Abstract
Hepatocellular carcinoma (HCC) is one of the common lethal types of tumor all over the world. The lethality of HCC accounts for many reasons. One of them, the lack of reliable diagnostic markers at the early stage, in this context, serum miRNAs became promising diagnostic biomarkers. Herein, we aimed to identify the predictive value of two miRNAs (miR-122 and miR-224) in plasma of patients with HCC preceded by chronic HCV infection. Taqman miRNA assays specific for hsa-miR-122 and hsa-miR-224 were used to assess the expression levels of the chosen miRNAs in plasma samples collected from three groups; 40 patients with HCC related to HCV, 40 with CHC patients and 20 healthy volunteers. This study revealed that the mean plasma values of miRNA-122 were significantly lower among HCC group when compared to CHC and control groups (P < 0.001). Whereas, miR-224 mean plasma values were significantly higher among HCC group when compared to both CHC group and control group. Moreover, it was found that miR-122 can predict development of HCC at cut-off value <0.67 (RQ) and (AUC = 0.98, P < 0.001). As regards miR-224, it can predict development of HCC at cut-off value >1.2 (RQ) and (AUC = 0.93, P < 0.001), while the accuracy of AFP to diagnose HCC was (AUC: 0.619; P = 0.06). In conclusion, the expression plasma of miR-122 and miR-224 could be used as noninvasive biomarkers for the early prediction of developing HCC at the early stage.
Keywords: Diagnosis, Hepatocellular carcinoma, miR-122, miR-224, Sensitivity
List of Abbreviation: ADAM17, A disintegrin and metalloprotease domain-containing protein 17; AFP, Alpha-fetoprotein; AKT, AKT/Protein kinase B; ALP, Alkaline phosphatase; ALT, Alanine aminotransferase; ANOVA, Analysis of variance; API-5, Apoptosis inhibitor-5; AST, Aspartate aminotransferase; AUC, Area under the curve; Bcl-2, B cell leukemia/lymphoma 2 like protein; BCLC, Barcelona Clinic Liver Cancer; Ccgn1, Cyclin G1 protein; CT, Computed tomography; Ct, Cycle threshold; CTP, Child-Turcotte-Pugh; ELISA, Enzyme-linked immunosorbent assay; has-miR-122, Homo sapien-micro RNA-122; HCC, Hepatocellular carcinoma; HCV, Hepatitis C virus; miRNA/miR, Micro-RNA; mRNA, Messenger RNA; NF-κβ, nuclear factor kappa-light-chain-enhancer of activated B cells; PCR, Polymerase chain reaction; RNA, Ribonucleic acid; ROC, Receiver operating characteristic; RQ, Relative quantity; SE, standard error
Introduction
Hepatocellular carcinoma represents the 6th most common cancer worldwide and the 3rd most common cause of cancer death.1 Infection with hepatitis C virus (HCV) is a leading etiological factor for the developing HCC,2 especially in Egypt.3
The early screening of HCC depends on imaging techniques including mainly ultrasonography and laboratory tests involving mainly serum alpha-fetoprotein (AFP).4 However, ultrasonography is an operator-dependent procedure with varied diagnostic accuracy; in addition, it fails to detect small tumors.5 As well, the accuracy of AFP as a diagnostic biomarker for the screening of HCC patients at the early stage is modest with the sensitivity of 60%–80% and with the specificity of 70%–90%.4, 6 The lack of good diagnostic biomarkers for early-stage HCC accounts for low five-years survival rate from the time of diagnosis.4, 5 Thus, discovering minimally invasive sensitive and specific biomarkers for the early detection of HCC improving the prognosis of HCC patients is recommended.4
MicroRNAs are small, non-coding RNA molecules; act as post-transcriptional regulators for expression of genes involved in diverse biological processes that underlie physiological and pathological conditions.7
The differential expression of microRNAs deregulation has been reported in the development of many cancer types including HCC.8 Many studies have reported a number of circulating miRNAs as potential biomarkers for HCC diagnosis and/or good prognosis.9, 10, 11, 12, 13 For example, MiR-224 was reported to be specifically up regulated in HCV-related HCC liver tissues, compared to healthy livers.14 It acts as an oncomir in HCC cells and its up-regulation promotes malignant hepatocyte proliferation and migration via activating AKT signaling pathway,15 whereas miR-122, a liver-specific miRNA, may function as a tumor suppressor gene and its expression is commonly down or lost in HCC cells contributing to the tumorigenic phenotype of these cells.16 We began to study the involvement of miRNAs as noninvasive biomarkers in developing HCC of Egyptian patients.11
In this context, our study aimed to identify the performance of two plasma HCC-related miRNAs (miR-122 and miR-224), compared with the conventional marker serum AFP, in early prediction of primary HCC associated with chronic HCV infection.
