virological response (SVR) in HCV genotype-4 patients. (A/A, A/B, B/B); represent C/C, C/T, T/T genotypes of IL28B & G/G, G/C, CC genotypes of P53 & A/A, A/G, G/G genotypes of CD95 & G/G, G/A, A/A genotypes of BCL-2, respectively. a; represents high significant difference (p < 0.01), b; represents significant difference (p < 0.05), and c; represents no significant difference (p > 0.05).
ROC curve analyses
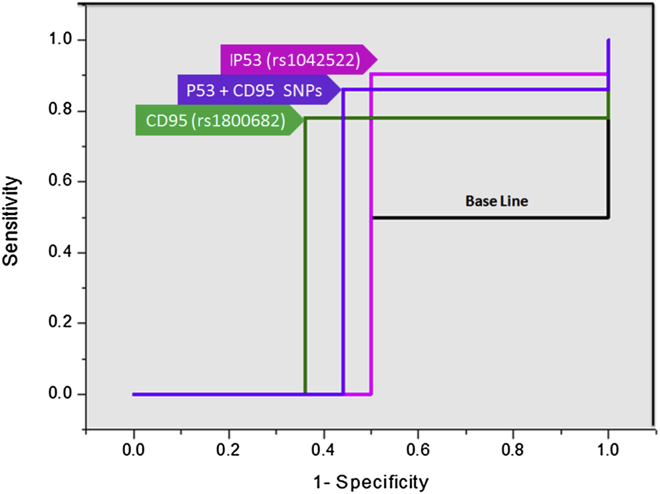
The significance of P53 rs1042522 Arg/Pro, CD95 rs1800682 A/G, and P53 rs1042522 Arg/Pro plus CD95 rs1800682 A/G polymorphisms as prognostic biomarkers for the HCV treatment response (responders, n = 86; non-responders, n = 74) was assessed using a receiver operating characteristic (ROC) curve. The P53 rs1042522 Arg/Pro, CD95 rs1800682 A/G, and the P53 rs1042522 Arg/Pro plus CD95 rs1800682 A/G genetic polymorphisms could be used to distinguish between the SVR and NR patients with area under the curve (AUC) of 0.702, 0.707, and 0.710 respectively (Odds ratio: 9.49, 4.61, and 5.60; 95% CI: 0.594–0.811, 0.582–0.832, and 0.630–0.790, respectively), sensitivity of 90.5%, 78%, and 86% as well as specificity of 50%, 36%, and 44% respectively (p = 0.001, 0.004, and 0.000, respectively) as illustrated in Fig. 4.
Figure 4.
ROC curve of P53 rs1042522, CD95 rs1800682, and P53 rs 1042522 plus CD95 rs 1800682 polymorphisms.
Sensitivity and specificity of P53, CD95, IL28B, IP-10, and IRRDR
A comparison between IL28B rs12979860, interferon gamma-induced protein-10 (IP-10 < 150 pg/mL), IL28B rs12979860 plus IP-10 < 150 pg/mL,40 IFN-RBV resistance-determining region (IRRDR/NS5A of HCV-4a),41 P53 rs1042522, CD95 rs1800682, and P53 rs1042522 plus CD95 rs1800682 was illustrated in Fig. 5 in terms of sensitivity and specificity according to sustained virological response rate among patients infected with HCV genotype-4a. By considering IL28B rs12979860 plus IP-10 < 150 pg/mL have the highest specificity,30 there were high significant increases when comparing this marker of response with the remaining ones in term of specificity (p < 0.01). Whereas, by considering P53 rs1042522 has the highest sensitivity, there were no significant increases when comparing this response marker with IRRDR/NS5A of HCV-4a and P53 rs1042522 plus CD95 rs1800682 (p > 0.05), while there were significant increases when comparing P53 rs1042522 with IL28B rs12979860, IP-10 < 150 pg/mL, IL28B rs12979860 plus IP-10 < 150 pg/mL, P53 rs1042522 (p < 0.01), and CD95 rs1800682 (p < 0.05).
Figure 5.
Comparative sensitivity and specificity of IL28B (rs12979860), IP-10<150 pg/mL, IL28B (rs12979860) plus IP-10<150 pg/mL, IRRDR (HCV-4a), P53 rs1042522, CD95 rs1800682, and P53 rs1042522 plus CD95 rs 1800682 in order to discriminate between the responders and non-responders among the chronic HCV genotype-4a patients.
