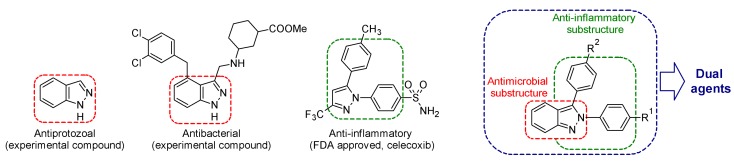
Abstract
Indazole is considered a very important scaffold in medicinal chemistry. It is commonly found in compounds with diverse biological activities, e.g., antimicrobial and anti-inflammatory agents. Considering that infectious diseases are associated to an inflammatory response, we designed a set of 2H-indazole derivatives by hybridization of cyclic systems commonly found in antimicrobial and anti-inflammatory compounds. The derivatives were synthesized and tested against selected intestinal and vaginal pathogens, including the protozoa Giardia intestinalis, Entamoeba histolytica, and Trichomonas vaginalis; the bacteria Escherichia coli and Salmonella enterica serovar Typhi; and the yeasts Candida albicans and Candida glabrata. Biological evaluations revealed that synthesized compounds have antiprotozoal activity and, in most cases, are more potent than the reference drug metronidazole, e.g., compound 18 is 12.8 times more active than metronidazole against G. intestinalis. Furthermore, two 2,3-diphenyl-2H-indazole derivatives (18 and 23) showed in vitro growth inhibition against Candida albicans and Candida glabrata. In addition to their antimicrobial activity, the anti-inflammatory potential for selected compounds was evaluated in silico and in vitro against human cyclooxygenase-2 (COX-2). The results showed that compounds 18, 21, 23, and 26 display in vitro inhibitory activity against COX-2, whereas docking calculations suggest a similar binding mode as compared to rofecoxib, the crystallographic reference.
Keywords: anticandidal, indazole, Entamoeba histolytica, Giardia intestinalis, rational drug design, Trichomonas vaginalis
1. Introduction
Infectious diseases caused by protozoa, bacteria, and yeasts have a major impact on human health. Enteric pathogenic protozoa and bacteria are a frequent cause of intestinal disease which, in turn, is an important cause of morbidity and mortality around the world [1]. Two important etiological agents of intestinal parasitic diseases are the protozoa Giardia intestinalis and Entamoeba histolytica, which have been estimated to affect 280 million and 50 million people worldwide each year, respectively [2,3]. Furthermore, some bacterial strains have been identified as responsible for severe intestinal illness. Examples of these are pathogenic strains of Escherichia coli, e.g., enterohemorrhagic E. coli (EHEC) and enteroaggregative E. coli (EAEC), and Salmonella enterica serovar Typhi [1,4]. Intestinal diseases caused by protozoa and bacteria affect persons of all ages, but have a high incidence in children [1,5]. Even though infections associated with each pathogen display particular clinical symptoms, all of them are causal agents of infectious diarrhea with severe health consequences and that could lead to death [1,4,5].
On the other hand, Trichomonas vaginalis and Candida albicans are two of the major etiological agents of vaginitis. According to the World Health Organization (WHO), 276 million new cases of trichomoniasis have been estimated [6]. Infection by T. vaginalis can cause severe inflammation of the genital tract, which has been associated with preterm labor, low-birth weight, sterility, cervical cancer, and predisposition to HIV infection [5,6,7]. In addition, it has been reported that 75% of women have at least one vaginal yeast infection during their lifespan [8]. Infections by Candida usually cause swelling, itching, and irritation and can turn into a very serious problem for pregnant and immunocompromised women [8,9].
Although some antimicrobial drugs are currently available for treatment of intestinal or vaginal infections, it has been reported that resistant strains of these microbes to the current therapies are emerging and that patients’ responses to the available chemotherapeutic agents vary [5,7,8,10]. Therefore, it is important to develop new active molecules to address these current health problems.
The indazole nucleus is a very important heterocyclic framework in medicinal chemistry. This scaffold is present in a large number of compounds with a wide range of biological activities [11]. Some indazole derivatives have recently been reported as antiprotozoals, with activity against E. histolytica and T. vaginalis [12,13]. Furthermore, indazole derivatives have been synthesized and tested against several Gram-positive and Gram-negative bacterial strains [11,14,15]. Particularly, 3-phenyl-1H-indazole, and some derivatives, have been identified as DNA gyrase B inhibitors [16]. Although, these reports give an insight of the potential of indazole derivatives as antiprotozoal and antibacterial agents, the information available is still limited. Therefore, it is necessary to synthesize new indazole derivatives to obtain more information about their antimicrobial potential. Considering a multitarget design approach [17], the derivatives presented in this work were designed from a combination of cyclic systems found in antiprotozoal [12,13], antibacterial [11,14,15,16,18], and anti-inflammatory compounds (Figure 1) [19,20,21]. This strategy was chosen because an inflammatory response is commonly found in infectious and parasitic diseases (e.g., amebiosis and trichomonosis). Moreover, previous studies showed that amebic infections induce host cyclooxygenase-2 (COX-2) and consequently the production of prostaglandin PGE2. Therefore, it has been suggested that PGE2 could play a major role in pathogenesis of E. histolytica [22,23]. Eighteen compounds including 2-phenyl-2H-indazole and 2,3-diphenyl-2H-indazole derivatives were synthesized and tested against the protozoa G. intestinalis, E. histolytica, and T. vaginalis. The more active antiprotozoal compounds were tested against some bacterial and yeast strains including enterohemorrhagic and enteroaggregative E. coli (strain 933/EHEC and strain 042/EAEC), S. enterica serovar Typhi, C. albicans, and C. glabrata. Additionally, six selected compounds were assayed in vitro and in silico as potential anti-inflammatory agents using the COX-2 inhibition assay. To shed light on the potential toxic effects, the cytotoxicity of a selected group of compounds was assessed on HaCaT (aneuploid immortal keratinocytes) and HeLa (human epitheloid cervix carcinoma) cells.
Figure 1.
Design of the 2,3-diphenyl-2H-indazole derivatives.
2. Results and Discussion
2.1. Chemical Synthesis
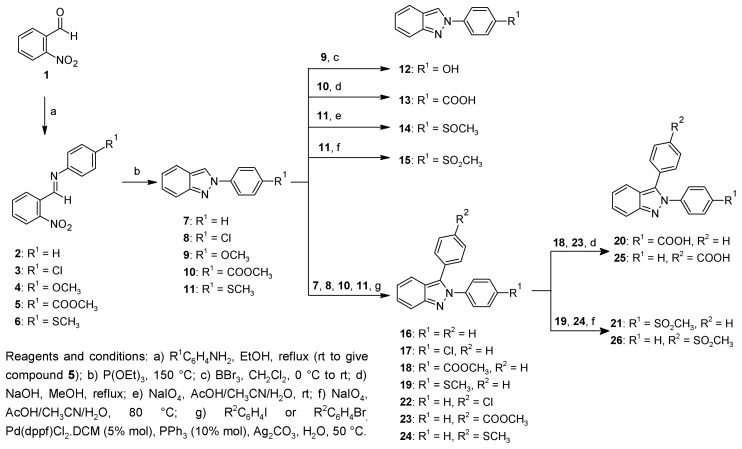
The 2-phenyl-2H-indazole and 2,3-diphenyl-2H-indazole derivatives (7–26) were synthesized as illustrated in Scheme 1. The commercially-available 2-nitrobenzaldehyde (1) was heated with aniline or p-substituted aniline under reflux conditions to afford the corresponding Schiff bases (2–4, 6); only the reaction to afford compound 5 was conducted at room temperature to achieve better results. Compounds 2–6 were reduced and cyclocondensed with P(OEt)3 to give the 2-phenyl-2H-indazole derivatives (7–11) by the Cadogan reaction [24]. Compound 12 was synthesized by o-demethylation of 9 with boron tribromide [25]. Hydrolysis of 10 with NaOH afforded the carboxylic acid 13. Compounds 14 and 15 were obtained by S-oxidation of 11 with sodium metaperiodate [26]. Compounds 16–19 and 22–24 were synthesized by a palladium-catalyzed arylation of the corresponding 2-phenyl-2H-indazole derivative with a variety of aryl iodides or bromides as previously reported [27]. Chemical yields for the palladium catalyzed arylation were slightly lower as compared to previously reported data by Ohnmacht et al. These results can be explained since the original reported methodology was scaled up tenfold to achieve the needed quantity of products for the biological assays. With the same procedure of hydrolysis and S-oxidation described above, compounds 20 and 25 were obtained from 18 and 23, respectively, whereas 19 and 24 were used as starting material to produce 21 and 26.
Scheme 1.
Synthesis of indazole derivatives 7–26.
All synthesized compounds have sharp melting points and were characterized by using 1H-NMR (nuclear magnetic resonance) and 13C-NMR spectra. The data on previously-reported structures were consistent with literature reports. Eight of the synthetized indazole derivatives resulted in new structures, which were also characterized by high-resolution mass spectrometry. The nuclear magnetic resonance and mass spectra of compounds can be found in Figures S1–S49 in the Supporting Information.