Subjects and methods
This study was conducted on eighty adult chronic HCV patients (age range: 34–55 years) who were recruited from the outpatient clinic of Medical Services Unit at National Research Centre (NRC), from October 2015 to March 2016. 20 healthy subjects were involved in the current study as a control group. This study was approved by the NRC Ethical Committee on Human Research, and all patients signed consent documents allowing their clinical information to be gathered and analyzed for research purposes. The patients were categorized into two groups; CHC group (n = 40): patients with chronic hepatitis C infection (>6 months infection). HCC group (n = 40): patients with HCC related to chronic HCV evident by triphasic spiral Computed tomography (CT) abdomen. The selection of HCC patients at the early stage was done by using Okuda17 and Child-Turcotte-Pugh (CTP)18, 19 as well Barcelona clinic liver cancer (BCLC)20 staging systems.
Blood samples (10 ml) were withdrawn from enrolled subjects. Three ml were collected in tubes containing EDTA for processing total RNA extraction and miRNA. The remaining were left to clot at the temperature of a room then centrifuged and sera were separated for determination of biochemical parameters.
Serum alpha-fetoprotein (AFP) assessment
Sera from the three studied groups were used for estimation of AFP level by sandwich enzyme-linked immunosorbent assay using washer (State fax ®) reader (state fax chromate-3033®) and kit for AFP (Pointe Scientific, Catalog No. TM 1009).
Expression of Micro-RNA 122 and Micro-RNA 224 by RNA extraction and RT-quantitative PCR
RNA was isolated using RNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions for copurification of miRNA, then stored at −80 °C. MicroRNAs expression (miR-122 and miR-224) was determined by applying the TaqMan MicroRNA Assays (Applied Biosystems, Carlsbad, CA, USA). The extracted miRNA was reverse transcribed in the reaction mixture containing miR-specific stem-loop RT primers for each using Reverse Transcription Kit (Applied Biosystems). Master Mix of TaqMan Universal PCR without AmpErase UNG (Applied Biosystems) was applied for real-time PCR. The reaction was run on an ABI PRISM 7000 system (Applied Biosystems). The resulted miRNA data are calculated in relative to a RUN6B. All samples were measured in duplicates and Relative quantity (RQ) of miRNAs 122 and 224 was calculated by the formula (RQ = 2−ΔΔCt).
Statistical methods
Sigma Plot® 12.5 software was used for analysis of data. Quantitative data were demonstrated as mean ± standard error (SE). Analysis of data was done by one-way ANOVA test followed by Tukey's multiple comparison tests. Student's t-test was used to compare two quantitative variables. The relationship between miRNAs expressions and other variables was determined by Pearson correlation test. Receiver Operating Characteristic (ROC) curve analysis was used to determine the cutoff points that yielded ‘the highest sensitivity, specificity, and the diagnostic accuracy. Multiple logistic regressions were applied to evaluate how well combinations of the laboratory tests discriminate between patients and controls.
Results
The results of demographic data indicated male predominance among HCC patients and HCV patients (80.5% and 84.6% respectively). Highest mean values of age were found among patients with HCC (P = 0.31).
The clinical data were represented in Table 1 and revealed a highly statistically significant increase in the serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), total bilirubin and alpha-fetoprotein (AFP) in both HCC and CHC groups compared to controls. Also, highly statistically significant increases in the serum levels of AST, ALT, ALP and total bilirubin in HCC group compared with CHC group. While, serum albumin, prothrombin concentration and platelets count were significantly decreased in both CHC and HCC groups compared to control group. Moreover, the serum levels of albumin and platelets count were significantly lower among HCC patients than in CHC patients.
Table 1.