Discussion
Several demographic, clinical, virological, and histological factors were studied among responder and non-responder patients infected with HCV genotype-4a. In addition, the association between these factors and the response to the antiviral therapy of pegylated-interferon and ribavirin was investigated. This study found that there was a potential impact of non-responsiveness to antiviral treatment on elevating frequencies of post-treatment ALT, AST, total bilirubin, direct bilirubin, and albumin. The statistical differences of these hepatic tests between responders and non-responders were supported by previous observations.42, 43 Thus, the early normalization of abnormal ALT and AST levels of HCV infection may indicate response to antiviral treatment which agrees with numerous studies.44, 45, 46, 47, 48 In addition, the present study has demonstrated that AFP was negatively associated with treatment response in 160 HCV genotype-4a infected patients, which supported by a previously reported study of 100 Egyptian HCV infected patients.49
On the virological and histological side, some studies have shown that high viral load of HCV-RNA is negatively associated with sustained virological response to treatment.50, 51, 52 This observation was confirmed by the current finding which indicates that pre-treatment response prediction was not associated with the HCV-RNA viral loads. On the contrary, there were high statistically significant differences of the low, moderate, and high viral loads when comparing post-treated responders with non-responders. In addition, there were high statistically significant differences of pre- and post-treatment hepatic activity and fibrotic scores when comparing responders with non-responders. This finding agreed with a previous observation indicates that high baseline viral load and high fibrosis stage were associated with non-responsiveness to antiviral treatment.53 On the other hand, previous studies found that non-responders have non-significant higher frequencies of viral load as well as higher grade of hepatic activity and fibrotic stages than in responders.17, 54
The present study found that Pro variant of P53 rs1042522 was high frequently found in non-responders compared to responders, while P53 recessive genetic model (Arg/Arg + Arg/Pro) was frequently found in responders compared to non-responders. Therefore, Pro variant and recessive model of P53 rs1042522 may be used as sensitive genetic biomarkers for non-responsiveness and responsiveness respectively to the antiviral dual therapy. Numerous studies have investigated the association between P53 rs1042522 and disease severity, while there was a lack of observations associated with treatment response. Very recently, it was investigated that P53 rs1042522 Pro/Pro and Arg/Arg genotypes may be potentially used as sensitive genetic markers for HCV genotype-4a susceptibility.38 Authors suggested that there was a frequent loss of proline allele in HCV-positive carriers which in turn plays critical role in hepatocarcinogenesis.10 Others investigated that P53 genetic polymorphisms were frequently found in patients with cirrhotic livers compared to patient with chronic hepatitis, suggesting that P53 polymorphisms at this stage may be a causative factor that may potentially leads to HCC.55 On the contrary, it was investigated that there was no association between codon +72 genotypes and HCV genotypes 2a and 2b infections,56 disease severity,20 or hepatocellular carcinoma19 compared to controls. Finally, there was a significant correlation between male homozygotes for P53 72Pro with HCV type 1b infection.56
In the current study, it was investigated that A/A variant of CD95 rs1800682 was high frequently found in responders compared to non-responders. Therefore, A/A variant of CD95 rs1800682 may be used as a strong polymorphic marker for responsiveness to the interferon-based therapy. Numerous studies have investigated the association between CD95 rs1800682 and disease severity, while there was a lack of observations associated with the treatment response. Recently, it was found that CD95 rs1800682 A/G polymorphism was potentially associated with significant fibrosis and cirrhosis in chronic HCV patients.23, 38
Reference to the current study and our recently published article,38 we can deduced that the Pro variant of P53 rs1042522 may be used as a sensitive genetic biomarker for non-responsiveness to the antiviral dual therapy (NR: SVR = 85.7%: 14.3%), HCV genotype-4a susceptibility (HCV: controls = 17.5%: 6.25%), and severity of inflammatory-based disease (Advanced: mild fibrosis = 28.95%: 12.5%). Meanwhile, the A/A variant of CD95 rs1800682 may be used as a strong polymorphic marker for responsiveness to the antiviral therapy (SVR: NR = 77.8%: 22.2%) and HCV clearance (Controls: HCV = 36.25%: 22.5%; Mild: advanced fibrosis = 29.1%: 13.2%).
