Abstract
OBJECTIVE
Dysbiosis of the gut microbiota has been linked to disease pathogenesis in type 1 diabetes, yet the functional consequences to the host of this dysbiosis are unknown. We investigated the functional interactions between the microbiota and the host associated with type 1 diabetes disease risk.
RESEARCH DESIGN AND METHODS
We performed a cross-sectional analysis of stool samples from subjects with recent-onset type 1 diabetes (n = 33), islet autoantibody–positive subjects (n = 17), low-risk autoantibody-negative subjects (n = 29), and healthy subjects (n = 22). Metaproteomic analysis was used to identify gut- and pancreas-derived host and microbial proteins, and these data were integrated with sequencing-based microbiota profiling.
RESULTS
Both human (host-derived) proteins and microbial-derived proteins could be used to differentiate new-onset and islet autoantibody–positive subjects from low-risk subjects. Significant alterations were identified in the prevalence of host proteins associated with exocrine pancreas output, inflammation, and mucosal function. Integrative analysis showed that microbial taxa associated with host proteins involved in maintaining function of the mucous barrier, microvilli adhesion, and exocrine pancreas were depleted in patients with new-onset type 1 diabetes.
CONCLUSIONS
These data support that patients with type 1 diabetes have increased intestinal inflammation and decreased barrier function. They also confirmed that pancreatic exocrine dysfunction occurs in new-onset type 1 diabetes and show for the first time that this dysfunction is present in high-risk individuals before disease onset. The data identify a unique type 1 diabetes–associated signature in stool that may be useful as a means to monitor disease progression or response to therapies aimed at restoring a healthy microbiota.
Introduction
Type 1 diabetes is caused by T-cell–mediated destruction of insulin-producing cells and is influenced by both genetic susceptibility and the environment. Dysbiosis of the gut microbiota has been implicated in disease pathogenesis of many immune-mediated diseases, including type 1 diabetes (1). The composition of the gut microbiota is shaped by both genetics and environmental agents, such as diet and hygiene (2,3). Environmental factors such as infant diet and exposure to microbial agents have been linked to type 1 diabetes risk (4). How changes in the gut microbiota alter the intestinal environment and immune function in type 1 diabetes is unknown.
Disparate findings among studies have not led to a clear consensus of what constitutes a type 1 diabetes–associated microbiota (5). Dysbiosis of the gut microbiota is hypothesized to exacerbate gut inflammation before disease onset (1). Evidence suggests that individuals with islet autoimmunity have increased intestinal permeability (6), and studies of duodenal biopsy specimens from individuals with type 1 diabetes have found evidence of mild inflammation (7,8). An understanding of the functional properties of the microbiota and the interactions between the gut microbes and the host is needed to provide insight into the role of the intestinal microbiome in type 1 diabetes development. Metaproteomics of stool proteins is a relatively new approach that moves beyond compositional analysis of the microbiota to provide a noninvasive means to assay intestinal function and examine host-microbiota relationships (9).
In this study, we performed a metaproteomic analysis of stool samples from 101 individuals, including patients with new-onset type 1 diabetes, high- or low-risk first-degree relatives (FDRs), and healthy control subjects. We have uncovered a network of human and microbial stool proteins linked to epithelial barrier function as well as exocrine pancreas that is disturbed both in patients with new-onset type 1 diabetes and in high-risk individuals with ongoing islet autoimmunity and linked these changes to specific microbial taxa.
Research Design and Methods
Subject Characteristics
All subjects resided in the Denver, Colorado, area. Stool samples were collected either at home or at the Barbara Davis Center for Diabetes as previously described (10). Samples collected at home were stored at −20°C before delivery to the center on ice. Individuals with gastrointestinal disease or known infections or who had taken antibiotics in the 4 weeks prior were excluded. Subject characteristics are summarized in Table 1. Subjects included patients with newly diagnosed type 1 diabetes (<6 months, new-onset patients) recruited during their visits to the Barbara Davis Center for Childhood Diabetes. Individuals with one to four autoantibodies and seronegative FDRs of patients with type 1 diabetes were recruited from the Type 1 Diabetes TrialNet Pathway to Prevention study. Individuals without any family history of autoimmunity were recruited from University of Colorado employees and children of employees. Three seronegative subjects and one seropositive subject were vegetarian, and one seronegative subject, one seropositive subject, and two new-onset patients consumed a gluten-free diet. All subjects gave informed consent according to the study protocol approved by the institutional review board, University of Colorado Denver. The study was approved as an ancillary study by Type 1 Diabetes TrialNet.
Table 1.