Demographic; biochemical data and miRNAs expression of the studied groups.
| Variables mean ± SE |
HCC group (n = 40) | CHC group (n = 40) | Control group (n = 20) | (P-value) |
|---|---|---|---|---|
| Gender | ||||
| Female n (%) | 7 (17.5%) | 6 (15%) | 4 (20%) | 0.88 |
| Male n (%) | 33 (82.5%) | 34 (85%) | 16 (80%) | |
| Age (years) | 52.03 ± 1.55 | 48.94 ± 1.34 | 50.75 ± 1.8 | 0.31 |
| AFP (ng/ml) | 228.3 ± 42.5a | 17.07 ± 2.64a | 6.5 ± 0.69 | 0.008 |
| AST (U/L) | 108.9 ± 7.9a,b | 44.3 ± 5.6a | 13.85 ± 0.76 | 0.006 |
| ALT (U/L) | 113.9 ± 9.1a,b | 75.1 ± 3.23a | 21.5 ± 1.64 | 0.005 |
| ALP (U/L) | 250.7 ± 15.2a,b | 98.8 ± 4.38a | 60.6 ± 2.01 | <0.001 |
| PC (%) | 64.7 ± 1.23a | 70.4 ± 2.62a | 92.35 ± 2.45 | <0.001 |
| Albumin (g/dl) | 2.3 ± 0.08a,b | 2.85 ± 0.13a | 4.43 ± 0.12 | <0.001 |
| T. Bilirubin (mg/dl) | 2.5 ± 0.19a,b | 1.4 ± 0.038a | 0.65 ± 0.053 | 0.003 |
| PLT (×103/μL) | 137.7 ± 4.82a,b | 193.3 ± 6.09a | 304 ± 14.58 | <0.001 |
| MiR-122 (RQ) | 0.13 ± 0.05a,b | 12.93 ± 1.8a | 1.02 ± 0.04 | <0.001 |
| MiR-224 (RQ) | 22.9 ± 2.1a,b | 1.43 ± 0.14 | 1.01 ± 0.05 | <0.001 |
SE: stander error, AFP: alpha fetoprotein, ALT: alanine aminotransferase, AST: aspartate aminotransferase, ALP: alkaline phosphatase, PC: prothrombin concentration, PLT: platelets Count.
Significant difference from control group.
Significant difference from CHC group.
Regarding to Child-Turcotte-Pugh staging (CTP), 30% of HCC patients were at stage A and 70% of them at stage B. Also, regarding to Barcelona clinic liver cancer (BCLC) staging all HCC patients were at the early stage of the tumor (A4). Moreover, 35% of them were with single focal lesion and 65% with multiple (2-3) focal lesions. As regards the tumor size, 75% of HCC patients with ≤2 cm and 25% of them with >2 cm as demonstrated in Table 2.
Table 2.
Tumor-related characteristics (n = 40).
| Number of focal lesions | |
| Single n (%) | 14 (35%) |
| Multiple (2-3) n (%) | 24 (26%) |
| Site of focal lesions | |
| Rt. Lobe n (%) | 19 (47.5%) |
| Lt. Lobe n (%) | 14 (35%) |
| Both n (%) | 7 (17.5) |
| Tumor Size by CT (cm) | 2.3 ± 0.09 |
| ≤ 2 cm, n (%) | 30 (75%) |
| > 2 cm, n (%) | 10 (25%) |
| Ascites | |
| No | 34 (85%) |
| Yes (mild) | 6 (15%) |
| CTP | |
| Stage | 2 (30%) |
| Stage B | 8 (70%) |
| BCLC | |
| A4 (Early HCC) n (%) | 40 (100%) |
CT: Computed tomography; CTP: Child-Turcotte-Pugh staging; BCLC: Barcelona clinic liver cancer staging classification.
The results of miRNAs expression showed that miRNA-122 was significantly lower in HCC group than both control and CHC groups by (7.6 and 99.2-fold change) respectively at P < 0.001. While the expression of miR-224 was significantly higher in HCC group than in both control and CHC groups by (22.9 and 16-fold change respectively) at P < 0.001 as demonstrated in Table 1.
In CHC group, expression of miR-122 showed significant direct correlation with AFP, ALT, AST, and ALP. While it was significantly inversely correlated with PC, albumin and platelets count. MiR-122 expression in HCC group showed inverse significant correlations with the AFP, AST, ALT, ALP level and with the size of the tumor. Meanwhile, it showed direct significant correlations with albumin and prothrombin concentration. On the other hand, miR-224 expression showed direct significant correlations with the AFP, ALT, AST, ALP levels and the size of the tumor. Meanwhile, it showed inverse significant correlation with albumin and prothrombin concentration Table 3.