The present finding was agreed with Ksiaa's findings who found a higher frequency of the A/A genotype of the CD95 gene in spontaneously recovered patients from HCV than in patients with persistent HCV infection.24 They concluded that the A/A genotype of the CD95 gene influences the outcome of HCV infection in patients on hemodialysis. In addition, Aguilar-Reina group investigated that there was an association between the CD95 rs1800682 A/G polymorphic spot and the grade of necrosis in chronic HCV patients.57 Conversely, others demonstrated that there was no correlation between CD95 promoter genotypes and fibrotic stages as well as CD95 gene polymorphism may account for some of the histopathological variability in HCV infection.25
Viral eradication from infected cells through apoptosis16 and inhibiting viral replication through enhancing immune response10 are two crucial processes associated with achieving viral clearance. It was suggested that IFN-α can induce apoptosis through BCL2 family members. Thus, BCL2 may be used as a prognostic predictor for IFN-α sensitivity of the disease.58 Recently, the potential role of the BCL2 rs1800477 polymorphism was examined among HCV patients treated with PEG-IFN-α and RBV antiviral therapy.17, 59 Authors concluded that polymorphism in BCL2 rs1800477 polymorphism can augment the current array of predictors of therapeutic response to PEG-IFN-α/RBV in patients infected with HCV genotype-4. On the other hand, IFN-l3, coded by the IL28B gene, involved in the antiviral immune response and inhibited HCV replication in vitro.10 In addition, the impact of the IL28B polymorphisms on HCV treatment was investigated in genome wide association study11 and it has been studied in several ethnicities and various viral genotypes.12, 13, 60 Moreover, IL28B rs12979860 C/C genotype,39 P53 rs1042522 G/G genotype, CD95 rs1800682 A/A genotype, BCL2 rs1800477 A/A genotype17 were associated with better treatment response rates compared to the remaining genotypes of the same polymorphic points.
Intriguingly, it was found that HCV genotype-4 patients who have normal growth hormone concentrations and BCL2 43Ala genotype can successfully achieve response to interferon-based therapy.61 In addition, the hormonal profile (prolactin, total testosterone, growth hormone) provides promising biomarkers for HCV treatment response when associated with P53 rs1042522 and CD95 rs1800682 genetic polymorphisms; where sustained virological responders have low prolactin levels and either P53 rs1042522 C/C genotype or CD95 rs1800682 A/A genotype, high total testosterone concentrations and P53 rs1042522 C/C genotype, as well as high growth hormone levels and either P53 rs1042522 G/G genotype or CD95 rs1800682 A/A genotype compared to non-responders.
Finally, we concluded that Pro variant of P53 rs1042522 may be used as a polymorphic predictor for non-responsiveness to antiviral therapy of HCV genotype-4a infection, while A/A variant of CD95 rs1800682 and recessive model of P53 rs1042522 may be used as genetic biomarkers for responsiveness to antiviral therapy of HCV genotype-4a infection among Egyptian population. In addition, the combination between the P53 rs1042522 and CD95 rs1800682 may be used as a powerful prognostic predictor for the HCV genotype-4a treatment response. Intriguingly, low prolactin, high total testosterone, and high growth hormone levels may provide promising biomarkers for early prediction of HCV treatment response when associated with P53 rs1042522 C/C and CD95 rs1800682 A/A genotypes.
Conflicts of interest
No conflict of interests.
Acknowledgments
This work was supported by Medical Research Division of the National Research Centre (NRC), Cairo, Egypt and the Egyptian Science and Technology Development Fund (STDF) [Grant number 1512 to Olfat G. Shaker].