Cohort characteristics
| Cohort |
||||
|---|---|---|---|---|
| Healthy control (n = 22) | Seronegative (n = 29) | Seropositive (n = 17) | New-onset (n = 33) | |
| Sex | ||||
| Female, n | 8 | 14 | 9 | 15 |
| Male, n | 14 | 15 | 8 | 18 |
| Age (years), median (range) | 11.5 (4.0–45.0) | 12.0 (3.0–45.0) | 9.0 (4.0–36.0) | 11.0 (2.0–20.0) |
| HLA, n (%) | ||||
| 3 alone | 4 (18.2) | 11 (37.9) | 4 (23.5) | 10 (30.3) |
| 3 plus 4 | 3 (13.6) | 9 (31.0) | 4 (23.5) | 8 (24.2) |
| 4 alone | 4 (18.2) | 7 (24.1) | 9 (52.9) | 12 (36.4) |
| x/x | 11 (50.0) | 2 (6.9) | 0 (0.0) | 3 (9.1) |
| Autoantibodies, n (%) | ||||
| 0 | 22 (100.0) | 29 (100.0) | 0 (0.0) | 1 (3.0) |
| 1 | 0 (0.0) | 0 (0.0) | 3 (17.6) | 12 (36.4) |
| 2–4 | 0 (0.0) | 0 (0.0) | 14 (82.4) | 20 (60.6) |
| HbA1c (%), median (range) | — | — | 4.9 (4.2–5.3) | 7.9 (5.5–15.0) |
| Age at onset (years), median (range) | — | — | — | 10.0 (2.0–20.0) |
| Disease duration (weeks), median (range) | — | — | — | 5.1 (0.3–17.3) |
| Impaired glucose metabolism, n (%) | 4 (23.5) | |||
Protein Extraction and Digestion
Samples were resuspended in potassium phosphate buffer containing protease inhibitors (phenylmethylsofonyl fluoride, leupeptin, aprotinin) before centrifugation to remove debris. Total protein from the supernatant was deglycosylated with PNGase F (Promega, Madison, WI). Proteins were solubilized, reduced, and alkylated followed by digestion with modified pig trypsin (Promega). Detergents were precipitated and salts removed using C18 tips (Glygen, Columbia, MD). Tryptic peptides were resuspended in 0.1% formic acid for mass spectrometry analysis.
Mass Spectrometry
Tryptic peptides were used for liquid chromatography–tandem mass spectrometry analysis on a Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Peptides were separated on an EASY-Spray analytical column (Thermo Fisher Scientific). A 90-min gradient from 3 to 25% acetonitrile was performed over 60 min followed by 25–40% acetonitrile over 12 min and 95% acetonitrile for 15 min containing 0.1% formic acid at a flow rate of 250 nL/min. Survey mass spectrometric scans from charge/mass ratio 350–1,400 were acquired in the Orbitrap analyzer (Thermo Fisher Scientific) with resolution r = 70,000, automatic gain control of 3 × 106, and maximum injection time of 100 ms. The top 20 most intense ions were selected for tandem mass spectrometry analysis with a normalized collision energy of 29%. Dynamic exclusion was enabled for 30 s, with a mass exclusion width of 10 parts per million. Second stage of mass spectrometry was acquired with a mass/charge ratio 200–2,000 at a resolution r = 17,500, automatic gain control of 5 × 105, and maximum injection time of 55 ms.
Database Searching and Processing
Peptide spectrum matching, protein inference, grouping, and quantitation were performed using the MetaPro-IQ strategy (11), with exceptions as follow. The X! Tandem algorithm was implemented using rTANDEM, and the final database search was performed with Spectrum Mill (Agilent, Santa Clara, CA). For the final search, carbamidomethylation of cysteine was included as a fixed modification. Oxidation of methionine and deamidation of asparagine were considered as variable modifications. A maximum of two missed cleavages was allowed. A decoy database, prepared by reversing the search database with a false discovery rate of 0.01, was used. Proteins that shared one or more peptides were grouped. Proteins with only a single detected peptide were discarded. One sample with <500 proteins identified and 1 outlier were excluded on the basis of a very high ratio of human/microbial proteins present (8 SDs above the mean) (Supplementary Fig. 1). Raw spectra, peak lists, the custom sequence database, and quantification results have been deposited in the PRIDE (Proteomics Identifications) partner repository with the identifier PXD008870.
Microbiome Analysis
Bacterial abundance in stool was determined by 16S rRNA gene sequencing of the V4 variable region and was previously described (10).
Statistical Analyses
Proteins detected in >40% of any single subject group were included in univariate and multivariate analyses. All proteins were considered for functional pathway assignments. Human and nonhuman proteins were scaled to the sum of the total human or total nonhuman intensity, respectively. Protein intensities were normalized by constant log-ratio transformation, with an offset corresponding to the minimum detected intensity across the data set. One-way ANOVA was performed after adjustment for age and sex. Correction for multiple hypotheses used the Benjamini and Hochberg procedure. Proteins with adjusted P < 0.05 were further investigated with post hoc Tukey tests.
Supervised multivariate models were developed using supervised sparse partial least squares discriminant analysis (sPLS-DA) with the mixOmics R package (12). Differences between groups were evaluated by permutation ANOVA (PERMANOVA) using the vegan package in R and 100,000 permutations. Generalized linear models were developed using the 10 proteins with the lowest univariate P values (combined control and seronegative subjects vs. new-onset patients) and evaluated using leave-one-out cross validation. Integration of proteomic and 16S rRNA sequencing data were performed using DIABLO in mixOmics with a value of 0.5 in the design matrix (12). Relevance association networks were plotted in Cytoscape. Pearson correlations and hierarchical clustering were performed using R. Kegg ortholog (KO) assignments of microbial proteins were retrieved from the Integrated Reference Catalog of the Human Gut Microbiome (http://meta.genomics.cn/meta/home). The intensity of proteins that shared a KO were summed then normalized, filtered, and evaluated in the same way as individual proteins. KOs were assigned to gut metabolic modules (GMMs) and evaluated using GOmixer (13). The last common ancestor of each peptide was determined using Unipept (14).