Table 3.
Correlation of miR-122 and miR-224 level with clinical variables in CHC and HCC groups.
| CHC group |
HCC group |
|||||||
|---|---|---|---|---|---|---|---|---|
| MiR-122 |
MiR-224 |
MiR-122 |
MiR-224 |
|||||
| R | P-value | R | P-value | R | P-value | R | P-value | |
| AFP | 0.7 | <0.001 | 0.22 | 0.81 | −0.66 | <0.001 | 0.44 | 0.004 |
| ALT | 0.53 | <0.001 | −0.02 | 0.89 | −0.67 | <0.001 | 0.44 | 0.004 |
| AST | 0.57 | <0.001 | 0.06 | 0.69 | −0.61 | <0.001 | 0.52 | <0.001 |
| ALP | 0.47 | 0.002 | −0.07 | 0.69 | −0.66 | <0.001 | 0.37 | 0.02 |
| PC | −0.56 | <0.001 | −0.01 | 0.95 | 0.55 | <0.001 | −0.46 | 0.002 |
| PLT | −0.44 | 0.005 | 0.08 | 0.62 | −0.09 | 0.39 | −0.16 | 0.32 |
| Albumin | −0.54 | <0.001 | −0.15 | 0.35 | 0.54 | <0.001 | −0.5 | 0.001 |
| Bilirubin | −0.15 | 0.36 | −0.2 | 0.43 | −0.03 | 0.41 | 0.02 | 0.92 |
| Tumor Size | −0.35 | 0.03 | 0.87 | <0.001 | ||||
| MiR-122 | – | – | 0.04 | 0.81 | – | – | −0.34 | 0.03 |
| MiR-224 | 0.04 | 0.81 | – | – | −0.34 | 0.033 | – | – |
AFP: alpha fetoprotein, ALT: alanine aminotransferase, AST: aspartate aminotransferase, ALP: alkaline phosphatase, PC: prothrombin concentration, PLT: platelets Count, CT: Computed tomography.