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Choo Q.L., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Zidan A., Scheuerlein H., Schule S., Settmacher U., Rauchfuss F. Epidemiological pattern of hepatitis B and hepatitis C as etiological agents for hepatocellular carcinoma in Iran and worldwide. Hepat Mon. 2012;12:e6894. doi: 10.5812/hepatmon.6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohd Hanafiah K., Groeger J., Flaxman A.D., Wiersma S.T. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. J Hepatol. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 4.Belousova V., Abd-Rabou A.A., Mousa S.A. Recent advances and future directions in the management of hepatitis C infections. Pharmacol Ther. 2015;145:92–102. doi: 10.1016/j.pharmthera.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 5.U.S. Food and Drug Administration Approves Gilead's Sovaldi™ (Sofosbuvir) for the Treatment of Chronic Hepatitis C [press release] 2013. [Google Scholar]
- 6.Organization WH . 2002. Global Alert and Response (GAR), Hepatitis C.http://www.who.int/csr/disease/hepatitis/whocdscsrlyo2003/en/index2.html Available from: [Google Scholar]
- 7.Kamal S.M., El Kamary S.S., Shardell M.D. Pegylated interferon alpha-2b plus ribavirin in patients with genotype 4 chronic hepatitis C: the role of rapid and early virologic response. Hepatology. 2007;46:1732–1740. doi: 10.1002/hep.21917. [DOI] [PubMed] [Google Scholar]
- 8.Simmonds P., Bukh J., Combet C. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 9.Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y., Yang H., Borg B.B. A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection. Proc Natl Acad Sci USA. 2007;104:985–990. doi: 10.1073/pnas.0609954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge D., Fellay J., Thompson A.J. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 12.Afdhal N.H., McHutchison J.G., Zeuzem S. Hepatitis C pharmacogenetics: state of the art in 2010. J Hepatol. 2011;53:336–345. doi: 10.1002/hep.24052. [DOI] [PubMed] [Google Scholar]
- 13.Sarrazin C., Susser S., Doehring A. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415–421. doi: 10.1016/j.jhep.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 14.Akuta N., Suzuki F., Hirakawa M. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatol. 2010;52:421–429. doi: 10.1002/hep.23690. [DOI] [PubMed] [Google Scholar]
- 15.Rauch A., Kutalik Z., Descombes P., Swiss Hepatitis CCS, Swiss HIVCS Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 16.Teodoro J., Branton P. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaker O.G., Eskander E.F., Yahya S.M., Mohamed M.S., Abd-Rabou A.A. Genetic variation in BCL-2 and response to interferon in hepatitis C virus type 4 patients. Clin Chim Acta. 2011;412:593–598. doi: 10.1016/j.cca.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Matlashewski G.J., Tuck S., Pim D., Lamb P., Schneider J., Crawford L.V. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anzola M., Cuevas N., Lopez-Martinez M., Saiz A., Burgos J.J., de Pancorbo M.M. Frequent loss of p53 codon 72 pro variant in hepatitis C virus-positive carriers with hepatocellular carcinoma. Cancer Lett. 2003;193:199–205. doi: 10.1016/s0304-3835(03)00046-6. [DOI] [PubMed] [Google Scholar]
- 20.Leveri M.G.C., Rossi L., Zavaglia C., Civardi E., Mondelli M.U. Codon 72 polymorphism of P53 gene does not affect the risk of cirrhosis and hepatocarcinoma in HCV-infected patients. Cancer Lett. 2004;208:75–79. doi: 10.1016/j.canlet.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu N., Hayashi N., Katayama K. Immunohistochemical detection of Fas antigen in liver tissue of patients with chronic hepatitis C. J Hepatol. 1994;19:1354–1359. [PubMed] [Google Scholar]
- 22.Fischer R., Baumert T., Blum H.E. Hepatitis C virus infection and apoptosis. World J Gastroent. 2007;13:4865–4872. doi: 10.3748/wjg.v13.i36.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deghady A., Abdou A., El-Neanaey W.A., Diab I. Association of genetic polymorphism -670A>G in the Fas gene and serum markers AST platelet ratio index, AST/ALT with significant fibrosis and cirrhosis in chronic hepatitis C. Genet Test Mol Biomarkers. 2012;16:531–535. doi: 10.1089/gtmb.2011.0098. [DOI] [PubMed] [Google Scholar]