2.2. Antiprotozoal Activity
The in vitro antiprotozoal assays against E. histolytica, G. intestinalis, and T. vaginalis of the 2-phenyl-2H-indazole and 2,3-diphenyl-2H-indazole derivatives were carried out following the method previously described [28,29]. 2-Phenyl-2H-indazole derivatives 7–15 were evaluated and the results are shown in Table 1 as IC50 values. Metronidazole (MTZ) and albendazole (ABZ) were used as reference drugs. The most active 2-phenyl-2H-indazole derivatives against the three protozoa were compounds 8 and 10, these compounds containing 4-chlorophenyl and 4-(methoxycarbonyl)phenyl groups at position 2. Similarly, compound 7, having a phenyl at position 2, has good activity against the three protozoa, ranking third in activity for T. vaginalis and forth against G. intestinalis and E. histolytica. Additionally, compound 15, with 4-(methylsulfonyl)phenyl at the same position, can be classified among the fourth most active 2-phenyl-2H-indazole derivatives, at least for two parasites evaluated (G. intestinalis and T. vaginalis). Considering these results, 2,3-diphenyl-2H-indazole derivatives 16, 17, 18, 21, 22, 23, and 26 were selected to be tested for their antiprotozoal activity. These derivatives had groups H, COOCH3, Cl, and SO2CH3 at positions 2 and 3, substituents that induced the best response in 2-phenyl-2H-indazole derivatives. In addition, carboxylic acids 20 and 25 were of interest for comparative purposes with ester derivatives. Comparison of the 2,3-diphenyl-2H-indazole derivatives (16, 17, 18, 20, and 21) with its parent analogs 2-phenyl-2H-indazole derivatives (7, 8, 10, 13, and 15) indicated that only compounds 16, 17, and 20, retained or increased the potency against at least two parasites; however, the improvement in activity was poor. Compound 16 increased only its potency two-fold against G. intestinalis and T. vaginalis relative to 7; whereas compound 17 also increased its potency against E. histolytica two-fold when compared with 8, whereas the activity against G. intestinalis and T. vaginalis is preserved. A similar two-fold improvement in potency was found for compound 20, when compared to 13, against G. intestinalis and E. histolytica and a three-fold improvement against T. vaginalis.
Table 1.
Antiprotozoal activity of 2-pheny-2H-indazole derivatives and 2,3-diphenyl-2H-indazole derivatives.
| Compound | R1 | R2 | IC50 (µM) G. intestinalis | IC50 (µM) E. histolytica | IC50 (µM) T. vaginalis |
|---|---|---|---|---|---|
| 7 | H | – | 0.1133 ± 0.0218 | 0.0798 ± 0.0036 | 0.1184 ± 0.0218 |
| 8 | Cl | – | 0.0634 ± 0.0031 | 0.0415 ± 0.0031 | 0.1071 ± 0.0031 |
| 9 | OCH3 | – | 0.2051 ± 0.0063 | 0.1538 ± 0.0158 | 0.3723 ± 0.0158 |
| 10 | COOCH3 | – | 0.0634 ± 0.0056 | 0.0218 ± 0.0028 | 0.1070 ± 0.0056 |
| 11 | SCH3 | – | 0.2185 ± 0.0088 | 0.0978 ± 0.0147 | 0.2725 ± 0.0147 |
| 12 | OH | – | 0.1189 ± 0.0067 | 0.0737 ± 0.0101 | 0.1570 ± 0.0135 |
| 13 | COOH | – | 0.1931 ± 0.0119 | 0.0965 ± 0.0059 | 0.3274 ± 0.0178 |
| 14 | SOCH3 | – | 0.1678 ± 0.0110 | 0.0878 ± 0.0083 | 0.3121 ± 0.0110 |
| 15 | SO2CH3 | – | 0.0900 ± 0.0234 | 0.1359 ± 0.0052 | 0.1450 ± 0.0026 |
| 16 | H | H | 0.0518 ± 0.0052 | 0.3033 ± 0.0105 | 0.0573 ± 0.0026 |
| 17 | Cl | H | 0.0607 ± 0.0023 | 0.0213 ± 0.0023 | 0.1034 ± 0.0023 |
| 18 | COOCH3 | H | 0.0959 ± 0.0022 | 0.0502 ± 0.0022 | 0.1020 ± 0.0151 |
| 20 | COOH | H | 0.0795 ± 0.0045 | 0.0445 ± 0.0045 | 0.1113 ± 0.0180 |
| 21 | SO2CH3 | H | 0.1242 ± 0.0122 | 0.2081 ± 0.0061 | 0.2138 ± 0.0101 |
| 22 | H | Cl | 0.1132 ± 0.0070 | 0.0394 ± 0.0000 | 0.1181 ± 0.0046 |
| 23 | H | COOCH3 | 0.1188 ± 0.0086 | 0.0731 ± 0.0086 | 0.1431 ± 0.0043 |
| 25 | H | COOH | 0.1209 ± 0.0090 | 0.0509 ± 0.0000 | 0.2402 ± 0.0067 |
| 26 | H | SO2CH3 | 0.1062 ± 0.0081 | 0.0459 ± 0.0081 | 0.1837 ± 0.0162 |
| MTZ | – | – | 1.2260 ± 0.1250 | 0.3798 ± 0.1461 | 0.2360 ± 0.0160 |
| ABZ | – | – | 0.0370 ± 0.0030 | 56.5334 ± 18.8445 | 1.5905 ± 0.0113 |
On the other hand, the influence of changing Cl, COOCH3, SO2CH3, and COOH substituents from the phenyl group at position 2 to the phenyl group at position 3, was studied (compounds 22, 23, 25, and 26). The results showed that activities for compounds 22, 23, 25, and 26 were equal, or even lower, in most cases than their respective analogs 17, 18, 20, and 21 against the three parasites evaluated. The only exception was 26, which improved four-fold its activity against E. histolytica compared to 21. Although 2,3-diphenyl-2H-indazole derivatives have good antiprotozoal activity, most of the compounds have equal or even lower activity than its corresponding 2-phenyl-2H-indazole analog. Nevertheless, all tested compounds behave as potent antiprotozoal agents, in almost all cases better than metronidazole, the drug of choice. Additionally, most compounds were slightly more potent against E. histolytica, compared with the other two evaluated parasites. Although, some indazole derivatives have been reported as active compounds against E. histolytica and T. vaginalis, the activity against G. intestinalis had not been previously reported for derivatives having an indazole nucleus. Therefore, 2-phenyl-2H-indazole and 2,3-diphenyl-2H-indazole are promising frameworks for the design of new antiprotozoal agents.
2.3. Antibacterial and Anticandidal Assays
The susceptibility assays against E. coli 933, E. coli 042, S. enterica serovar Typhi, C. albicans, and C. glabrata were carried out using the disk diffusion test, in accordance with the procedure outlined by The Clinical and Laboratory Standards Institute (CLSI) [30]. A selection of compounds based on the results from the antiprotozoal assays were tested at 5 mg/mL (Table S1), however, they were inactive or poorly active even at high concentration against the bacterial strains tested. Nevertheless, compounds 18 and 23 showed a notable inhibition zone against C. albicans (inhibition halos of 10 and 13 mm, respectively). Moreover, these same compounds showed activity against C. glabrata (inhibition halos of 3 and 4 mm, respectively), which is usually less sensitive to the commercial antimycotics. Based on these observations, the minimum inhibitory concentration (MIC) against C. albicans and C. glabrata was calculated for compounds 18 and 23, Table 2. The results showed that both compounds display activity in the low millimolar range, and are slightly more active against C. albicans as compared to C. glabrata.
Table 2.