Our results disclosed that miR-122 could predict HCC with sensitivity 87.5%, specificity 95%, accuracy 0.96, and cut of value <0.67 (RQ) while, miR-224 could predict HCC with sensitivity 92.5%, specificity 90%, accuracy 0.94, and cut of value >1.2 (RQ). However, the sensitivity and the specificity of AFP were 57.5% and 95% respectively as shown in Table 4.
Table 4.
Diagnostic performance of AFP and miRNAs for discriminating HCC patients from Control group and CHC patients.
| Sensitivity % | Specificity % | Cut-off | Accuracy | p-value | |
|---|---|---|---|---|---|
| HCC vs. control | |||||
| AFP | 57.5 | 95 | >10 (ng/ml) | 0.70 | 0.01 |
| MiR-122 | 87.5 | 95 | <0.67 (RQ) | 0.96 | <0.001 |
| MiR-224 | 92.5 | 90 | >1.2 (RQ) | 0.94 | <0.001 |
| HCC vs. CHC | |||||
| AFP | 58% | 100% | >85.9 (ng/ml) | 0.62 | 0.06 |
| MiR-122 | 87.5% | 97.5% | <0.21 (RQ) | 0.98 | <0.001 |
| MiR-224 | 87.5% | 97% | >3.9 (RQ) | 0.93 | <0.001 |
| AFP + miR-122 | 97.5% | 100% | – | 1 | <0.001 |
| AFP + miR-224 | 90% | 100% | – | 0.93 | <0.001 |
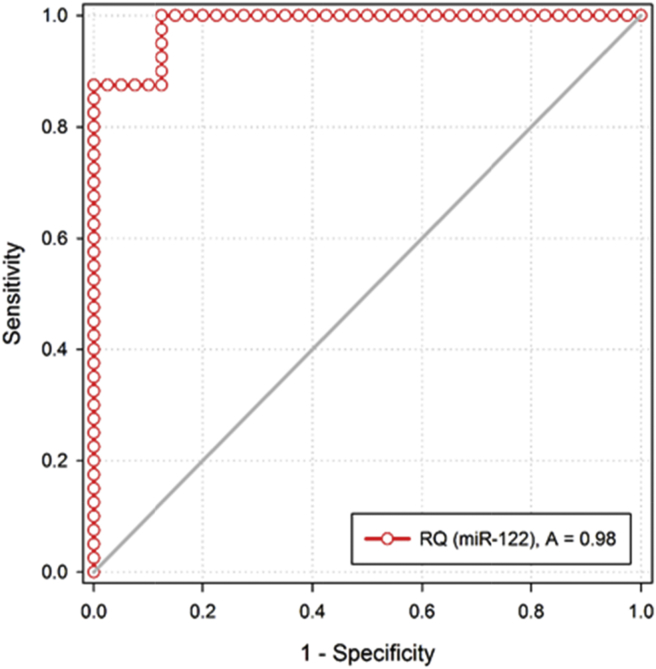
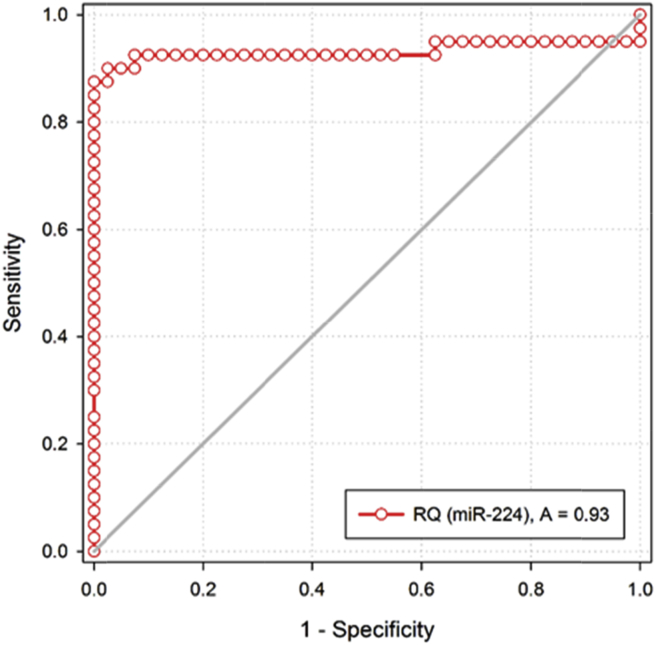
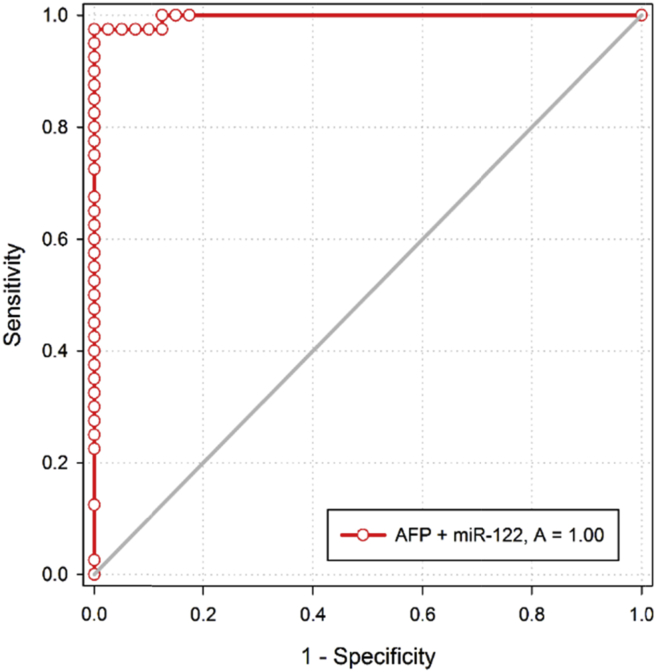
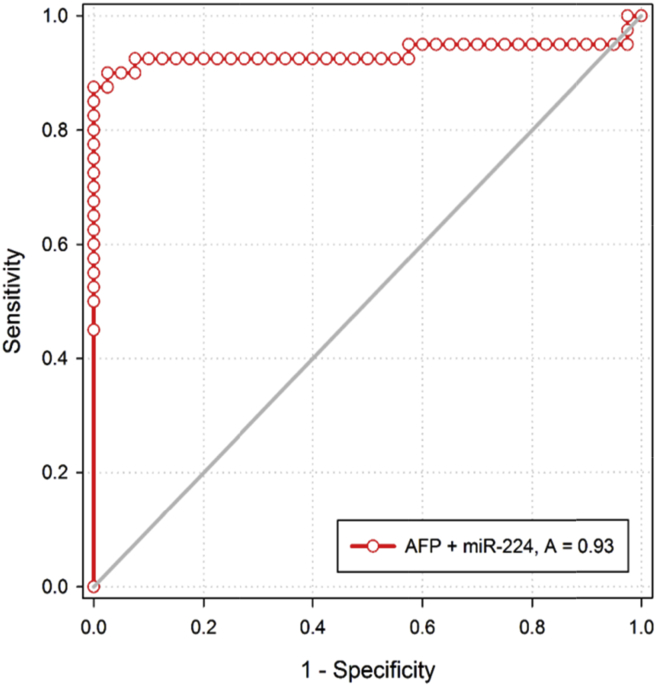
Both miR-122 and miR-224 showed higher diagnostic performance in distinguishing HCC patients from CHC patients (P < 0.001) as represented in Table 4 and Figure 1, Figure 2 and the combination of AFP with the two tested miRNAs as represented in Table 4 and Figure 3, Figure 4.