- 24.Ksiaa Cheikhrouhou L., Sfar I., Aounallah-Skhiri H. Cytokine and apoptosis gene polymorphisms influence the outcome of hepatitis C virus infection. Hepat Pancr Dis Int. 2011;10:280–288. doi: 10.1016/s1499-3872(11)60047-7. [DOI] [PubMed] [Google Scholar]
- 25.McIlroy D., Theodorou I., Ratziu V. FAS promoter polymorphisms correlate with activity grade in hepatitis C patients. Eur J Gastroenterol Hepatol. 2005;17:1081–1088. doi: 10.1097/00042737-200510000-00012. [DOI] [PubMed] [Google Scholar]
- 26.Liu N., Mertani H.C., Norstedt G., Tornell J., Lobie P.E. Mode of the autocrine/paracrine mechanism of growth hormone action. Exp Cell Res. 1997;237:196–206. doi: 10.1006/excr.1997.3789. [DOI] [PubMed] [Google Scholar]
- 27.Harvey S., Hull K.L. Growth hormone. A paracrine growth factor? Endocrine. 1997;7:267–279. doi: 10.1007/BF02801319. [DOI] [PubMed] [Google Scholar]
- 28.Nanbu-Wakao R., Fujitani Y., Masuho Y., Muramatu M., Wakao H. Prolactin enhances CCAAT enhancer-binding protein-beta (C/EBP beta) and peroxisome proliferator-activated receptor gamma (PPAR gamma) messenger RNA expression and stimulates adipogenic conversion of NIH-3T3 cells. Mol Endocrinol. 2000;14:307–316. doi: 10.1210/mend.14.2.0420. [DOI] [PubMed] [Google Scholar]
- 29.Horseman N.D. Kluwer Academic Publishers; Boston: 2001. Prolactin. [Google Scholar]
- 30.Ali A., Zein N.N. Hepatitis C infection: a systemic disease with extrahepatic manifestations. Clevel Clin J Med. 2005;72:1005–1008. doi: 10.3949/ccjm.72.11.1005. [DOI] [PubMed] [Google Scholar]
- 31.Kraus M.R., Schäfer A., Bentink T., Scheurlen M., Weissbrich B. Sexual dysfunction in males with chronic hepatitis C and antiviral therapy: interferon-induced functional androgen deficiency or depression? J Endocrinol. 2005;185:345–352. doi: 10.1677/joe.1.06007. [DOI] [PubMed] [Google Scholar]
- 32.Eskander E.F., Abd-Rabou A.A., Yahya S.M., El Sherbini A., Mohamed M., Shaker O. The impact of pegylated-interferon-α plus ribavirin on prolactinemia and testosteronemia among hepatitis C Genotype-4a patients. Open J Biochem. 2014;2 [Google Scholar]
- 33.Devaux A., Soula V., Sifer C. Hepatitis C virus detection in follicular fluid and culture media from HCV+ women, and viral risk during IVF procedures. Hum Reprod. 2003;18:2342–2349. doi: 10.1093/humrep/deg431. [DOI] [PubMed] [Google Scholar]
- 34.Plockinger U., Kruger D., Bergk A., Weich V., Wiedenmann B., Berg T. Hepatitis C patients have reduced Growth hormone secretion which improves during long term therapy with pegylated interferon alpha. Am J Gastroent. 2007;102:2724–2731. doi: 10.1111/j.1572-0241.2007.01445.x. [DOI] [PubMed] [Google Scholar]
- 35.Eskander E.F., Abd-Rabou A.A., Yahya S.M., Shaker O.G., Mohamed M.S. Does interferon and ribavirin combination therapy ameliorate growth hormone deficiency in HCV genotype-4 infected patients? Clin Biochem. 2012;45:3–6. doi: 10.1016/j.clinbiochem.2011.08.1145. [DOI] [PubMed] [Google Scholar]
- 36.Bedossa P., Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. J Hepatol. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 37.Ohno O., Mizokami M., Wu R.R. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201–207. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eskander E.F., Abd-Rabou A.A., Mohamed M.S., Yahya S.M.M., El Sherbini A., Shaker O.G. The potential impact of P53 and APO-1 genetic polymorphisms on hepatitis C genotype 4a susceptibility. Gene. 2014;550:40–45. doi: 10.1016/j.gene.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Asselah T., Muynck S.D., Broët P. IL28B polymorphism is associated with treatment response in patients with genotype 4 chronic hepatitis C. J Hepatol. 2012;56:527–532. doi: 10.1016/j.jhep.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Lagging M., Askarieh G., Negro F. Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-Related single nucleotide polymorphisms. PLoS ONE. 2011;2:e17232. doi: 10.1371/journal.pone.0017232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Shamy A., Shoji I., El-Akel W. NS5A sequence heterogeneity of hepaptitis C virus genotype 4a predicts clinical outcome of pegylated-interferon-ribavirin therapy in Egyptian patients. J Clin Micro. 2012;50:3886–3892. doi: 10.1128/JCM.02109-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis G.L., Balart L.A., Schiff E.R. Treatment of chronic hepatitis C with recombinant interferon alfa. A multicenter randomized, controlled trial. N Engl J Med. 1989;321:1501–1506. doi: 10.1056/NEJM198911303212203. [DOI] [PubMed] [Google Scholar]
