Abstract
OBJECTIVE
Lipodystrophy syndromes are a heterogeneous group of disorders associated with selective absence of fat. Currently, the diagnosis is established only clinically.
RESEARCH DESIGN AND METHODS
We developed a new method from DXA scans called a “fat shadow,” which is a color-coded representation highlighting only the fat tissue. We conducted a blinded retrospective validation study to assess its usefulness for the diagnosis of lipodystrophy syndromes.
RESULTS
We evaluated the fat shadows from 16 patients (11 female and 5 male) with generalized lipodystrophy (GL), 57 (50 female and 7 male) with familial partial lipodystrophy (FPLD), 2 (1 female and 1 male) with acquired partial lipodystrophy, and 126 (90 female and 36 male) control subjects. FPLD was differentiated from control subjects with 85% sensitivity and 96% specificity (95% CIs 72–93 and 91–99, respectively). GL was differentiated from nonobese control subjects with 100% sensitivity and specificity (95% CIs 79–100 and 92–100, respectively).
CONCLUSIONS
Fat shadows provided sufficient qualitative information to infer clinical phenotype and differentiate these patients from appropriate control subjects. We propose that this method could be used to support the diagnosis.
Introduction
Lipodystrophy syndromes are a heterogeneous group of disorders causing atypical diabetes, classified into four broad subtypes depending on the etiology and the extent of fat loss (1–4). The diagnoses of all subtypes currently depend on the clinical acumen of the physician (3,5). With niche therapies either approved or under investigation, there is a need for the development of objective diagnostic tools to reassure both regulatory bodies and third-party payors that correct patients are getting therapies. In this study, we describe a simple and reproducible method to obtain a “fat shadow” from a DXA body composition scan, which gives an overall impression of fat distribution throughout the body and may be useful as an additional clinical tool to diagnose lipodystrophy and document fat distribution.
Research Design and Methods
DXA scans were acquired from subjects who participated in lipodystrophy or healthy and obese volunteer studies (5–8). Patients with lipodystrophy and control subjects were all previously enrolled into various clinical studies in which measurement of adiposity was of interest, and total body composition scans were obtained. All participants were seen at either the Michigan Clinical Research Unit (Michigan cohort) or the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Intramural Research Program (National Institutes of Health [NIH] cohort) from 2007 to 2017 and had consented to participate in the research studies with permission to use their data in future studies. Patients with lipodystrophy were diagnosed clinically, which is the reference standard (3). Control subjects were selected to match age and fat-free mass index from the Michigan archives. At the NIDDK, control subjects were selected to match age, sex, and BMI.
Developing the Fat Shadow Method
Our objective was to find a way to visualize the information on fat distribution captured by DXA in a clinically useful way that conveys the overall fat distribution pattern. Body composition scans were acquired on a Prodigy table at the Michigan site and an iDXA table at the NIH site, both of which were DXA systems manufactured by GE Lunar (Chicago, IL). Scan files (.dfb for Prodigy and .meb for iDXA) were analyzed using enCORE v14.10. Visualization options available through the built-in functions of the software were evaluated in depth to find potentially useful modalities (Supplementary Fig. 1). A composition map color-coded to render out the nonfat tissues highlighted the overall fat distribution (Supplementary Fig. 2). The resulting image was termed a fat shadow, as it corresponded to the X-ray shadow of fat tissues.
Evaluation of Fat Shadows and Validation of Diagnostic Accuracy
First, the fat shadows available at Michigan were explored qualitatively in an unblinded fashion to identify clinically relevant features and count their frequencies in different subtypes. The method was then validated in a blinded, retrospective study. Both the Michigan and NIDDK sites compiled their scans for case and control subjects and removed all identifiers. For the first step of validation in familial partial lipodystrophy (FPLD), scan files from NIDDK were transferred to Michigan, where the images were rendered. For the second step, rendered images from the Michigan cohort were sent to NIDDK to be assessed by NIH investigators after reviewing a training deck. For the third step of validation, an independent investigator evaluated all cases from both cohorts after first reviewing the training deck. For validation of the method in GL, the independent investigator evaluated a compilation of all GL scans and nonobese control subjects. Training cases were excluded from all assessments. The only information shown in addition to the fat shadow was sex. In steps one and two, two investigators evaluated the images independently and reached a consensus in case of disagreement. Investigators identified the FPLD type 2 (FPLD2) from other FPLD (FPLDX) as an exploratory end point. CIs for sensitivity and specificity were derived with the Clopper-Pearson method. Agreement rates were calculated using Cohen κ.