Figure 1.
ROC curve of RQ of miR-122 in discriminating HCC group from CHC group.
Figure 2.
ROC curve of RQ of miR-224 in discriminating HCC group from CHC group.
Figure 3.
ROC curve of AFP and RQ of miR-122 combined in discriminating HCC group from CHC group.
Figure 4.
ROC curve of AFP and RQ of miR-224 combined in discriminating HCC group from CHC group.
Discussion
HCC is a complex disease with multiple underlying pathogenic mechanisms caused by a variety of risk factors and it is difficult to characterize it with a single biomarker. AFP has mainly been used for diagnosis of primary HCC; however, its sensitivity is not satisfactory.21 Thus, signatures of combined biomarkers may be more valuable for the diagnosis, staging, and prognosis of HCC.
MiR-122 is liver specific miRNA and plays a central role in hepatocyte development and differentiation.22 Here, Plasma miR-122 was reported to be significantly down-expressed in HCC patients compared to healthy controls and CHC patients supporting its function as a tumor suppressor gene.23, 24 MiR-122 has a central role in the suppression of HCC,25 the role of miR-122 was suppression of oncogenic genes involved in diverse HCC hallmarks. Among these genes, Bcl-2 which can inhibit tumor cells apoptosis, Wnt1 which is responsible for the proliferation of cells, ADAM17 which is responsible for the metastasis and Ccgn1 which is responsible for the progression of the cell cycle.26 Moreover, miR-122 can inhibit angiogenesis and intrahepatic metastasis by suppressing the expression of tumor necrosis factor-α-converting enzyme.27
In contrary to these finding, circulated miR-122 was found to be up regulated in HCC in two studies by Varnholt et al28 and El-Garem et al.29 They suggested that miR-122 may down-regulate target mRNA of obscure tumor suppressor genes and in this way prompt further tumor development.
In our study, miR-122 expression was reported to be significantly higher in CHC patients when compared to healthy controls. That could be explained by the leakage of this miRNA from apoptotic or necrotic cells into the blood. MiR-122 may contribute to the pathogenesis of chronic HCV due to its function in HCV replication, translation, and inflammation.30, 31
Many studies have reported that miR-224 is one of the most commonly over expressed miRNAs that affect diverse crucial cellular pathways in HCC pathogenesis.32, 33 In the present work, over expression of plasma miR-224 was statistically significant in HCC patients compared to CHC and healthy controls reflecting liver damage. Wang and his colleagues,34 reported that miR-224 was up-regulated in HCC patients and HCC cell lines inhibiting tumor cell apoptosis by targeting apoptosis inhibitor 5 (API-5) and promoting cell growth. In addition, miR-224 can act as an onco-miRNA in HCC through activating the AKT signaling pathway and promoting malignant hepatocyte proliferation and migration. Therefore, the multiple roles of miR-224 were supported its involvement in the pathogenesis of liver cancer and its elevation to >20-fold.15, 35
The inflammatory pathways, for example, p65/NF-κβ is one of the ordinary mechanisms which induce liver damage, was identified as a direct transcriptional regulator of miR-224 expression and the link of miR-224 with cell proliferation, migration and invasion in HCC.32, 36 The usefulness of miR-224 as miRNA biomarker for clinical diagnosis is due to its multiple roles in the pathogenesis of liver cancer.