Antimycotic effect for selected compounds.
| Compound | MIC (mM) C. albicans | MIC (mM) C. glabrata |
|---|---|---|
| 18 | 3.807 | 15.227 |
| 23 | 3.807 | 15.227 |
| Ketoconazole | 0.045 | 0.079 |
2.4. In Vitro and In Silico Studies on Cyclooxygenase-2
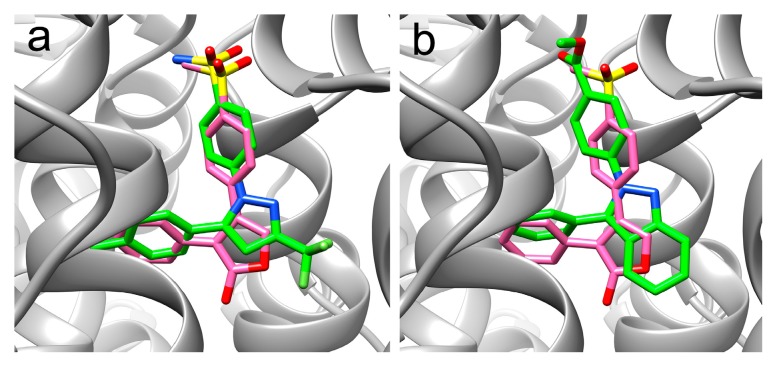
Considering that inflammatory response is associated with parasitic infections and the suggested role of PGE2, and therefore of the host COX-2 in the pathogenesis of E. histolytica [22], in vitro assays on human recombinant COX-2 were carried out. Additionally, molecular docking studies using AutoDock Vina software (TSRI, La Jolla, CA, USA) were performed to aid in the interpretation of the experimental results [31]. Compounds 18 and 23 were evaluated because of their strong antiprotozoal activity and their moderate anticandidal effects. Moreover, compounds 21 and 26 were of interest because of their methylsulfonyl group, which is commonly found in COX-2 inhibitors. Additionally, compounds 7 and 16 were considered as unsubstituted references. The results for in vitro assays and docking calculations are shown in Table 3. Compounds 7, 16, 18, 21, 23, and 26 were tested at 10 µM, whereas the positive reference celecoxib was used at 1 µM, as previously described [32]. The results showed in vitro COX-2 inhibition by compounds 18, 21, 23, and 26 (36–50%, at 10 µM); however, they are still weak inhibitors (see Table 3). Nevertheless, these compounds represent an interesting starting point towards the design of new antiparasitic compounds with an additional COX-2 inhibitory property. Docking studies suggest a similar binding mode of compounds 18, 21, 23, and 26 against human COX-2 as compared to celecoxib and the crystallographic ligand rofecoxib [33]. The predicted binding mode of the reference celecoxib and 18 are shown in Figure 2. Additionally, better docking scores were found for the 2,3-diphenyl-2H-indazole derivatives 16, 18, 21, 23, and 26 as compared to the 2H-indazole derivatives (e.g., compound 7).
Table 3.
Results for the in silico and in vitro evaluations against human COX-2 for selected compounds.
| Compound | Docking Score (Lowest Energy Conformation) | % of Inhibition of COX-2 |
|---|---|---|
| 7 1 | −8.0 | Inactive |
| 16 | −9.7 | Inactive |
| 18 | −9.5 | 50.01 ± 9.49 |
| 21 | −10.1 | 44.45 ± 2.65 |
| 23 | −10.0 | 36.35 ± 1.7 |
| 26 | −11.1 | 41.22 ± 5.93 |
| Celecoxib 2 | −11.7 | 64.92 ± 2.36 |
1 Compounds 7, 16, 18, 21, 23, and 26 were tested at 10 µM. 2 Reference tested at 1 µM.
Figure 2.
Predicted binding modes on human COX-2 for celecoxib (panel (a)) and 18 (panel (b)). Docked compounds are shown in green, whereas the crystallographic reference rofecoxib is in pink.
2.5. Cytotoxicity Assays
Biological assays on HaCaT and HeLa cell lines were conducted to gain insight into the cytotoxic effects of these derivatives on human cells, as compared with the effects observed on protozoa. Cellular viability was determined by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay [34,35]. Compounds 18 and 23 were chosen considering their antiprotozoal and anticandidal effect, in addition to their COX-2 inhibitory activity. Additionally, compound 16 was considered as an unsubstituted reference. The results of the cytotoxicity assays are shown in Table 4.
Table 4.
Results for cytotoxicity assays in HaCaT and HeLa cell lines.
| Compound | % Viability (10 µM) HaCaT Cells 1 | % Viability (10 µM) HeLa Cells | IC50 (µM) HaCaT cells 2 | IC50 (µM) HeLa Cells |
|---|---|---|---|---|
| 16 | 95.01 ± 2.44 | 93.04 ± 4.57 | 93.65 ± 17.30 | 125.00 ± 29.60 |
| 18 | 96.25 ± 4.14 | 94.14 ± 3.31 | - | - |
| 23 | 97.83 ± 5.19 | 93.72 ± 7.48 | - | - |
1 Percent of viability for selected compounds at 10 μM. 2 Half maximal inhibitory concentration for a selected compound.
Although the IC50 determinations were limited by the low solubility of the compounds in the cell culture medium at higher concentrations than 10 µM, they did not exhibit important cytotoxic effect at 10 µM in either of the cell lines (% viability > 90%). Therefore, the IC50 values are higher than 10 µM in all cases. Since all tested compounds showed antiprotozoal activity (IC50 values) lower than one micromolar (high nanomolar range), the results indicated that compounds 16, 18, and 23 are selective antiprotozoal compounds. Only compound 16 was soluble enough for IC50 determination on HaCaT and HeLa cell lines, having values of 93.65 and 125.00 µM, respectively.
In summary, nine 2-phenyl-2H-indazole derivatives (7–15) and eleven 2,3-diphenyl-2H-indazole derivatives (16–26) were synthesized. Eight compounds resulted in new structures (14, 15, 18–21, 25, and 26). Biological evaluations revealed that 2-phenyl-2H-indazole and 2,3-diphenyl-2H-indazole derivatives have giardicidal, amebicidal, and trichomonicidal activity lower than one micromolar and, in most cases, are more potent than the drug of choice metronidazole. Although the compounds are mainly inactive against the used bacterial strains, a major finding was that most of the compounds are selective antiprotozoal agents. In addition, compounds 18 and 23 inhibit in vitro growth of C. albicans and C. glabrata. Furthermore, compounds 18, 21, 23, and 26 showed inhibition of COX-2 at 10 µM, which adds an interesting property to these 2,3-diphenyl-2H-indazole derivatives, since COX-2 inhibition has been suggested to be beneficial on E. histolytica infections. Assays in HaCaT and HeLa cells revealed low cytotoxicity in human cells for a selection of these derivatives. These results suggest that 2-phenyl-2H-indazole and 2,3-diphenyl-2H-indazole are promising scaffolds for the design of new compounds against intestinal and vaginal pathogens, such as protozoa and yeasts. The mechanisms of action of indazole derivatives synthesized in this work as antiprotozoal and anticandidal agents are still unknown and constitutes a further research topic to be addressed in future research.
3. Materials and Methods
3.1. Chemicals and Instruments
All chemicals and starting materials were obtained from Sigma-Aldrich (Toluca, MC, Mexico). Reactions were monitored by TLC on 0.2 mm percolated silica gel 60 F254 plates (Merck, Darmstadt, Germany) and visualized by irradiation with a UV lamp. Silica gel 60 (70–230 mesh) was used for column chromatography. Melting points were determined in open capillary tubes with a Büchi M-565 melting point apparatus (Flawil, Switzerland) and are uncorrected. 1H-NMR and 13C-NMR spectra were measured with an Agilent DD2 spectrometer (Santa Clara, CA, USA), operating at 600 MHz and 151 MHz for 1H and 13C, respectively. Chemical shifts are given in parts per million relative to tetramethylsilane (Me4Si, δ = 0); J values are given in Hz. Splitting patterns are expressed as follow: s, singlet; d, doublet; q, quartet; dd, doublet of doublet; t, triplet; m, multiplet; bs, broad singlet. High-resolution mass spectra were recorded on a Bruker ESI/APCI-TOF, MicroTOF-II-Focus spectrometer (Billerica, MA, USA) by electrospray ionization (ESI). All compounds were named using the automatic name generator tool implemented in ChemBioDraw Ultra 13.0 software (PerkinElmer, Waltham, MA, USA), according IUPAC rules.
3.2. Chemical Synthesis
General procedure for the synthesis of 1-(2-nitrophenyl)-N-phenylmethanimines (2–6). 2-Nitrobenzaldehyde (5 g, 33.08 mmol) and aniline or the corresponding substituted aniline (33.08 mmol, 1 eq) were dissolved in ethanol (12–40 mL; the minimum quantity to dissolve the starting materials) and stirred at reflux temperature for 1–4 h to yield compounds 2–4, 6. Finally, the mixture was cooled to induce crystallization and the solid formed was separated using vacuum filtration and washed with cold ethanol. This same reaction was carried out at room temperature to yield compound 5.
General procedure for the synthesis of 2-phenyl-2H-indazole derivatives (7–11). 2-Phenyl-2H-indazole derivatives were synthesized employing a slight modification of the Cadogan method [24]. The corresponding imine 2–6 (20 mmol) was heated in triethyl phosphite (60 mmol) at 150 °C (0.5–2 h) until the starting material was totally consumed. Then, phosphite and phosphate were separated using vacuum distillation and the residue was purified using column chromatography with hexane–ethyl acetate (90:10) as a mobile phase to give the respective 2-phenyl-2H-indazole derivatives 7–9 and 11. A slightly more polar mobile phase was used for the purification of the compound 10, hexane-ethyl acetate (80:20).
4-(2H-indazol-2-yl) phenol (12). Compound 9 (4 mmol) was dissolved in dichloromethane (12 mL) and cooled to 0 °C under N2 atmosphere. Then, boron tribromide (12 mL of 1 M solution in dichloromethane, 12 mmol) was added and the reaction mixture was warmed to room temperature and stirred overnight. After completion of the reaction, a saturated sodium bicarbonate solution was added and the solid formed was filtered under vacuum. The crude product was purified using a short column packed with silica gel and ethyl acetate-hexanes (6:4) as a mobile phase to give compound 12.