Results
Metaproteomic Analysis of Stool Samples
We used metaproteomics of fecal proteins to test the hypothesis that intestinal inflammation is associated with type 1 diabetes progression and to determine whether the functional characteristics of the microbiota are altered in individuals with islet autoimmunity or clinical disease. To this end, we analyzed soluble fecal proteins, which are enriched for human proteins but also contain soluble microbial proteins. The four subject groups included new-onset patients, seropositive individuals, seronegative FDRs, and unrelated healthy control subjects (Table 1). A total of 470 human protein and 10,908 nonhuman protein clusters were identified, with a mean of 1,566 ± 305 protein clusters identified per sample. Although a smaller proportion of human protein identifications were made, human origin proteins represented 52 ± 22% of the total intensity. No significant difference was found in the total number of human and microbial proteins identified or the proportion of human-to-bacterial protein intensity present among the groups (Supplementary Fig. 1). No individual proteins correlated significantly with age or sex after adjustment for multiple testing. We then investigated whether the abundance of specific proteins differed between individuals with and without ongoing islet autoimmunity.
Multivariate Analysis of Stool Proteins Differentiates Individuals With Islet Autoimmunity From Low-Risk and Healthy Individuals
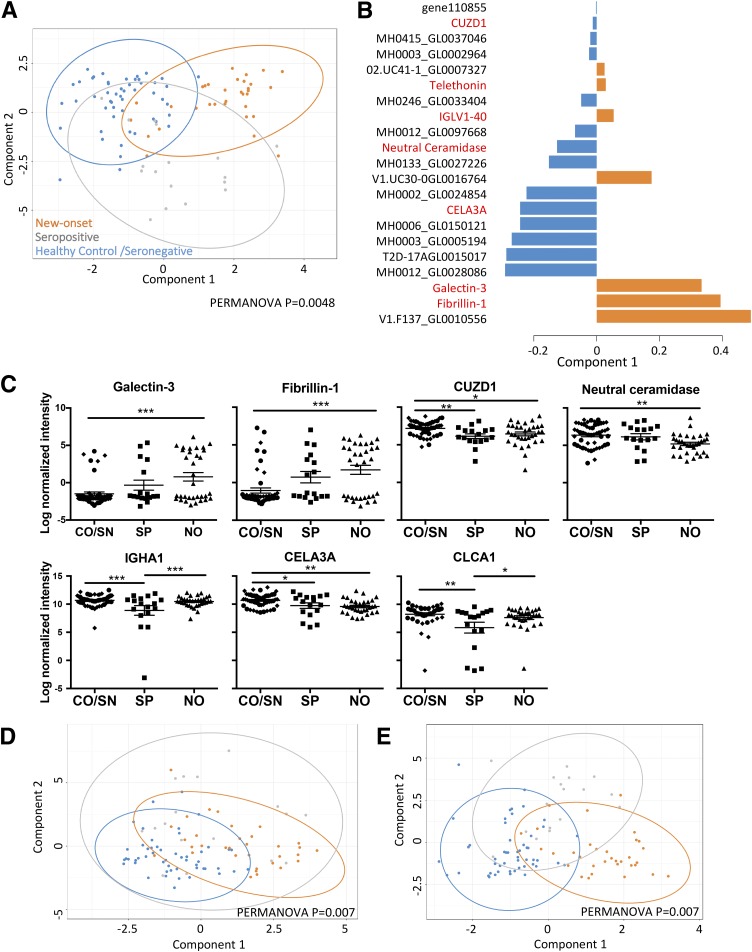
sPLS-DA was used to identify proteins that may discriminate the four groups (15). This analysis showed that healthy control subjects and seronegative individuals overlapped, whereas the new-onset patients and seropositive individuals separated from these groups (Supplementary Fig. 2A). A statistical comparison found that although overall the four subject groups differed (P = 0.014), the healthy control versus seronegative groups did not (P = 0.08). On the basis of this result, we pooled the healthy control and seronegative groups to increase the power to detect differences in individuals with ongoing islet autoimmunity from low-risk individuals. sPLS-DA of these three groups clustered the majority of the new-onset patients away from the combined control group on component 1, whereas the majority of the seropositive group separated from the control group on both components 1 and 2 (Fig. 1A) (PERMANOVA P = 0.0048). Both microbial and human proteins contributed to segregating these three groups (Fig. 1B). Two-way sPLS-DA showed that all new-onset patients and almost all seropositive individuals could be separated from the control subjects (P = 0.036 and P = 0.021) (Supplementary Fig. 2B and C). Hence, the stool metaproteome was able to distinguish individuals with islet autoimmunity from low-risk healthy individuals in this cohort.
Figure 1.
The fecal meta-proteome distinguishes individuals with islet autoimmunity and new-onset (NO) patients from control subjects. A: sPLS-DA of all human and microbial proteins comparing NO, seropositive (SP), and combined healthy control and seronegative (CO/SN) groups. B: Component 1 contributing variables. Red text, human proteins; black text, microbial proteins; 158742018-stool1_revised_scaffold9355_1_gene110855 abbreviated as gene11085. C: Log-normalized intensity of significant proteins comparing NO, SP, and CO/SN subject groups by one-way ANOVA (adjusted P < 0.05). Data are mean ± SEM. D and E: sPLS-DA of human and microbial proteins, respectively, comparing NO, SP, and CO/SN subject groups. A PERMANOVA comparing the three groups is shown. *P < 0.05; **P < 0.01; ***P < 0.001.