Interestingly, the statistical ROC curve analysis showed that miR-122 and miR-224 as potential biomarkers could predict HCC at the early stage (Table 4) these findings were in similar with two studies which confirmed that they were to be better than the sensitivity and accuracy of AFP in the diagnosis of HCC, especially at the early stage.31, 37
In the current work, serum miR-122 and miR-224 expressions showed higher diagnostic performance in distinguishing HCC from CHC patients (Figure 1, Figure 2). However, the conventional marker AFP for HCC failed to do this discrimination. The combination of AFP with each miR individually could increase the sensitivity (for miR-122 with 97.5% and for miR-224 with 90%) (Figure 3, Figure 4).
These results supported by Muawia and his colleagues38 which revealed that miR-122 presented a significant (AUC) of 0.705, sensitivity (63.64%) and specificity (75%) in distinguishing HCC patients from CHC patients.
It was evident from results of the current study that mean values of miR-224 were significantly increased among HCC patients when compared to patients with chronic HCV patients without HCC. At the same time it was significantly increased among patients with CHC without HCC when compared to controls.
This gradual increase from healthy livers to livers with HCC passing through CHC patients could allowing the dependence on miR-224 as early detector biomarker of HCC as it was low among controls, then slightly increased among CHC patients who suffer from liver cirrhosis which is pre malignant lesion and it was highly increased among HCC patients.
MiR-122 can easily discriminate HCC among patients with chronic HCV infection as suggested by findings of the current research. It was found that the mean values of miR-122 were significantly increased among CHC patients.
Conclusions
Plasma miR-122 and miR-224can be used as feasible noninvasive early detectors biomarkers for HCC at the early stage among patients with chronic HCV infection because of their sensible sensitivity and specificity for HCC, however, larger patient cohort analysis is recommended for clinical value.
We suggest paying more endeavors in the exploration zone of microRNAs which are identified with HCC planning to recognize the genetic mark of this cancerous disease; to investigate the riddle of HCC pathogenesis and to make ready for improvement of custom fitted treatment for HCC relying upon the contributing microRNAs.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Venook A.P., Papandreou C., Furuse J., Ladrón De Guevara L. The incidence and epidemiology of hepatocellular carcinoma: AGlobal and regional perspective. Oncol. 2010;15(Suppl 4):5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- 2.Vescovo T., Refolo G., Vitagliano G., Fimia G.M., Piacentini M. Molecular mechanisms of hepatitis C virus-induced hepatocellular carcinoma. Clin Microbiol Infect. 2016;22(10):853–861. doi: 10.1016/j.cmi.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita T., Honda M., Kaneko S. Molecular mechanisms of hepatocarcinogenesis in chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2011;26(6):960–964. doi: 10.1111/j.1440-1746.2011.06723.x. [DOI] [PubMed] [Google Scholar]
- 4.Huang J.T., Liu S.M., Ma H. Systematic review and meta-analysis: circulating miRNAs for diagnosis of hepatocellular carcinoma. J Cell Physiol. 2016;231(2):328–335. doi: 10.1002/jcp.25135. [DOI] [PubMed] [Google Scholar]
- 5.Qi J., Wang J., Katayama H., Sen S., Liu S. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60(2):135–142. doi: 10.4149/neo_2013_018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu M., Kong X., Wang H., Huang G., Ye C. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(5):8775–8784. doi: 10.18632/oncotarget.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macfarlane L.A., Murphy P.R. MicroRNA: biogenesis, function, and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong J., He X.X., Tian D.A. Emerging role of microRNA in hepatocellular carcinoma (Review) Oncol Lett. 2014:1027–1033. doi: 10.3892/ol.2014.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Abd N.E., Fawzy N.A., El-Sheikh S.M., Soliman M.E. Circulating miRNA-122, miRNA-199a, and miRNA-16 as biomarkers for early detection of hepatocellular carcinoma in egyptian patients with chronic hepatitis C virus infection. Mol Diagn Ther. 2015;19(4):213–220. doi: 10.1007/s40291-015-0148-1. [DOI] [PubMed] [Google Scholar]
- 10.Wen Y., Han J., Chen J. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137(7):1679–1690. doi: 10.1002/ijc.29544. [DOI] [PubMed] [Google Scholar]