Results
The Michigan cohort included 48 patients with lipodystrophy (41 female and 7 male, aged 13–65 years), 43 of whom had FPLD, and 70 control subjects (42 female and 28 male, aged 19–65 years). The NIH cohort included 13 patients with GL (9 female and 4 male, aged 14–40 years), 14 patients with FPLD (12 female and 2 male, aged 18–65 years), and 56 control subjects (48 female and 8 male, aged 18–65 years). Supplementary Tables 1 and 2 list all of the subtypes, demographic characteristics, and average body composition characteristics of both cohorts.
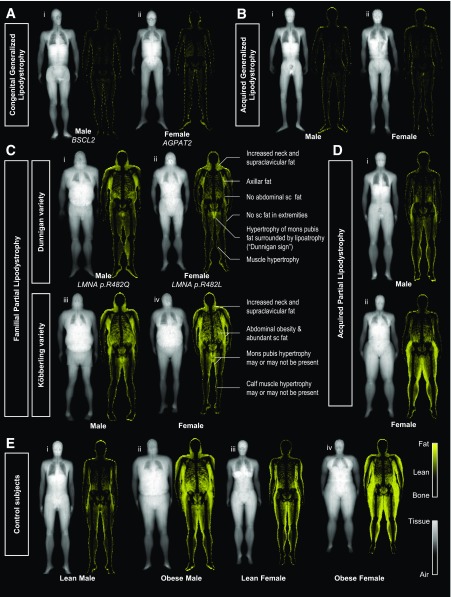
Overall, patients with lipodystrophy had uniquely different fat shadows compared with control subjects. Patients with GL had a striking appearance with near-complete absence of fat signal (Fig. 1A and B). Residual fat depots were seen in the periauricular area, soles, and pubic region in some patients with congenital generalized lipodystrophy (CGL) type 1. Patients with acquired GL (two females and two males) had no visible residual fat. Figure 1C compares the typical presentations of FPLD2 and FPLDX (specifically, FPLD1, Köbberling variety [9]). Presentation of FPLD2 followed a relatively consistent pattern (Supplementary Fig. 3), with subcutaneous fat absent throughout the body in most patients, but with hypertrophic fat in the neck and pubic region. In contrast, the FPLDX population was very heterogeneous (Supplementary Fig. 4). Images from patients with acquired partial lipodystrophy showed very little fat in the upper half of the body, whereas the fat depots in the hips and lower extremities were preserved or even hypertrophic (Fig. 1D). Obese and lean control subjects are shown in Fig. 1E for comparison. Supplementary Tables 3 and 4 show the frequency of different features in each subtype organized into different sex categories. The Fat Shadow Atlas in the Supplementary Data demonstrates the fat shadows of all participants used in this study.
Figure 1.
Fat shadows derived from DXA scans of patients with lipodystrophy and selected control subjects. A: Patients with CGL have minimal fat signal on the fat shadow. CGL type 2 (i) was associated with no visible residual fat depots, whereas CGL type 1 (ii) is seen to have residual fat in the periauricular area and soles and in females the labial region. B: Male (i) and Female (ii) acquired GL patients are shown. They have no subcutaneous fat depots throughout their body, with minimal fat signal visible in the scan. C: The two classic presentations of different FPLD subtypes. FPLD2 (Dunnigan variety; i and ii) typically presents with the same signature phenotype with absence of subcutaneous fat in the extremities, hips, and abdomen (flanks), along with a hypertrophy in the neck, axillae, and mons pubis. Dunnigan sign was defined as the isolated high signal area corresponding to the mons pubis, surrounded by profound subcutaneous (sc) lipoatrophy. This sign was found to be helpful in identifying patients with FPLD2 and also evident in the one male with FPLD2 present in this cohort (i). Increased fat signal around the mons pubis region was present in all 21 (100%) FPLD2 females versus only 10 out of 29 (34%) FPLDX females and 0 out of 90 (0%) control females (95% CIs 84–100%, 18–54%, and 0–4%, respectively). FPLD1 (Köbberling variety; iii and iv) had neck hypertrophy and abdominal obesity. Calf muscle hypertrophy was present in 8 out of 35 patients with FPLDX (23%; 95% CIs 10–40). FPLDX population in its entirety was very heterogeneous, possibly due to heterogeneity in etiology (15). See Supplementary Fig. 4 and the Fat Shadow Atlas in the Supplementary Data for details. D: Acquired partial lipodystrophy: patients have acquired loss of fat from the upper half of the body with unknown cause. Fat depots in the lower half are spared and may be normal (i) or hypertrophied (ii). E: A selection of lean (i, iii) and obese (ii, iv) control subjects with sex-representative android/gynoid ratios and without metabolic disease. AGPAT2, 1-Acylglycerol-3-phosphate O-acyltransferase 2; BSCL2, Berardinelli-Seip congenital lipodystrophy 2; FPLDX, a broad group that includes FPLD1, FPLD3, and other types not numbered in standard nomenclature system; LMNA, lamin A/C gene.