General procedure for the synthesis of derivatives 13, 20, and 25. The appropriate methyl ester derivative (10, 18, and 23, 1.2 mmol) was dissolved in methanol (7.5 mL) and an aqueous solution of NaOH (3.6 mmol in 3 mL of water) was added. The reaction mixture was heated under reflux for five hours. After completion of the reaction, the mixture was cooled on ice and acidified to pH 1 with HCl to induce precipitation. The solid was separated using vacuum filtration and dried.
2-(4-(Methylsulfinyl) phenyl)-2H-indazole (14). To a solution of compound 11 (0.8 mmol) in 28 mL of CH3CN/CH3COOH (1:1), NaIO4 (0.8 mmol) dissolved in 2 mL of H2O/AcOH (4:1) was added. The reaction mixture was stirred at room temperature for 24 h. Then, the reaction was neutralized with a saturated solution of sodium bicarbonate and the product was extracted with dichloromethane (3 × 50 mL). The organic phase was dried with anhydrous sodium sulfate and concentrated under vacuum. The evaporation residue was purified by column chromatography using dichloromethane/methanol (98:2) as a mobile phase to give compound 14.
General procedure for the synthesis of derivatives 15, 21, and 26. NaIO4 (5 mmol) dissolved in 5 mL of H2O/AcOH (4:1) were added to a solution of the proper indazole derivative 11, 19, or 24 (2 mmol) in 28 mL of CH3CN/CH3COOH (1:1). The reaction mixture was stirred at reflux temperature for 12 h. Then, the mixture was neutralized with a saturated solution of sodium bicarbonate and brine solution was added until complete precipitation. The solid was separated using vacuum filtration and dried. The crude product was purified by column chromatography using dichloromethane as a mobile phase.
General procedure for the synthesis of 2,3-diphenyl-2H-indazole derivatives 16–19 and 22–24. Compounds 16–19 and 22–24 were synthesized by a palladium catalyzed arylation as previously described by Ohnmacht et al. [27]. It is worth mentioning that the previously-reported methodology was scaled up to 0.5 g of starting 2-phenyl-2H-indazole. Whereas compounds 16–19, 22, and 23, were synthesized using the proper 2-phenyl-2H-indazole and the substituted 4-iodobenzene, only compound 24 was synthesized from 2-phenyl-2H-indazole and 4-bromothioanisole.
1-(2-Nitrophenyl)-N-phenylmethanimine (2). Yellow solid (93% yield); m.p.: 64.1–64.9 °C (lit [24]: 63–64 °C); 1H-NMR (600 MHz, CDCl3) δ 8.94 (s, 1H), 8.31 (dd, J = 7.8, 1.4 Hz, 1H), 8.07 (dd, J = 8.2, 1.1 Hz, 1H), 7.74 (t, J = 7.6 Hz, 1H), 7.64–7.60 (m, 1H), 7.45–7.40 (m, 2H), 7.31–7.27 (m, 3H); 13C-NMR (151 MHz, CDCl3) δ 155.84, 151.07, 149.34, 133.58, 131.18, 131.12, 129.75, 129.28, 126.92, 124.54, 121.18.
N-(4-Chlorophenyl)-1-(2-nitrophenyl) methanimine (3). Dark yellow solid (91% yield); m.p.: 91.2–92.2 °C (lit [36]: 91–92 °C). 1H-NMR (600 MHz, CDCl3) δ 8.93 (s, 1H), 8.29 (dd, J = 7.8, 1.5 Hz, 1H), 8.08 (dd, J = 8.2, 1.2 Hz, 1H), 7.78–7.72 (m, 1H), 7.67–7.61 (m, 1H), 7.41–7.36 (m, 2H), 7.25–7.20 (m, 2H); 13C-NMR (151 MHz, CDCl3) δ 156.24, 149.49, 149.32, 133.64, 132.58, 131.40, 130.87, 129.72, 129.40, 124.61, 122.54.
N-(4-Methoxyphenyl)-1-(2-nitrophenyl) methanimine (4). Yellow solid (92% yield); m.p.: 79.1–79.9 °C (lit [36]: 81–82 °C); 1H-NMR (600 MHz, CDCl3) δ 8.97 (s, 1H), 8.32 (dd, J = 7.8, 1.4 Hz, 1H), 8.06 (dd, J = 8.2, 1.1 Hz, 1H), 7.75–7.70 (m, 1H), 7.62–7.57 (m, 1H), 7.35–7.29 (m, 2H), 6.98–6.94 (m, 2H), 3.85 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 159.09, 153.31, 143.88, 133.48, 131.36, 130.81, 129.55, 124.53, 122.78, 114.50, 55.53.
Methyl 4-((2-nitrobenzylidene) amino)benzoate (5) Pale yellow solid (73% yield); m.p.: 122.7–124.4 °C; 1H-NMR (600 MHz, CDCl3) δ 8.93 (s, 1H), 8.30 (dd, J = 7.7, 1.0 Hz, 1H), 8.10 (d, J = 8.4 Hz, 3H), 7.76 (t, J = 7.6 Hz, 1H), 7.68–7.63 (m, 1H), 7.30–7.25 (m, 2H), 3.93 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 166.66, 157.50, 155.14, 149.39, 133.71, 131.66, 130.95, 130.70, 129.84, 128.26, 124.65, 120.93, 52.15; MS (HR-ESI) for C15H12N2O4 [M + H]+, calcd: m/z 285.0870, found: m/z 285.0861.
N-(4-(Methylthio)phenyl)-1-(2-nitrophenyl)methanimine (6). Burnt orange solid (92% yield); m.p.: 69.3–70.4 °C; 1H-NMR (600 MHz, CDCl3) δ 8.96 (s, 1H), 8.31 (dd, J = 7.8, 1.4 Hz, 1H), 8.07 (dd, J = 8.2, 1.1 Hz, 1H), 7.73 (t, J = 7.5 Hz, 1H), 7.63–7.58 (m, 1H), 7.33–7.22 (m, 4H), 2.52 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 154.86, 149.25, 148.06, 137.45, 133.51, 131.07, 129.63, 127.37, 124.53, 121.89, 16.06; MS (HR-ESI) for C14H12N2O2S [M + H]+, calcd: m/z 273.0692, found: m/z 273.0683.
2-Phenyl-2H-indazole (7). White solid (64% yield); m.p.: 81.2–81.6 °C (lit [24]: 81–82 °C); the spectroscopic data matched previously reported data [37]: 1H-NMR (600 MHz, CDCl3) δ 8.40 (d, J = 0.9 Hz, 1H), 7.91–7.88 (m, 2H), 7.79 (dd, J = 8.8, 0.9 Hz, 1H), 7.70 (dt, J = 8.5, 1.0 Hz, 1H), 7.54–7.50 (m, 2H), 7.41–7.37 (m, 1H), 7.32 (ddd, J = 8.8, 6.6, 1.0 Hz, 1H), 7.11 (ddd, J = 8.4, 6.6, 0.7 Hz, 1H); 13C-NMR (151 MHz, CDCl3) δ (ppm): 149.78, 140.52, 129.54, 127.88, 126.81, 122.76, 122.44, 120.99, 120.39, 120.37, 117.94.
2-(4-Chlorophenyl)-2H-indazole (8). White solid (57% yield); m.p.: 143.0–145.5 °C (lit [38]: 138–140 °C); the spectroscopic data matched previously reported data [38]: 1H-NMR (600 MHz, CDCl3) δ 8.37 (d, J = 1.0 Hz, 1H), 7.87–7.82 (m, 2H), 7.77 (dq, J = 8.8, 0.9 Hz, 1H), 7.69 (dt, J = 8.5, 1.0 Hz, 1H), 7.51–7.47 (m, 2H), 7.33 (ddd, J = 8.8, 6.6, 1.1 Hz, 1H), 7.12 (ddd, J = 8.5, 6.6, 0.8 Hz, 1H); 13C-NMR (151 MHz, CDCl3) δ 149.89, 139.02, 133.55, 129.67, 127.09, 122.87, 122.71, 122.00, 120.29, 117.90.
2-(4-Methoxyphenyl)-2H-indazole (9). Beige solid (56 % yield); m.p.: 133.2–135.8 °C (lit [39]: 130–131 °C); the spectroscopic data matched previously reported data [40]: 1H-NMR (600 MHz, CDCl3) δ 8.30 (d, J = 0.9 Hz, 1H), 7.82–7.76 (m, 3H), 7.69 (dt, J = 8.4, 1.0 Hz, 1H), 7.31 (ddd, J = 8.7, 6.6, 1.0 Hz, 1H), 7.10 (ddd, J = 8.4, 6.6, 0.8 Hz, 1H), 7.05–6.99 (m, 2H), 3.86 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 159.28, 149.58, 134.12, 126.53, 122.70, 122.41, 122.22, 120.30, 120.25, 117.77, 114.63, 55.60.