Identification of Individual Human Proteins in Stool That Differ Among Subject Groups
We next assessed whether individual human proteins distinguish subjects with disease or islet autoantibody positivity from control subjects. Comparison of the new-onset, seropositive, and the combined control groups by one-way ANOVA (Supplementary Table 1) resulted in 24 proteins with a P < 0.05, with seven significant after multiple testing adjustment (galectin-3, fibrillin-1, CUZD1, neutral ceramidase, CELA3A, CLCA1, and IGHA1) (Fig. 1C). Galectin-3 and fibrillin-1 were increased in new-onset patients, whereas CUZD1, neutral ceramidase, CELA3A, CLCA1, and IGHA1 were decreased in new-onset patients and/or seropositive subjects compared with control subjects. sPLS-DA analysis showed that the three subject groups could be differentiated by using human proteins alone (P = 0.007) (Fig. 1D). The differentially abundant proteins included three produced by the exocrine pancreas (CELA3A, CUZD1, and neutral ceramidase). The CELA3A protein was grouped with the highly homologous CELA3B, and the two are both known as pancreatic elastase 1. Other differentially abundant proteins were involved with IgA antibody production (IGHA1), inflammation and neutrophil activation (galectin-3), extracellular matrix (fibrillin-1), tumor growth factor-β (TGF-β) bioavailability (fibrillin-1), and goblet cell mucous production (CLCA1). These observations confirm the findings of others that exocrine pancreas output is decreased in individuals with type 1 diabetes (16). The current findings suggest for the first time in our knowledge that reduced output of exocrine enzymes begins in islet autoantibody–positive individuals. Increased abundance of galectin-3 and fibrillin-1 in new-onset patients raises the hypothesis that proteins associated with inflammation are released into the stool in type 1 diabetes.
Microbial Protein Identification in the Stool of Individuals With Islet Autoimmunity
We next identified individual microbial proteins associated with type 1 diabetes. Although 34 proteins had a P < 0.01 and 143 proteins had a P < 0.05, none were significant after multiple testing adjustment (Supplementary Table 2). The five microbial proteins with the lowest unadjusted P values (P < 0.001) are shown in Supplementary Fig. 3. The KO assignments for these five proteins were phosphotransferase system, sugar-specific enzyme IIA component [EC:2.7.1.-], elongation factor thermo unstable, basic membrane protein A and related proteins, unassigned and ferredoxin hydrogenase [EC:1.12.7.2]. The taxonomic assignments of these five proteins were Faecalibacterium prausnitzii, Clostridiales, and unassigned. Although individual microbial proteins did not significantly differentiate subject groups, multivariate sPLS-DA using microbial proteins only was able to separate new-onset, seropositive, and control subjects (P = 0.007) (Fig. 1E). These results indicate that collectively, bacterial-derived proteins discriminate subjects with islet autoimmunity from control subjects.
Functional Changes in the Stool Metaproteome May Be Associated With Islet Autoimmunity
To probe the biological pathways associated with the microbial proteins driving the differences observed, we collapsed the microbial proteins by KO. However, one-third of microbial proteins identified did not have an assigned KO. Of the 545 KOs represented, none were significantly different between groups after adjusting for multiple testing (Supplementary Table 3). sPLS-DA showed that KO drivers were close to differentiating subject groups (P = 0.06) (Supplementary Fig. 4). As an alternative approach, the package GOmixer was used to infer GMMs associated with the identified microbial proteins. A comparison of new-onset patients and combined control subjects showed that mucin degradation was increased in the new-onset patients (P = 0.02). A comparison of seropositive individuals with combined control subjects showed that five GMMs had a P < 0.05, including butyrate production through transferase (P = 0.023), which was reduced in seropositive individuals. All identified GMMs are shown in Supplementary Table 4. Although none of these differences were significant after adjusting for multiple testing, they suggest that mucin degradation and butyrate production may be altered in individuals with islet autoimmunity.