- 43.Davis G.L., Lau J.Y. Factors predictive of a beneficial response to therapy of hepatitis C. J Hepatol. 1997;26:1225–1275. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 44.Roudot-Thoraval F., Bastie A., Pawlotsky J.-M., Dhumeaux D. Epidemiological factors affecting the severity of hepatitis C virus-related disease: a French survey of 6,664 patients. J Hepatol. 1997;26:485–490. doi: 10.1002/hep.510260233. [DOI] [PubMed] [Google Scholar]
- 45.Niederau C., Lange S., Heintges T. Prognosis of chronic hepatitis C: results of a large prospective cohort study. J Hepatol. 1998;28:1687–1695. doi: 10.1002/hep.510280632. [DOI] [PubMed] [Google Scholar]
- 46.Marcellin P. Hepatitis C: the clinical spectrum of the disease. J Hepatol. 1999;31:9–16. doi: 10.1016/s0168-8278(99)80368-7. [DOI] [PubMed] [Google Scholar]
- 47.Seeff L.B. Natural history of hepatitis C. J Hepatol. 2002;36:S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 48.Taketa K. Alfafetoprotein: reevaluation in hepatology. J Hepatol. 1998;12:1420–1432. [Google Scholar]
- 49.Males S., Raafat Gad R., Esmat G., Abobakr H., Anwar M. Serum alpha foetoprotein level predicts treatment outcome in chronic hepatitis C. Antivir Ther. 2007;12:797–803. [PubMed] [Google Scholar]
- 50.Hasan F., Asker H., Al-Khaldi J. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C genotype 4. Am J Gastroent. 2004;99:1733–1737. doi: 10.1111/j.1572-0241.2004.40077.x. [DOI] [PubMed] [Google Scholar]
- 51.Kamal S., El Tawil A., Nakano T. Peginterferon {alpha}-2b and ribavirin therapy in chronic hepatitis C genotype 4: impact of treatment duration and viral kinetics on sustained virological response. Gut. 2005;54:858–866. doi: 10.1136/gut.2004.057182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akuta N., Suzuki F., Sezaki H. Predective factors of virologic response to interferon- ribavirin combination therapy for patients infected with hepatitis C virus of genotype 1b and high viral load. J Med Virol. 2006;78:83–90. doi: 10.1002/jmv.20507. [DOI] [PubMed] [Google Scholar]
- 53.Gao B., Hong F., Radaeva S. Host factors and failure of interferon-alpha treatment in hepatitis C virus. J Hepatol. 2004;39:880–890. doi: 10.1002/hep.20139. [DOI] [PubMed] [Google Scholar]
- 54.Derbala M., Amer A., Bener A., Lopez A., Omar M., El-Ghannam M. Pegylated interferon-alpha 2b-ribavirin combination in Egyptian patients with genotype 4 chronic hepatitis. J Viral Hepatol. 2005;12:380–385. doi: 10.1111/j.1365-2893.2005.00604.x. [DOI] [PubMed] [Google Scholar]
- 55.Okada F., Shiraki T., Maekawa M., Sato S. A p53 polymorphism associated with increased risk of hepatitis C virus infection. Cancer Lett. 2001;172:137–142. doi: 10.1016/s0304-3835(01)00653-x. [DOI] [PubMed] [Google Scholar]
- 56.Minouchi K., Kaneko S., Kobayashi K. Mutation of p53 gene in regenerative nodules in cirrhotic liver. J Hepatol. 2002;37:231–239. doi: 10.1016/s0168-8278(02)00144-7. [DOI] [PubMed] [Google Scholar]
- 57.Aguilar-Reina J., Ruiz-Ferrer M., Pizarro M.A., Antinolo G. The -670A > G polymorphism in the promoter region of the FAS gene is associated with necrosis in periportal areas in patients with chronic hepatitis C. J Viral Hepatol. 2005;12:568–573. doi: 10.1111/j.1365-2893.2005.00639.x. [DOI] [PubMed] [Google Scholar]
- 58.Imam H., Gobl A., Eriksson B., Oberg K. Interferon-alpha induces bcl-2 proto-oncogene in patients with neuroendocrine gut tumor responding to its antitumor action. Anticancer Res. 1997;17:4659–4665. [PubMed] [Google Scholar]
- 59.Abd-Rabou A.A., Eskander E.F., Yahya S.M. LAMBERT Academic Publishing; 2012. p. vol. 168. (BCL2 Gene Polymorphisms and hGH Levels in HCV Patients). ISBN: 10365911832X. [Google Scholar]
- 60.Tanaka Y., Nishida N., Sugiyama M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 61.Eskander E.F., Abd-Rabou A.A., Yahya S.M., Mohamed M.S., Shaker O.G. Does HCV patients who have BCL2 43Ala genotype and Normal GH1 levels can achieve response to IFN based therapy? Ind J Clin Biochem. 2012;27:344–350. doi: 10.1007/s12291-012-0219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]