Accuracy of the method is presented in Supplementary Table 5. FPLD was differentiated from control subjects with 85% sensitivity and 96% specificity (95% CIs 72–93 and 91–99, respectively). GL was differentiated from nonobese control subjects with 100% sensitivity and specificity (95% CIs 79–100 and 92–100, respectively). The κ for interobserver agreement rates ranged between 0.73 and 0.96 (Supplementary Table 6). Some subtypes were recognizable from their fat distribution patterns (Supplementary Table 7).
Conclusions
This study shows that fat shadows derived from DXA scans are simple tools to diagnose lipodystrophy syndromes. Various quantitative measurements from DXA scans have been used in lipodystrophy research (5,10–12); however, these measurements were done in regions of interest that were too broad and not necessarily designed to be clinically relevant in the context of lipodystrophy (Supplementary Fig. 5). This study is the first to use DXA body composition scans to diagnose disease in a pixel-based approach. Using this technique, investigators were able to identify patients with both GL and partial lipodystrophy accurately.
Metreleptin was approved in 2014 as the first therapy for GL (13). This therapy, although very helpful for the treatment of difficult metabolic complications, is currently very expensive in the U.S., and third-party payers require verification of the diagnosis with genotype even when insurance coverage for genetic testing may not be provided. When genetic testing is not feasible or negative, having the ability to document the body fat pattern with the fat shadow method may aid complicated therapeutic decisions. FPLD has also recently attracted attention from industry with development of novel therapeutic agents; however, the U.S. Food and Drug Administration has expressed concern about whether this phenotype is sufficiently distinct from truncal obesity or common metabolic syndrome (14). We believe that our data add to the evidence that FPLD covers a spectrum of heterogeneous diseases with unique fat distribution patterns that are distinct from common forms of truncal obesity. It may prove useful not only in drug reimbursement decisions, but also for adjudication of cases for drug trials or other different clinical settings.
Our study had a number of limitations listed in Supplementary Table 8 and is only the first step to bring this tool to clinic. However, the tool has substantial strengths over other methods, which are also listed. This method can be further enhanced with standardization and by adding lateral views, as proposed in Supplementary Fig. 6. With further studies, it may be possible to use fat shadows for classification of not only lipodystrophy but also other metabolic diseases.
Supplementary Material
Article Information
Acknowledgments. The authors thank all of the patients and control subjects for participation in the study and Lipodystrophy United for help in patient recruitment into various lipodystrophy trials. The authors also thank Laura Foess-Wood for performing all of the DXA scans at the Michigan Clinical Research Unit and for the positive personality that cheers us up every time and Dr. Kong Y. Chen and Dr. Robert J. Brychta for performing the DXA scans at the Metabolic Clinical Research Unit at the NIDDK. The authors dedicate this article to those patients whom we have lost since 2009 and especially to Natalie K. Hunt-Embry, who fought the disease with great courage and passion.
Funding. This work was supported by NIH grants R01-DK-088114, P30-DK-089503, and P30-DK-034933, which allowed the DXA scans presented in this study to be performed and the Intramural Research Program of the NIH, NIDDK. B.J.R. was supported by NIH grant T32-DK-007245. This work was also supported by generous gifts to the Lipodystrophy Fund at the University of Michigan made by the Sopha family, Ionis Pharmaceuticals, and the White Point Foundation of Turkey. The fund is used specifically to cover the salary of R.Me.
These funding sources had no interference in the study design or direction.