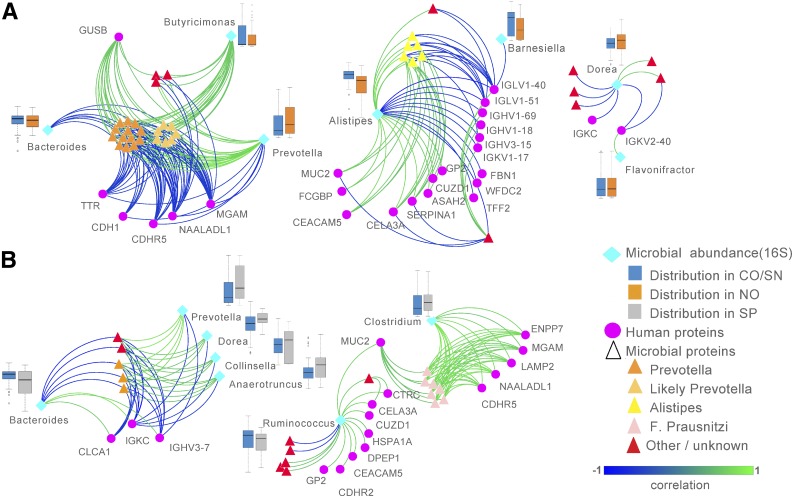
Integrative Network Analysis Reveals Correlations Between Differentially Expressed Proteins and Bacterial Taxa Associated With Disease Risk
We next performed an integrative analysis to explore correlations between the human and microbial proteins and bacterial taxa that best discriminated each subject group with other highly correlated features in the data by using the newly developed DIABLO method (12). The abundance of bacterial taxa was previously determined by 16S rRNA gene sequencing analysis (10). We first compared combined control subjects with new-onset patients, and the selected features and similarity scores were used to generate a similarity matrix visualized in a network diagram (Fig. 2A). This analysis resulted in three clusters, the first driven by Prevotella, which was negatively correlated with Bacteroides and a cluster of human proteins, including adhesion molecules CDHR5 and CDH1 and brush border enzymes MGAM and NAALADL1. The second cluster was driven by Alistipes, which was positively correlated with mucous layer proteins MUC2 and FCGBP and adhesion molecule CEACAM5 and a cluster of exocrine pancreas–produced proteins (CUZD1, CELA3A, GP2, and neutral ceramidase) but negatively correlated with fibrillin-1 and a cluster of heavy- and light-chain antibody variable regions. Because Alistipes was previously found to be significantly more abundant in seronegative individuals compared with new-onset patients (10), these data suggest that Alistipes may promote mucous production and a healthy epithelial barrier, whereas the presence of certain antibody specificities, which were abundant in new-onset patients, may oppose this interaction.
Figure 2.
Integrative multivariate analyses identify multiomics signatures associated with features that discriminate new-onset (NO) patients or seropositive (SP) individuals from seronegative (SN) and healthy control (CO) individuals. DIABLO method was used to integrate three data sets: microbial taxonomic abundance data, microbial protein abundance, and human protein abundance. This model was used to identify features highly correlated with variables that discriminate NO patients from combined CO/SN subjects (A) and SP individuals from CO/SN subjects (B). Similarity matrix among the identified features were obtained from DIABLO and represented using relevance networks. Relative bacterial abundance of each microbial taxon is shown in box-and-whisker plots. Microbial proteins have been manually grouped according to taxonomic identity.
We then repeated the DIABLO analysis by comparing combined control subjects with seropositive individuals, resulting in two clusters (Fig. 2B). One cluster again contained Prevotella, which negatively correlated with Bacteroides, goblet cell–produced CLCA1, and two antibody components. The second cluster linked a group of F. prausnitzii–derived proteins with MUC2 and Clostridium. This cluster positively correlated with the adhesion molecule CDHR5 and brush border enzymes MGAM and NAALADL1. MUC2 also was linked to Ruminococcus, which was positively correlated with adhesion molecules CDHR2 and CEACAM5 and exocrine pancreas proteins CELA3A, CUZD1, GP2, and DPEP1. These data suggest that proteins that were significantly increased in control subjects (e.g., CELA3A, CUZD1) are positively correlated with proteins and taxa that have been previously linked with gut health, such as MUC2 and F. prausnitzii (17).
We next calculated Pearson correlation coefficients for each pair of proteins or taxa selected in the two DIABLO networks by using all subject group data and grouped these using hierarchal clustering (Supplementary Fig. 5). This clustering analysis clearly identified three major groupings linked to 1) Prevotella, 2) Alistipes and F. prausnitzii, and 3) Bacteroides. This showed that Alistipes and F. prausnitzii proteins both positively correlated with mucin-layer components, adhesion molecules, and exocrine pancreas–derived proteins, suggesting that these taxa may occupy similar functional niches. We conclude from these data that proteins and taxa that are increased in healthy control subjects and seronegative individuals are associated with functions and taxa linked to anti-inflammatory effects and improved barrier function.
Evaluation of the Diagnostic Performance of Stool Proteins as Biomarkers
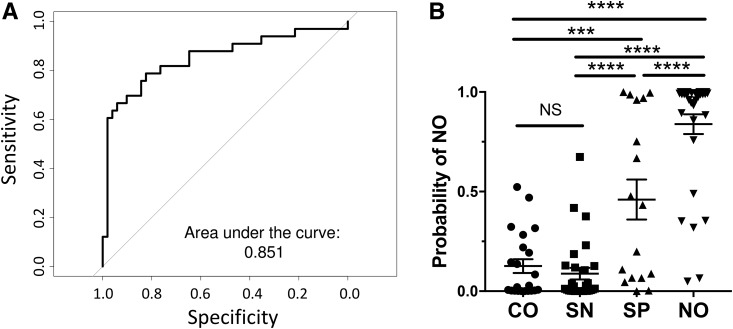
Finally, we assessed how the top 10 differentially abundant proteins performed as a potential biomarker signature. A general linear model was developed using combined control subjects versus new-onset patients. Receiver operating characteristic analysis with leave-one-out cross validation resulted in an area under the curve of 0.85 (Fig. 3A). We used this model to predict the probability of seropositive individuals identifying as developing new-onset diabetes (Fig. 3B) and found that seropositive individuals were significantly more likely to be identified as new-onset than seronegative or healthy control individuals (P < 0.001). Our model did not significantly correlate with sex (P = 0.22) or HbA1c in new-onset patients (P = 0.968), indicating that it was unlikely to have been induced by dysglycemia. Of note, a significant negative correlation with age was found (Supplementary Fig. 6A). However, if the older subjects were excluded, the correlation with age was lost (Supplementary Fig. 6B), but the model still significantly distinguished the subject groups (Supplementary Fig. 6C). These results demonstrate the potential diagnostic value of stool protein signatures in type 1 diabetes risk stratification and further suggest involvement of intestinal/stool proteins in disease progression.