- 11.Amr K.S., Ezzat W.M., Elhosary Y.A., Hegazy A.E., Fahim H.H., Kamel R.R. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene. 2016;575(1):66–70. doi: 10.1016/j.gene.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 12.Gougelet A., Colnot S. Hepatocellular carcinoma diagnosis: circulating microRNAs emerge as robust biomarkers. Clin Res Hepatol Gastroenterol. 2016;40(4):367–369. doi: 10.1016/j.clinre.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Okajima W., Komatsu S., Ichikawa D. Circulating microRNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget. 2016;7(33):53820–53836. doi: 10.18632/oncotarget.10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz G., Melis M., Tice A. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133(4):590–609. doi: 10.1002/ijc.28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma D., Tao X., Gao F., Fan C., Wu D. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol Lett. 2012;4(3):483–488. doi: 10.3892/ol.2012.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulouarn C., Factor V.M., Andersen J.B., Durkin M.E., Thorgeirsson S.S. Loss of miR 122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda K., Ohtsuki T., Obata H. Natural history of hepatocellular carcinoma and prognosis in relation to treatment study of 850 patients. Cancer. 1985;56(4):918–928. doi: 10.1002/1097-0142(19850815)56:4<918::aid-cncr2820560437>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 18.Child C.G., Turcotte J.G. Surgery and portal hypertension. Major Probl Clin Surg. 1964;1:1–85. [PubMed] [Google Scholar]
- 19.Pugh R.N., Murray-Lyon I.M., Dawson J.L., Pietroni M.C., Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60(8):1971–1974. doi: 10.1002/bjs.1800600817. [DOI] [PubMed] [Google Scholar]
- 20.Llovet J.M., Bru C., Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Seminars Liver Dis. 1999;19(3):329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 21.Zinkin N.T., Grall F., Bhaskar K. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 22.Morita K., Taketomi A., Shirabe K. Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 2011;31(4):474–484. doi: 10.1111/j.1478-3231.2010.02433.x. [DOI] [PubMed] [Google Scholar]
- 23.Girard M., Jacquemin E., Munnich A., Lyonnet S., Henrion-Caude A. MiR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48(4):648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 24.Luo J., Chen M., Huang H. Circulating microRNA-122a as a diagnostic marker for hepatocellular carcinoma. OncoTargets Ther. 2013;6:577–583. doi: 10.2147/OTT.S44215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zekri A., Youssef A., El-Desouky E. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumor Biol. 2016;37:12273–12286. doi: 10.1007/s13277-016-5097-8. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa T., Takata A., Otsuka M. Silencing of microRNA-122 enhances interferon-α signaling in the liver through regulating SOCS3 promoter methylation. Sci Rep. 2012;2:1–10. doi: 10.1038/srep00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai W.C., Hsu P.W., Lai T.C. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49(5):1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 28.Varnholt H., Drebber U., Schulze F. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47(4):1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 29.El-Garem H., Ammer A., Shehab H. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol. 2014;6(11):818–824. doi: 10.4254/wjh.v6.i11.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laterza O.F., Lim L., Garrett-Engele P.W. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55(11):1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 31.Xu J., Wu C., Che X. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50(2):136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Lee C.G. Role of miR-224 in hepatocellular carcinoma: a tool for possible therapeutic intervention? Epigenomics. 2011;3(2):235–243. doi: 10.2217/epi.11.5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Takahashi S., Tasaka A., Yoshima T., Ochi H., Chayama K. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol Aust. 2013;28(3):565–575. doi: 10.1111/j.1440-1746.2012.07271.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Lee A.T., Ma J.Z. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283(19):13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 35.Ladeiro Y., Couchy G., Balabaud C. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47(6):1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 36.Scisciani C., Vossio S., Guerrieri F. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol. 2012;56(4):855–861. doi: 10.1016/j.jhep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 37.Lin L., Lu B., Yu J., Liu W., Zhou A. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2015;40(4):397–404. doi: 10.1016/j.clinre.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Muawia S., El-Said H., Kamel T. Correlation of miR-122 with Bcl-w is a paradigm for the role of micro RNAs in the liver injury development. Int J Biol Sci Appl. 2015;2(6):86–96. [Google Scholar]