Methyl 4-(2H-indazol-2-yl) benzoate (10). White solid (52% yield); m.p.: 185.8–186.2 °C (lit [41]: 186–187 °C); the spectroscopic data matched previously reported data [40]: 1H-NMR (600 MHz, CDCl3) δ 8.47 (d, J = 0.7 Hz, 1H), 8.22–8.18 (m, 2H), 8.02–7.99 (m, 2H), 7.77 (dd, J = 8.8, 0.8 Hz, 1H), 7.69 (d, J = 8.5 Hz, 1H), 7.33 (ddd, J = 8.8, 6.6, 1.0 Hz, 1H), 7.14–7.10 (m, 1H), 3.95 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 166.19, 150.19, 143.64, 131.16, 129.27, 127.45, 123.01, 122.98, 120.47, 120.26, 118.06, 52.33.
2-(4-(Methylthio) phenyl)-2H-indazole (11). Pale yellow solid (61% yield); m.p.: 148.3–149.7 °C (lit [38]: 137–139 °C); the spectroscopic data matched previously reported data [38]: 1H-NMR (600 MHz, CDCl3) δ 8.35 (d, J = 0.8 Hz, 1H), 7.84–7.80 (m, 2H), 7.79–7.76 (m, 1H), 7.68 (dt, J = 8.5, 0.9 Hz, 1H), 7.39–7.35 (m, 2H), 7.31 (ddd, J = 8.7, 6.6, 1.0 Hz, 1H), 7.10 (ddd, J = 8.4, 6.6, 0.8 Hz, 1H), 2.53 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 149.72, 138.63, 137.78, 127.27, 126.82, 122.77, 122.46, 121.26, 120.30, 120.12, 117.84, 15.88.
4-(2H-Indazol-2-yl) phenol (12). Beige solid (64% yield); m.p.: 179–181 °C (lit [25]: 193–194 °C); the spectroscopic data matched previously reported data [42]: 1H-NMR (600 MHz, DMSO-d6) δ 9.85 (s, 1H), 8.91 (d, J = 0.9 Hz, 1H), 7.91–7.84 (m, 2H), 7.75 (dt, J = 8.4, 1.0 Hz, 1H), 7.69 (dq, J = 8.8, 0.9 Hz, 1H), 7.29 (ddd, J = 8.7, 6.6, 1.1 Hz, 1H), 7.08 (ddd, J = 8.3, 6.6, 0.8 Hz, 1H), 6.98–6.92 (m, 2H); 13C-NMR (151 MHz, DMSO-d6) δ 157.09, 148.47, 132.11, 126.10, 122.24, 121.75, 121.57, 120.78, 120.58, 117.12, 115.81.
4-(2H-Indazol-2-yl) benzoic acid (13). White solid (96% yield); m.p.: 288.3–288.5 °C (lit [41]: 286–288 °C); 1H-NMR (600 MHz, DMSO-d6) δ 9.23 (s, 1H), 8.29–8.23 (m, 2H), 8.18–8.12 (m, 2H), 7.79 (dt, J = 8.5, 1.0 Hz, 1H), 7.73 (dq, J = 8.8, 0.9 Hz, 2H), 7.35 (ddd, J = 8.8, 6.5, 1.1 Hz, 1H), 7.13 (ddd, J = 8.5, 6.6, 0.8 Hz, 1H); 13C-NMR (151 MHz, DMSO-d6) δ 166.46, 149.22, 142.83, 130.82, 129.65, 127.28, 122.54, 122.43, 122.04, 120.99, 119.86, 117.48.
2-(4-(Methylsulfinyl) phenyl)-2H-indazole (14). White solid (92% yield); m.p.: 150.1–152.7 °C; 1H-NMR (600 MHz, CDCl3) δ 8.47 (d, J = 0.9 Hz, 1H), 8.13–8.07 (m, 2H), 7.83–7.75 (m, 3H), 7.70 (dt, J = 8.5, 1.0 Hz, 1H), 7.34 (ddd, J = 8.8, 6.6, 1.1 Hz, 1H), 7.13 (ddd, J = 8.5, 6.6, 0.8 Hz, 1H), 2.78 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 150.14, 145.05, 142.47, 127.45, 125.03, 123.01, 121.49, 120.46, 120.43, 118.01, 44.10; MS (HR-ESI) for C14H12N2OS [M + Na]+, calcd: m/z 279.0562, found: m/z 279.0481.
2-(4-(Methylsulfonyl) phenyl)-2H-indazole (15). White solid (68% yield); m.p.: 200.6–201.5 °C; 1H-NMR (600 MHz, CDCl3) δ 8.50 (d, J = 0.8 Hz, 1H), 8.19–8.05 (m, 4H), 7.76 (m, 1H), 7.70 (m, 1H), 7.35 (ddd, J = 8.8, 6.6, 1.0 Hz, 1H), 7.14 (ddd, J = 8.5, 6.6, 0.7 Hz, 1H), 3.11 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 150.43, 144.23, 139.27, 129.18, 127.87, 123.36, 123.18, 120.99, 120.57, 120.54, 118.11, 44.62; MS (HR-ESI) for C14H12N2O2S [M + H]+, calcd: m/z 273.0692, found: m/z 273.0659.
2,3-Diphenyl-2H-indazole (16). White solid (77% yield); mp: 107.4–107.9 °C (lit [27]: 102–103 °C); 1H-NMR (600 MHz, CDCl3) δ 7.82–7.79 (m, 1H), 7.73–7.70 (m, 1H), 7.45–7.42 (m, 2H), 7.41–7.34 (m, 9H), 7.14 (ddd, J = 8.4, 6.6, 0.8 Hz, 1H); 13C-NMR (151 MHz, CDCl3) δ 148.99, 140.24, 135.41, 129.91, 129.69, 128.97, 128.76, 128.30, 128.25, 126.98, 126.02, 122.50, 121.74, 120.52, 117.76.
2-(4-Chlorophenyl)-3-phenyl-2H-indazole (17). White solid (45% yield); m.p.: 124.4–125.0 °C (lit [43]: 126 °C); 1H-NMR (600 MHz, CDCl3) δ 7.78 (dt, J = 8.8, 0.9 Hz, 1H), 7.68–7.69 (dt, J = 8.5, 0.9 Hz, 1H), 7.45–7.32 (m, 10H), 7.14 (ddd, J = 8.4, 6.6, 0.8 Hz, 1H); 13C-NMR (151 MHz, CDCl3) δ 149.12, 138.75, 135.47, 134.09, 129.67, 129.63, 129.18, 128.94, 128.55, 127.26, 127.10, 122.73, 121.86, 120.49, 117.72; MS (HR-ESI) for C19H13ClN2 [M + H]+, calcd: m/z 305.0840, found: m/z 305.0736.
Methyl 4-(3-phenyl-2H-indazol-2-yl) benzoate (18). Pale yellow solid (40% yield); m.p.: 152.4–154.9 °C; 1H-NMR (600 MHz, CDCl3) δ 8.07–8.04 (m, 2H), 7.80 (dt, J = 8.8, 0.8 Hz, 1H), 7.69 (dt, J = 8.6, 1.0 Hz, 1H), 7.55–7.52 (m, 2H), 7.44–7.34 (m, 6H), 7.15 (ddd, J = 8.5, 6.6, 0.8 Hz, 1H), 3.93 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 166.21, 149.34, 143.76, 135.69, 130.42, 129.70, 129.62, 128.98, 128.66, 127.46, 125.69, 122.89, 122.08, 120.53, 117.81, 52.33; MS (HR-ESI) for C21H16N2O2 [M + H]+, calcd: m/z 329.1285, found: m/z 329.1103.
2-(4-(Methylthio) phenyl)-3-phenyl-2H-indazole (19). Pale yellow solid (71% yield) m.p.: 87.7–89.0 °C; 1H-NMR (600 MHz, CDCl3) δ 7.79 (dt, J = 8.9, 1.0 Hz, 1H), 7.70 (dt, J = 8.6, 1.0 Hz, 1H), 7.43–7.34 (m, 8H), 7.24–7.21 (m, 2H), 7.13 (ddd, J = 8.4, 6.6, 0.8 Hz, 1H), 2.49 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 148.97, 139.16, 137.23, 135.26, 129.88, 129.68, 128.83, 128.35, 127.00, 126.40, 126.19, 122.50, 121.78, 120.46, 117.68, 15.58; MS (HR-ESI) for C20H16N2S [M + H]+, calcd: m/z 317.1107, found: m/z 317.1108.