Figure 3.
Diagnostic biomarker performance of a multivariate prediction model. A: Receiver operating characteristic curve of cross-validated linear models. Each sample was classified as combined control and seronegative (CO/SN) or new-onset (NO) using a generalized linear model developed without that sample. B: Probability of being classified as NO. Probabilities were determined using a linear model developed on all CO/SN subjects and NO patients. The 10 proteins selected for this model were MH0003_GL0005194, T2D.17A_GL0015017, V1.FI37_GL0010556, galectin-3, MH0133_GL0027226, fibrillin-1, CELA3A, neutral ceramidase, MH0012_GL0028086, and V1.UC30.0_GL0016764. NS, not significant. ***P < 0.001; ****P < 0.0001. SP, seropositive.
Conclusions
We have used metaproteomics to functionally characterize human and microbial fecal proteins linked to type 1 diabetes in the largest study of its kind to our knowledge. We have made the following observations: 1) Human and microbial proteins present in stool appear able to discriminate individuals with ongoing islet autoimmunity from low-risk and healthy individuals; 2) both patients with new-onset diabetes and islet autoantibody–positive individuals have reduced markers of exocrine pancreas output compared with control subjects; 3) proteins linked to inflammation are increased in abundance in the stool of patients with new-onset type 1 diabetes; and 4) a functional relationship exists between proteins and bacterial taxa, which are reduced in new-onset patients and seropositive individuals with mucosal barrier function and exocrine pancreas output.
Of the significantly altered human proteins, several had known immunomodulatory functions. Fibrillin-1 forms a key constituent of extracellular matrix microfibrils where it stores latent TGF-β, reducing TGF-β activity and signaling (18). Because regulatory T-cell induction in the intestine is TGF-β dependent (19), increased fibrillin-1 may reduce regulatory T-cell frequency. In addition, fibrillin-1 plays a role in macrophage chemotaxis (20). Galectin-3 is believed to be a proinflammatory mediator in a wide range of inflammation-associated disorders (21) and is chemotactic for inflammatory cells through reactive oxygen species production (22). Overproduction of these proteins supports our hypothesis that increased intestinal inflammation is associated with type 1 diabetes pathogenesis. However, the source of these inflammatory proteins is unknown and may be intestinal or pancreatic.
The abundance of the fecal inflammation marker calprotectin (comprising S100A8 and S100A9) was not altered. Calprotectin is produced by activated neutrophils (e.g., as in active inflammatory bowel disease). Mild intestinal inflammation has been reported in studies of duodenal and jejunal biopsy specimens from patients with type 1 diabetes (7,23). This included overexpression of HLA class II, ICAM-1 in the epithelium, and interleukin-1α and -4 in the lamina propria (7) and increased infiltration of macrophages and monocytes (8). Increased neutrophilic infiltration has not been described in the type 1 diabetes gut, which may explain the unchanged calprotectin. Studies of biopsy samples from the ileum or colon in type 1 diabetes have not been reported; therefore, the involvement of inflammation in these tissues is unknown.
Although IGHA1 (IgA1 heavy-chain constant region) was reduced in seropositive individuals, several Ig variable regions contributed to the variance among the groups. Some studies have reported fecal IgA levels to be lower in patients with type 1 diabetes (24), whereas others found no difference (25,26). The reduced soluble IgA levels possibly indicate increased coating of bacteria by IgA, as reported by others (27). Alternatively, reduced IgA levels could be related to the reduction in bacterial diversity reported to precede the onset of clinical disease in high-risk individuals (28).
In addition to immunomodulatory proteins, exocrine pancreas proteins were decreased in new-onset patients and seropositive individuals. Both ultrasound imaging and fecal pancreatic elastase-1 measurement have found a deficiency of exocrine pancreas function in patients with type 1 diabetes (16,29). Consistent with our findings, fecal elastase was reported as decreased in new-onset patients and continued to drop with disease duration (29). Fecal elastase positively correlated with C-peptide and negatively correlated with HbA1c (29). Our demonstration of reduced fecal elastase in islet antibody–positive individuals suggests that the process of exocrine dysfunction begins before diagnosis. The causes of exocrine pancreas dysfunction are unknown, with multiple hypotheses proposed (16). Studies have suggested a possible role for immune involvement in exocrine pancreas abnormality (30). Of note, our integrative analysis suggests that a relationship exists between reduced exocrine pancreas function in type 1 diabetes and the gut microbiota.
By using multivariate integrative analysis, we found that an increased abundance of exocrine pancreas proteins correlates with proteins associated with a healthy epithelial barrier (e.g., MUC2, FCGBP, CDHR5, CDH1) (31,32) and with Alistipes, which was significantly increased in seronegative individuals (10). This health-associated cluster contained butyrate producers F. prausnitzii and Clostridium as well as Alistipes, Ruminococcus, Barnesiella, and Dorea. Colonic microbial-produced butyrate is known to exert several beneficial effects (33). A recent meta-analysis of 3,048 data sets found that Barnesiella and Alistipes were universally associated with health (34).