Duality of Interest. E.A.O. received grant support from and served as an advisor to Amylin Pharmaceuticals LLC, Bristol-Myers Squibb, and AstraZeneca in the past and is currently receiving grant support from Gemphire Therapeutics, Aegerion Pharmaceuticals, Ionis Pharmaceuticals, and Akcea Therapeutics and serving as an advisor to Aegerion Pharmaceuticals, Akcea Therapeutics, and Regeneron Pharmaceuticals. E.A.O. recently completed grant support from GI Dynamics. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. R.Me. analyzed the DXA scans, developed the new visualization technique, and designed the validation experiment under the direction and supervision of E.A.O. R.Me., B.J.R., and N.M. gathered scan files from different sites. R.Me. and A.J. conducted the statistical analyses under the supervision of E.A.O. R.Me., N.M., B.A., R.J.B., and E.A.O. conducted the blinded assessments. Control scans were provided by R.Mu. and J.F.H. R.J.B. and E.A.O. established the clinical diagnoses of the patients. R.Me. and E.A.O. wrote the manuscript. R.Me., B.J.R., N.M., A.J., A.H.N., R.Mu., B.A., J.F.H., R.J.B., and E.A.O. approved the final version of the manuscript. E.A.O. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in poster form at the 78th Scientific Sessions of the American Diabetes Association, Orlando, FL, 22–26 June 2018.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc18-0978/-/DC1.
References
- 1.Capeau J, Magré J, Caron-Debarle M, et al. . Human lipodystrophies: genetic and acquired diseases of adipose tissue. In Adipose Tissue Development. Basel, Switzerland, Karger Publishers, 2010, p. 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang-Doran I, Sleigh A, Rochford JJ, O’Rahilly S, Savage DB. Lipodystrophy: metabolic insights from a rare disorder. J Endocrinol 2010;207:245–255 [DOI] [PubMed] [Google Scholar]
- 3.Brown RJ, Araujo-Vilar D, Cheung PT, et al. . The diagnosis and management of lipodystrophy syndromes: a multi-society practice guideline. J Clin Endocrinol Metab 2016;101:4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillín-Amarelle C, Sánchez-Iglesias S, Castro-Pais A, et al. . Type 1 familial partial lipodystrophy: understanding the Köbberling syndrome. Endocrine 2016;54:411–421 [DOI] [PubMed] [Google Scholar]
- 5.Ajluni N, Meral R, Neidert AH, et al. . Spectrum of disease associated with partial lipodystrophy: lessons from a trial cohort. Clin Endocrinol (Oxf) 2017;86:698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ajluni N, Dar M, Xu J, Neidert AH, Oral EA. Efficacy and safety of metreleptin in patients with partial lipodystrophy: lessons from an expanded access program. J Diabetes Metab 2016;7:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oral EA, Reilly SM, Gomez AV, et al. . Inhibition of IKKɛ and TBK1 improves glucose control in a subset of patients with type 2 diabetes. Cell Metab 2017;26:157–170.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahinoz M, Khairi S, Cuttitta A, et al. . Potential association of LMNA-associated generalized lipodystrophy with juvenile dermatomyositis. Clin Diabetes Endocrinol 2018;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köbberling J, Dunnigan MG. Familial partial lipodystrophy: two types of an X linked dominant syndrome, lethal in the hemizygous state. J Med Genet 1986;23:120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang JS, Rietschel P, Hadigan CM, Rosenthal DI, Grinspoon S. Increased abdominal visceral fat is associated with reduced bone density in HIV-infected men with lipodystrophy. AIDS 2001;15:975–982 [DOI] [PubMed] [Google Scholar]
- 11.Valerio CM, Zajdenverg L, de Oliveira JE, Mory PB, Moyses RS, Godoy-Matos AF. Body composition study by dual-energy x-ray absorptiometry in familial partial lipodystrophy: finding new tools for an objective evaluation. Diabetol Metab Syndr 2012;4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuvoli S, Caruana G, Babudieri S, et al. . Body fat changes in HIV patients on highly active antiretroviral therapy (HAART): a longitudinal DEXA study. Eur Rev Med Pharmacol Sci 2018;22:1852–1859 [DOI] [PubMed] [Google Scholar]
- 13.Center for Drug Evaluation and Research. Approval package for application number: 125390Orig1s000 [Internet], 2014. Available from https://www.accessdata.fda.gov/drugsatfda_docs/nda/2014/125390orig1s000approv.pdf. Accessed 23 March 2018
- 14.The Endocrinologic and Metabolic Drugs Advisory Committee meeting briefing document BLA 125390 Myalept (metreleptin for injection) [Internet], 2013. Available from https://wayback.archive-it.org/7993/20170405215819/https://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/Drugs/EndocrinologicandMetabolicDrugsAdvisoryCommittee/UCM377928.pdf. Accessed 23 March 2018
- 15.Lotta LA, Gulati P, Day FR, et al.; EPIC-InterAct Consortium; Cambridge FPLD1 Consortium . Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet 2017;49:17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.