4-(3-Phenyl-2H-indazol-2-yl) benzoic acid (20). White solid (70% yield); m.p.: 129.2–130.1 °C; 1H-NMR (600 MHz, DMSO-d6) δ 8.04–7.99 (m, 2H), 7.77 (d, J = 8.8 Hz, 1H), 7.69 (d, J = 8.5 Hz, 1H), 7.59–7.55 (m, 2H), 7.51–7.37 (m, 6H), 7.18 (dd, J = 8.4, 6.6 Hz, 1H); 13C-NMR (151 MHz, DMSO-d6) δ 166.41, 148.44, 143.00, 135.18, 130.45, 130.07, 129.44, 128.95, 128.87, 128.63, 127.18, 125.91, 122.73, 121.30, 120.32, 117.41; MS (HR-ESI) for C20H14N2O2 [M + H]+, calcd: m/z 315.1128, found: m/z 315.1142.
2-(4-(Methylsulfonyl) phenyl)-3-phenyl-2H-indazole (21). Pale yellow solid (77% yield), m.p.: 101.8–102.7 °C; 1H-NMR (600 MHz, CDCl3) δ 7.98–7.94 (m, 2H), 7.78 (dt, J = 8.9, 0.8 Hz, 1H), 7.70–7.66 (m, 3H), 7.48–7.42 (m, 3H), 7.39 (ddd, J = 8.8, 6.5, 1.0 Hz, 1H), 7.38–7.35 (m, 2H), 7.16 (ddd, J = 8.4, 6.5, 0.7 Hz, 1H), 3.08 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 149.58, 144.52, 139.70, 135.94, 129.70, 129.28, 129.23, 129.00, 128.41, 127.85, 126.45, 123.23, 122.29, 120.58, 117.81, 44.52; MS (HR-ESI) for C20H16N2O2S [M + H]+, calcd: m/z 349.1005, found: m/z 349.1005.
3-(4-Chlorophenyl)-2-phenyl-2H-indazole (22). White solid (67% yield); m.p.: 141.1–142.8 °C (lit [27]: 134–135 °C); the spectroscopic data matched previously reported data [27,44]: 1H-NMR (600 MHz, CDCl3) δ 7.80 (dt, J = 8.8, 0.8 Hz, 1H), 7.67 (dt, J = 8.6, 1.0 Hz, 1H), 7.44–7.35 (m, 8H), 7.30–7.27 (m, 2H), 7.16 (ddd, J = 8.4, 6.5, 0.7 Hz, 1H); 13C-NMR (151 MHz, CDCl3) δ 149.00, 139.98, 134.45, 134.08, 130.84, 129.14, 129.12, 128.48, 128.38, 127.08, 126.01, 122.86, 121.71, 120.11, 117.91.
Methyl 4-(2-phenyl-2H-indazol-3-yl) benzoate (23). Pale yellow solid (76% yield): m.p.: 164.5–166.3 °C; the spectroscopic data matched previously reported data [45]: 1H-NMR (600 MHz, CDCl3) δ 8.08–8.04 (m, 2H), 7.84–7.80 (m, 1H), 7.72 (dt, J = 8.5, 0.9 Hz, 1H), 7.45–7.37 (m, 8H), 7.19 (ddd, J = 8.5, 6.5, 0.6 Hz, 1H), 3.93 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 166.55, 149.08, 139.99, 134.37, 134.13, 129.97, 129.66, 129.49, 129.18, 128.59, 127.14, 126.04, 123.18, 121.90, 120.09, 118.02, 52.29.
3-(4-(Methylthio) phenyl)-2-phenyl-2H-indazole (24). White solid, (36% yield); m.p.: 119.3–121.4 °C; the spectroscopic data matched previously reported data [45]: 1H-NMR (600 MHz, CDCl3) δ 7.79 (dt, J = 8.8, 0.9 Hz, 1H), 7.70 (dt, J = 8.5, 1.0 Hz, 1H), 7.46–7.43 (m, 2H), 7.42–7.34 (m, 4H), 7.29–7.23 (m, 4H), 7.14 (ddd, J = 8.5, 6.6, 0.8 Hz, 1H), 2.50 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 149.01, 140.22, 139.32, 134.94, 129.90, 129.06, 128.29, 126.99, 126.27, 126.16, 126.02, 122.50, 121.66, 120.43, 117.80, 15.26.
4-(2-Phenyl-2H-indazol-3-yl) benzoic acid (25). White solid (87% yield); mp: 296.2–298.2 °C; 1H-NMR (600 MHz, DMSO-d6) δ 7.94–7.90 (m, 2H), 7.75 (d, J = 8.7 Hz, 1H), 7.71 (d, J = 8.5 Hz, 1H), 7.49–7.42 (m, 5H), 7.38 (ddd, J = 8.7, 6.6, 0.9 Hz, 1H), 7.30–7.26 (m, 2H), 7.16 (ddd, J = 8.4, 6.6, 0.6 Hz, 1H); 13C-NMR (151 MHz, DMSO-d6) δ 169.13, 148.11, 140.15, 139.79, 135.10, 129.32, 129.17, 128.98, 128.37, 128.26, 126.77, 125.88, 122.34, 121.02, 120.38, 117.29; MS (HR-ESI) for C20H14N2O2 [M + H]+, calcd: m/z 315.1128, found: m/z 315.1139.
3-(4-(Methylsulfonyl) phenyl)-2-phenyl-2H-indazole (26). Pale yellow solid (60% yield), mp: 206.9–208.8 °C; 1H-NMR (600 MHz, CDCl3) δ 7.98–7.94 (m, 2H), 7.84 (dt, J = 8.7, 0.9 Hz, 1H), 7.71 (dt, J = 8.5, 1.0 Hz, 1H), 7.57–7.54 (m, 2H), 7.45–7.39 (m, 6H), 7.22 (ddd, J = 8.5, 6.6, 0.9 Hz, 1H), 3.11 (s, 3H); 13C-NMR (151 MHz, CDCl3) δ 149.10, 139.83, 139.71, 135.46, 132.95, 130.24, 129.40, 128.90, 127.86, 127.27, 126.06, 123.70, 122.03, 119.64, 118.22, 44.42; MS (HR-ESI) for C20H16N2O2S [M + H]+, calcd: m/z 349.1005, found: m/z 349.1005.
3.3. Biological Assays
3.3.1. Antiprotozoal Activity Assays
Trichomonas vaginalis strain GT3, Giardia intestinalis isolate IMSS:0981:1, and Entamoeba histolytica strain HM1-IMSS were used. Trophozoites of G. intestinalis were maintained in a TYI-S-33 medium supplemented with 10% calf serum and bovine bile. E. histolytica and T. vaginalis trophozoites were maintained in TYI-S-33 medium supplemented with 10% bovine serum. Briefly, 5 × 104 trophozoites of G. intestinalis or T. vaginalis, or 6 × 103 trophozoites of E. histolytica were incubated for 48 h at 37 °C with different concentrations of the compound to be tested, each added as solutions in DMSO. As a negative control, parasite cultures received an equivalent amount of DMSO only, while albendazole and metronidazole were included as positive controls. At the end of the treatment period, the cells were washed and subcultured for another 48 h in a fresh medium to which no drug was added. The trophozoites were then counted with a haemocytometer and the 50% inhibitory concentration (IC50), together with the respective 95% confidence limit was calculated by Probit analysis. Experiments were carried out in triplicate and repeated at least twice.
3.3.2. Antibacterial and Anticandidal Assays
Escherichia coli strain EDL933 (EHEC), Escherichia coli strain 042 (EAEC), Salmonella enterica serovar Typhi, Candida albicans, and Candida glabrata were used. The susceptibility assays were carried out using the disk diffusion test, in accordance with the outlined by CLSI (M02-A12) [18]. The inoculum was adjusted to 1.5 × 108 CFU/mL (0.5 McFarland). Sensi-Discs (Becton Dickinson and Company, Sparks, MD, USA) were used as growth inhibition controls. Ciprofloxacin (10 µg) and ampicillin (25 µg) were used as antibiotics for the positive control; whereas ketoconazole (50 μg) was used as antimycotic. Each compound was tested at 5 mg per disc. Petri dishes with the compounds and the positive control were incubated at 37 °C for 24 h. The degree of effectiveness was measured by determining the zone of inhibition in millimeters resulting from the compounds. Experiments were carried out in triplicate and the results are reported as average values.
3.3.3. Cytotoxicity Assays in Human Cells
HeLa (human cervical carcinoma) and HaCaT (immortalized human keratinocytes) cells were grown in DMEM (Invitrogen Corporation, Carlsbad, CA, USA) supplemented with 10% FBS (BioWest, Riverside, MO, USA), and maintained in standard culture conditions (37 °C, 95% humidity, and 5% CO2). Cells were allowed to grow to a density of 80% and then were harvested using sterile PBS/EDTA (pH 7.4) before starting every experiment. Cells were seeded in 96-well plates (7000 cells/well in 200 µL of DMEM). After 24 h the cells were exposed to test compounds dissolved in DMSO (J.T. Baker, Phillipsburg, NJ, USA) at different concentrations and diluted in 50 µL of DMEM, to reach 250 µL in the well. The exposure time was 48 h, and then viability was determined by MTT assay. The absorbance of formazan was determined for each well and its viability was related to the vehicle (100%). The IC50 was calculated from dose-response curve by non-linear fit using OriginPro 7.0 software (RockWare, Golden, CO, USA).