In antagonism to the health-associated cluster were Bacteroides- and Prevotella-associated clusters. Several studies have reported an increased abundance of Bacteroides species either in islet autoantibody–positive individuals or around disease onset (26,35,36). Prevotella is known to predominate in individuals who consume a plant-rich diet, whereas Bacteroides is associated with high-fat, high-protein western diets (3). Despite this association of Prevotella with a plant-based diet, many studies have found an increased abundance of Prevotella species in inflammatory diseases (37). These seemingly contradictory findings may be due to the large diversity within the Prevotella genus. The current analyses do not support a protective role for Prevotella in preventing progression to islet autoimmunity or type 1 diabetes.
Seropositive individuals can be classified into disease stages by number of islet autoantibodies and the presence of dysglycemia (38). Stage 1 is two or more autoantibodies, stage 2 is two or more autoantibodies and dysglycemia, and stage 3 is hyperglycemia. Of the 17 seropositive individuals in this study, 3 had one autoantibody; however, 1 of these had dysglycemia and could not be staged by this method, and another 3 were stage 2. Because of the low numbers, we were unable to determine whether our model fitted with disease stage. Longitudinal studies will be needed to determine definitively whether proteins in stool predict disease progression.
Although we have detected a striking disease-associated signature in our samples, the differences in protein abundance need to be validated in a larger, independent cohort. Furthermore, we did not measure C-peptide, so we were not able to make correlations between β-cell function and stool proteins. Shotgun proteomics is limited by a lack of sensitivity for detecting low-abundance proteins and by low reproducibility compared with other methods. Therefore, the changes we have observed should be validated with other methods. A reduction in CUZD1 and CELA3A in patients with type 1 diabetes has been reported in other studies (39,40), which suggests that the current findings are reproducible. Another limitation was that we analyzed only soluble stool proteins. Although this analysis resulted in an enrichment of human proteins, it biased the microbial protein detection toward those that are secreted or released from lysis-prone bacteria, such as gram-negative species. In addition, many identified bacterial proteins were uncharacterized and could not be included in our functional analyses. The subjects in our study were of a relatively wide age range. Because disease pathogenesis mechanisms may vary between children and adults, it will be necessary to validate our findings separately in these populations. We also did not collect detailed information on dietary intake, which may be an additional confounder to our data.
Despite these limitations, we were able to describe novel associations among gut-, pancreas-, and microbiota-derived proteins related to type 1 diabetes. These studies identify a negative correlation between the presence of specific stool antibodies and taxa associated with gut health and demonstrate an association between the abundance of exocrine pancreatic proteins and anti-inflammatory microbial taxa and mucosal barrier function. This study establishes for the first time in our knowledge the utility of using stool-based metaproteomics to link changes in the gut microbiota with the health of the intestinal mucosa and exocrine pancreas in type 1 diabetes.
Supplementary Material
Article Information
Acknowledgments. The authors thank the Translational Research Institute Proteomics Core facility, the individuals who participated in this study, Cini James (The University of Queensland, Brisbane, Australia) and Casey Wright (The University of Queensland, Brisbane, Australia) for technical assistance, Nicholas Matigian (The University of Queensland, Brisbane, Australia) for statistical advice, and Ranjeny Thomas (The University of Queensland, Brisbane, Australia) and Mark Harris (Lady Cilento Children’s Hospital, Brisbane, Australia) for helpful discussions.
Funding. J.A.M. was funded by a JDRF postdoctoral fellowship (PDF-2014-222-A-N). The study was funded by JDRF grant 2-SRA-2015-306-Q-R (to D.Z.). E.E.H.-W. is funded by a JDRF career development fellowship (2-2013-34). Type 1 Diabetes TrialNet is funded by National Institute of Diabetes and Digestive and Kidney Diseases grant U01-DK-85509.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. P.G.G. performed the data analysis, statistical analysis, and visualization and revised the manuscript. J.A.M. designed the approach, developed the methodology, performed experiments, performed the data analysis, supervised students, and revised the manuscript. D.L. performed experiments and provided technical advice. K.-A.L.C. provided statistical oversight. P.A.G. and D.Z. provided resources and revised the manuscript. M.M.H. provided technical advice, supervised students, and revised the manuscript. E.E.H.-W. conceptualized and led the project, designed experiments, supervised students, and wrote the manuscript. E.E.H.-W. is the guarantor for this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Data Availability. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository with the data set identifier PXD008870.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc18-0777/-/DC1.
J.A.M. is currently affiliated with AgResearch, Grasslands Research Centre, Palmerston North, New Zealand.
K.-A.L.C. is currently affiliated with Melbourne Integrative Genomics, School of Mathematics and Statistics, University of Melbourne, Parkville, Victoria, Australia.
M.M.H. is currently affiliated with QIMR Berghofer Medical Research Institute, Brisbane, Queensland, Australia.
D.Z. is currently affiliated with Innate Biotechnologies, LLC, Denver, CO.
References
- 1.Atkinson MA, Chervonsky A. Does the gut microbiota have a role in type 1 diabetes? Early evidence from humans and animal models of the disease. Diabetologia 2012;55:2868–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullaney JA, Stephens JE, Costello ME, et al. Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome 2018;6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet 2016;387:2340–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardissone A, Kemppainen KM, Triplett EW. Type 1 diabetes and intestinal microbiota: how geographic differences between human cohorts can influence interpretation of associations. Diabetes Case Rep 2017;2:128 [Google Scholar]
- 6.Bosi E, Molteni L, Radaelli MG, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia 2006;49:2824–2827 [DOI] [PubMed] [Google Scholar]
- 7.Westerholm-Ormio M, Vaarala O, Pihkala P, Ilonen J, Savilahti E. Immunologic activity in the small intestinal mucosa of pediatric patients with type 1 diabetes. Diabetes 2003;52:2287–2295 [DOI] [PubMed] [Google Scholar]
- 8.Pellegrini S, Sordi V, Bolla AM, et al. Duodenal mucosa of patients with type 1 diabetes shows distinctive inflammatory profile and microbiota. J Clin Endocrinol Metab 2017;102:1468–1477 [DOI] [PubMed] [Google Scholar]
- 9.Lichtman JS, Sonnenburg JL, Elias JE. Monitoring host responses to the gut microbiota. ISME J 2015;9:1908–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkanani AK, Hara N, Gottlieb PA, et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes 2015;64:3510–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Ning Z, Mayne J, et al. MetaPro-IQ: a universal metaproteomic approach to studying human and mouse gut microbiota. Microbiome 2016;4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohart F, Gautier B, Singh A, Lê Cao KA. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLOS Comput Biol 2017;13:e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darzi Y, Falony G, Vieira-Silva S, Raes J. Towards biome-specific analysis of meta-omics data. ISME J 2016;10:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mesuere B, Devreese B, Debyser G, Aerts M, Vandamme P, Dawyndt P. Unipept: tryptic peptide-based biodiversity analysis of metaproteome samples. J Proteome Res 2012;11:5773–5780 [DOI] [PubMed] [Google Scholar]
- 15.Lê Cao KA, Boitard S, Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 2011;12:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardt PD, Ewald N. Exocrine pancreatic insufficiency in diabetes mellitus: a complication of diabetic neuropathy or a different type of diabetes? Exp Diabetes Res 2011;2011:761950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeyer KA, Reinhardt DP. Fibrillin-containing microfibrils are key signal relay stations for cell function. J Cell Commun Signal 2015;9:309–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coombes JL, Siddiqui KR, Arancibia-Cárcamo CV, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med 2007;204:1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G, Booms P, Halushka M, et al. Induction of macrophage chemotaxis by aortic extracts of the mgR Marfan mouse model and a GxxPG-containing fibrillin-1 fragment. Circulation 2006;114:1855–1862 [DOI] [PubMed] [Google Scholar]
- 21.de Oliveira FL, Gatto M, Bassi N, et al. Galectin-3 in autoimmunity and autoimmune diseases. Exp Biol Med (Maywood) 2015;240:1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madrigal-Matute J, Lindholt JS, Fernandez-Garcia CE, et al. Galectin-3, a biomarker linking oxidative stress and inflammation with the clinical outcomes of patients with atherothrombosis. J Am Heart Assoc 2014;3:e000785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Badami E, Sorini C, Coccia M, et al. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes 2011;60:2120–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lassenius MI, Fogarty CL, Blaut M, et al.; FinnDiane Study Group . Intestinal alkaline phosphatase at the crossroad of intestinal health and disease - a putative role in type 1 diabetes. J Intern Med 2017;281:586–600 [DOI] [PubMed] [Google Scholar]
- 25.de Groot PF, Belzer C, Aydin Ö, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One 2017;12:e0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Goffau MC, Luopajärvi K, Knip M, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 2013;62:1238–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostic AD, Gevers D, Siljander H, et al.; DIABIMMUNE Study Group . The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015;17:260–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavalot F, Bonomo K, Perna P, et al. Pancreatic elastase-1 in stools, a marker of exocrine pancreas function, correlates with both residual beta-cell secretion and metabolic control in type 1 diabetic subjects. Diabetes Care 2004;27:2052–2054 [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Calvo T, Ekwall O, Amirian N, Zapardiel-Gonzalo J, von Herrath MG. Increased immune cell infiltration of the exocrine pancreas: a possible contribution to the pathogenesis of type 1 diabetes. Diabetes 2014;63:3880–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawley SW, Shifrin DA Jr., Grega-Larson NE, et al. Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell 2014;157:433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelaseyed T, Bergström JH, Gustafsson JK, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev 2014;260:8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–450 [DOI] [PubMed] [Google Scholar]
- 34.Mancabelli L, Milani C, Lugli GA, et al. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol Ecol 2017;93:fix153 [DOI] [PubMed] [Google Scholar]
- 35.Davis-Richardson AG, Ardissone AN, Dias R, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol 2014;5:678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murri M, Leiva I, Gomez-Zumaquero JM, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med 2013;11:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017;151:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Insel RA, Dunne JL, Atkinson MA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015;38:1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto E, Anselmo M, Calha M, et al. The intestinal proteome of diabetic and control children is enriched with different microbial and host proteins. Microbiology 2017;163:161–174 [DOI] [PubMed] [Google Scholar]
- 40.Heintz-Buschart A, May P, Laczny CC, et al. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat Microbiol 2016;2:16180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.