3.3.4. Cyclooxygenase Assays
The in vitro assays on human recombinant cyclooxygenase-2 were performed using the COX inhibitor screening assay kit manufactured by Cayman Chemical, catalog number 560131 (Ann Arbor, MI, USA). Assays were carried out by duplicate following the instructions provided by the manufacturer. Compounds were tested at 10 µM, whereas the reference compound celecoxib (purchased from Sigma-Aldrich, catalog number PHR1683) was tested at 1 µM because of its high inhibition at 10 µM.
3.4. Molecular Docking
The crystal structure of human COX-2 was retrieved from the Protein Data Bank (www.rcsb.org; www.wwpdb.org) [46,47], PDB ID: 5KIR [33]. The protein structure was prepared using Maestro 9.1 (Schrödinger, Cambridge, MA, USA) [48]; first, chain B was selected and solvent molecules were removed. Then, the pdb structure was submitted for minimization using the YASARA web server (YASARA, Vienna, Austria) [49]. The protein was exported to Autodock Tools 1.5.6 (TSRI, La Jolla, CA, USA) and the grid coordinates and the pdbqt files were generated [50,51,52]. Ligands were constructed and minimized using the universal force field implemented in Maestro 9.1 [53] and exported to Autodock Tools 1.5.6 to generate the pdbqt files. Docking calculations were carried out using Autodock Vina (TSRI, La Jolla, CA, USA) employing a grid box of 40 × 40 × 40 centered on the co-crystalized ligand binding site (rofecoxib) and an exhaustiveness value of 500 [31]. The docking protocol was validated by comparison of docked rofecoxib and the co-crystalized rofecoxib. Molecular graphics and analyses were performed with the UCSF Chimera package version 1.10.2 (RBVI, San Francisco, CA, USA) [54].
Acknowledgments
This work was supported by project UAM-PTC-503 (Apoyo a la Incorporación de Nuevos PTC) from Secretaría de Educación Pública (SEP). J.P-V and I.G-S also acknowledge the support provided by project 1238 from Consejo Nacional de Ciencia y Tecnologia (Cátedras CONACYT). The authors would like to express their sincere thanks to Amparo Tapia for the support with biological assays, and to Ernesto Sánchez Mendoza and Mónica A. Rincón for the analytical support. The authors gratefully acknowledge Schrödinger, LLC, for providing the academic version of Maestro; YASARA Biosciences GmbH for providing the YASARA Energy Minimization Server; the Scripps Research Institute for providing Autodock Tools and Autodock Vina; and the Resource for Biocomputing, Visualization, and Informatics (RBVI) for providing Chimera (Chimera is developed by the RBVI at the UCSF supported by NIGMS P41-GM103311).
Supplementary Materials
The following are available online. Figures S1–S49: 1H-NMR and 13C-NMR spectra of compounds 7–26, and HRMS spectra of compounds 14, 15, 17–21, 25, and 26. Table S1. Antibacterial and antimycotic effect for selected compounds.
Author Contributions
Jaime Pérez-Villanueva, Olivia Soria-Arteche, and Juan Francisco Palacios-Espinosa conceived and designed the experiments and wrote the paper; Jaime Pérez-Villanueva, Karen Rodríguez-Villar, Miguel Cortés-Gines, Zeltzin Custodio-Galván, and Dante B. Estrada-Castro performed the chemical synthesis; Lilián Yépez-Mulia performed the antiprotozoal activity assays; Ignacio González-Sánchez and Marco A. Cerbón, performed the cytotoxicity assays; Ignacio González-Sánchez and Juan Francisco Palacios-Espinosa performed the COX-2 inhibition assays; Teresita del Rosario Sainz-Espuñes and Ana Karina Rodríguez-Vicente performed the antibacterial and anticandidal assays; and Jaime Pérez-Villanueva performed the docking studies.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are available from the authors.
References
- 1.Hodges K., Gill R. Infectious diarrhea: Cellular and molecular mechanisms. Gut Microbes. 2010;1:4–21. doi: 10.4161/gmic.1.1.11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cacciò S.M., Sprong H. Epidemiology of giardiasis in humans. In: Luján H.D., Svärd S., editors. Giardia: A Model Organism. Springer; Vienna, Austria: 2011. pp. 17–28. [Google Scholar]
- 3.Ximenez C., Partida O., Nieves M., Hernandez E., Moran P., Valadez A., Gonzalez E., Cerritos R., Rojas L. Immune response in human amebiasis: A protective response? In: Nozaki T., Bhattacharya A., editors. Amebiasis: Biology and Pathogenesis of Entamoeba. Springer; Japan, Tokyo: 2015. pp. 497–519. [Google Scholar]
- 4.Parry C.M., Hien T.T., Dougan G., White N.J., Farrar J.J. Typhoid fever. N. Eng. J. Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 5.Upcroft P., Upcroft J.A. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin. Microbiol. Rev. 2001;14:150–164. doi: 10.1128/CMR.14.1.150-164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman L., Rowley J., Vander Hoorn S., Wijesooriya N.S., Unemo M., Low N., Stevens G., Gottlieb S., Kiarie J., Temmerman M. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS ONE. 2015;10:e0143304. doi: 10.1371/journal.pone.0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cudmore S.L., Delgaty K.L., Hayward-McClelland S.F., Petrin D.P., Garber G.E. Treatment of infections caused by metronidazole-resistant Trichomonas vaginalis. Clin. Microbiol. Rev. 2004;17:783–793. doi: 10.1128/CMR.17.4.783-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chew S.Y., Than L.T.L. Vulvovaginal candidosis: Contemporary challenges and the future of prophylactic and therapeutic approaches. Mycoses. 2016;59:262–273. doi: 10.1111/myc.12455. [DOI] [PubMed] [Google Scholar]
- 9.Cassone A. Vulvovaginal Candida albicans infections: Pathogenesis, immunity and vaccine prospects. BJOG-Int. J. Obstet. Gynaecol. 2015;122:785–794. doi: 10.1111/1471-0528.12994. [DOI] [PubMed] [Google Scholar]
- 10.Poole D.N., McClelland R.S. Global epidemiology of Trichomonas vaginalis. Sex. Transm. Infect. 2013;89:418–422. doi: 10.1136/sextrans-2013-051075. [DOI] [PubMed] [Google Scholar]
- 11.Thangadurai A., Minu M., Wakode S., Agrawal S., Narasimhan B. Indazole: A medicinally important heterocyclic moiety. Med. Chem. Res. 2012;21:1509–1523. doi: 10.1007/s00044-011-9631-3. [DOI] [Google Scholar]
- 12.Lopez-Vallejo F., Castillo R., Yepez-Mulia L., Medina-Franco J.L. Benzotriazoles and indazoles are scaffolds with biological activity against Entamoeba histolytica. J. Biomol. Screen. 2011;16:862–868. doi: 10.1177/1087057111414902. [DOI] [PubMed] [Google Scholar]
- 13.Marrero-Ponce Y., Meneses-Marcel A., Castillo-Garit J.A., Machado-Tugores Y., Escario J.A., Barrio A.G., Pereira D.M., Nogal-Ruiz J.J., Arán V.J., Martínez-Fernández A.R., et al. Predicting antitrichomonal activity: A computational screening using atom-based bilinear indices and experimental proofs. Bioorg. Med. Chem. 2006;14:6502–6524. doi: 10.1016/j.bmc.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Minu M., Thangadurai A., Wakode S.R., Agrawal S.S., Narasimhan B. Synthesis, antimicrobial activity and QSAR studies of new 2,3-disubstituted-3,3a,4,5,6,7-hexahydro-2H-indazoles. Bioorg. Med. Chem. Lett. 2009;19:2960–2964. doi: 10.1016/j.bmcl.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 15.Tanitame A., Oyamada Y., Ofuji K., Kyoya Y., Suzuki K., Ito H., Kawasaki M., Nagai K., Wachi M., Yamagishi J.-I. Design, synthesis and structure–activity relationship studies of novel indazole analogues as DNA gyrase inhibitors with Gram-positive antibacterial activity. Bioorg. Med. Chem. Lett. 2004;14:2857–2862. doi: 10.1016/j.bmcl.2004.03.044. [DOI] [PubMed] [Google Scholar]
- 16.Boehm H.-J., Boehringer M., Bur D., Gmuender H., Huber W., Klaus W., Kostrewa D., Kuehne H., Luebbers T., Meunier-Keller N., et al. Novel inhibitors of DNA gyrase: 3D structure based biased needle screening, hit validation by biophysical methods, and 3D guided optimization. A promising alternative to random screening. J. Med. Chem. 2000;43:2664–2674. doi: 10.1021/jm000017s. [DOI] [PubMed] [Google Scholar]
- 17.Méndez-Lucio O., Naveja J.J., Vite-Caritino H., Prieto-Martínez F.D., Medina-Franco J.L. Review. One drug for multiple targets: A computational perspective. J. Mex. Chem. Soc. 2016;60:168–181. [Google Scholar]
- 18.Ni H., Wendoloski J. Structure-based design of new antibacterial agents. In: David C.S., editor. Annual Reports in Computational Chemistry. Volume 2. Elsevier; Amsterdam, The Netherlands: 2006. pp. 279–295. [Google Scholar]
- 19.Pérez-Villanueva J., Medina-Franco J.L., Méndez-Lucio O., Yoo J., Soria-Arteche O., Izquierdo T., Lozada M.C., Castillo R. CASE plots for the chemotype-based activity and selectivity analysis: A CASE study of cyclooxygenase inhibitors. Chem. Biol. Drug Des. 2012;80:752–762. doi: 10.1111/cbdd.12019. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Villanueva J., Méndez-Lucio O., Soria-Arteche O., Medina-Franco J.L. Activity cliffs and activity cliff generators based on chemotype-related activity landscapes. Mol. Divers. 2015;19:1021–1035. doi: 10.1007/s11030-015-9609-z. [DOI] [PubMed] [Google Scholar]
- 21.Zarghi A., Arfaei S. Selective COX-2 inhibitors: A review of their structure-activity relationships. Iran. J. Pharm. Res. 2011;10:655–683. [PMC free article] [PubMed] [Google Scholar]
- 22.Stenson W.F., Zhang Z., Riehl T., Stanley S.L. Amebic infection in the human colon induces cyclooxygenase-2. Infect. Immun. 2001;69:3382–3388. doi: 10.1128/IAI.69.5.3382-3388.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rub A., Arish M., Husain S.A., Ahmed N., Akhter Y. Host-lipidome as a potential target of protozoan parasites. Microbes Infect. 2013;15:649–660. doi: 10.1016/j.micinf.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Cadogan J.I.G., Mackie R.K. 2-Phenylindazole. Org. Synth. 1968;48:113. doi: 10.15227/orgsyn.048.0113. [DOI] [Google Scholar]
- 25.De Angelis M., Stossi F., Carlson K.A., Katzenellenbogen B.S., Katzenellenbogen J.A. Indazole estrogens: Highly selective ligands for the estrogen receptor β. J. Med. Chem. 2005;48:1132–1144. doi: 10.1021/jm049223g. [DOI] [PubMed] [Google Scholar]
- 26.Soria-Arteche O., Castillo R., Hernández-Campos A., Hurtado-de la Peña M., Navarrete-Vázquez G., Medina-Franco J.L., Gómez-Flores K. Studies on the selective S-oxidation of albendazole, fenbendazole, triclabendazole, and other benzimidazole sulfides. J. Mex. Chem. Soc. 2005;49:353–358. [Google Scholar]
- 27.Ohnmacht S.A., Culshaw A.J., Greaney M.F. Direct arylations of 2H-indazoles on water. Org. Lett. 2010;12:224–226. doi: 10.1021/ol902537d. [DOI] [PubMed] [Google Scholar]
- 28.Cedillo-Rivera R., Chávez B., González-Robles A., Tapia A., Yépez-Mulia L. In vitro effect of nitazoxanide against Entamoeba histolytica, Giardia intestinalis and Trichomonas vaginalis trophozoites. J. Eukaryot. Microbiol. 2002;49:201–208. doi: 10.1111/j.1550-7408.2002.tb00523.x. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Villanueva J., Hernández-Campos A., Yépez-Mulia L., Méndez-Cuesta C., Méndez-Lucio O., Hernández-Luis F., Castillo R. Synthesis and antiprotozoal activity of novel 2-{[2-(1H-imidazol-1-yl)ethyl]sulfanyl}-1H-benzimidazole derivatives. Bioorg. Med. Chem. Lett. 2013;23:4221–4224. doi: 10.1016/j.bmcl.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards . Document: M02-A12. National Committee for Clinical Laboratory Standards; Wayne, PA, USA: 2015. [Google Scholar]
- 31.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lill A., Scholich K., Stark H. Synthesis of novel dansyl-labeled celecoxib derivatives. Tetrahedron Lett. 2013;54:6682–6686. doi: 10.1016/j.tetlet.2013.09.025. [DOI] [Google Scholar]
- 33.Orlando B.J., Malkowski M.G. Crystal structure of rofecoxib bound to human cyclooxygenase-2. Acta Crystallogr. F. 2016;72:772–776. doi: 10.1107/S2053230X16014230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quintero A., Pelcastre A., Solano J.D. Antitumoral activity of new pyrimidine derivatives of sesquiterpene lactones. J. Pharm. Pharm. Sci. 1999;2:108–112. [PubMed] [Google Scholar]
- 35.Loza-Mejía M.A., Olvera-Vázquez S., Maldonado-Hernández K., Guadarrama-Salgado T., González-Sánchez I., Rodríguez-Hernández F., Solano J.D., Rodríguez-Sotres R., Lira-Rocha A. Synthesis, cytotoxic activity, DNA topoisomerase-II inhibition, molecular modeling and structure-activity relationship of 9-anilinothiazolo[5,4-b]quinoline derivatives. Bioorg. Med. Chem. 2009;17:3266–3277. doi: 10.1016/j.bmc.2009.03.052. [DOI] [PubMed] [Google Scholar]
- 36.Johnston D., Smith D.M., Shepherd T., Thompson D. o-Nitrobenzylidene compounds. Part 3. Formation of 4-arylamino-3-methoxycinnoline 1-oxides from N-o-nitrobenzylideneanilines, cyanide ion, methanol: The intermediacy of 2-aryl-3-cyano-2H-indazole 1-oxides. J. Chem. Soc. Perk. Trans. 1987;1:495–500. doi: 10.1039/p19870000495. [DOI] [Google Scholar]
- 37.Fang Y., Wu C., Larock R.C., Shi F. Synthesis of 2H-indazoles by the [3 + 2] dipolar cycloaddition of sydnones with arynes. J. Org. Chem. 2011;76:8840–8851. doi: 10.1021/jo201605v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar M.R., Park A., Park N., Lee S. Consecutive condensation, C–N and N–N bond formations: A copper-catalyzed one-pot three-component synthesis of 2H-indazole. Org. Lett. 2011;13:3542–3545. doi: 10.1021/ol201409j. [DOI] [PubMed] [Google Scholar]
- 39.Cadogan J.I.G., Searle R.J.G. Cyclizations induced by triethyl phosphite. Convenient new route to indazoles and triazoles. Chem. Ind. 1963:1282–1283. [Google Scholar]
- 40.Genung N.E., Wei L., Aspnes G.E. Regioselective synthesis of 2H-indazoles using a mild, one-pot condensation–Cadogan reductive cyclization. Org. Lett. 2014;16:3114–3117. doi: 10.1021/ol5012423. [DOI] [PubMed] [Google Scholar]
- 41.Zhou D., Sze J.Y.-C., Gross J.L., Robichaud A.J. Preparation of Azacyclylbenzamide Derivatives as Histamine-3 Antagonists for Treating CNS Disorders. 20080293771A1. U.S. Patent. 2008 Nov 27;
- 42.Aspnes G.E., Didiuk M.T., Filipski K.J., Guzman-Perez A., Lee E.C.Y., Pfefferkorn J.A., Stevens B.D., Tu M.M. Preparation of 3-[4-(Heterocyclyl-Substituted)-Benzamido]Propanoic Acids as Glucagon Receptor Modulators. 20120202834A1. U.S. Patent. 2012 Aug 9;
- 43.Lee J.H., Matsumoto A., Yoshida M., Simamura O. New routes to 1,2-diazoles with a fused ring system by reductive and oxidative cyclizations. Chem. Lett. 1974;3:951–954. doi: 10.1246/cl.1974.951. [DOI] [Google Scholar]
- 44.Hattori K., Yamaguchi K., Yamaguchi J., Itami K. Pd- and Cu-catalyzed C-H arylation of indazoles. Tetrahedron. 2012;68:7605–7612. doi: 10.1016/j.tet.2012.05.091. [DOI] [Google Scholar]
- 45.Geng X., Wang C. Rhenium-catalyzed [4 + 1] annulation of azobenzenes and aldehydes via isolable cyclic rhenium(I) complexes. Org. Lett. 2015;17:2434–2437. doi: 10.1021/acs.orglett.5b00938. [DOI] [PubMed] [Google Scholar]
- 46.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berman H., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 48.Maestro. Schrödinger, LLC; New York, NY, USA: 2010. version 9.1. [Google Scholar]
- 49.Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinform. 2009;77:114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.AutoDockTools. The Scripps Research Institute; La Jolla, CA, USA: 2013. version 1.5.6. [Google Scholar]
- 51.Sanner M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999;17:57–61. doi: 10.1016/S1093-3263(99)99999-0. [DOI] [PubMed] [Google Scholar]
- 52.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rappe A.K., Casewit C.J., Colwell K.S., Goddard W.A., Skiff W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992;114:10024–10035. doi: 10.1021/ja00051a040. [DOI] [Google Scholar]
- 54.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.