Abstract
We describe an efficient and mild method for the synthesis of macrocyclic peptides via nitrogen arylation from unprotected precursors. Various electro-philes and lysine-based nucleophiles were investigated and showed high-yielding product formation, even for a macrocyclization scan with 14 variants. We found that nitrogen-linked aryl products were more stable to base and oxidation when compared to thiol arylated species, thereby highlighting the utility of this methodology. Finally, N-aryl macrocyclization was performed on a p53 peptide inhibitor of MDM2 and resulted in identification of a nanomolar binder with improved proteolytic stability and cell permeability.
The N-aryl bond is often found in pharmaceuticals and biologically active molecules.1 Over the past century, extensive research and development has been devoted to discover efficient chemical methods to produce N−C(aryl) bonds. Such bonds are typically formed by nucleophilic aromatic substitution (SN Ar).2 Widely used methods include transition-metal-catalyzed cross-coupling reactions with copper-catalyzed Ullman-type N-arylation or palladium-catalyzed Buchwald−Hartwig coupling.1c,3 However, N−C(aryl) bond formation often requires high temperatures and/or expensive and oxygen-sensitive catalysts. Therefore, these conditions have precluded their use with unprotected peptides because rapid degradation will occur. In contrast to N-arylation, highly efficient and selective cysteine arylations have been reported and applied to peptide macrocyclization and bioconjugation.4
Peptide macrocyclization, also known as peptide stapling,5 isa useful strategy to design inhibitors of protein−protein interactions6 and, in some cases, display enhanced cell permeability and proteolytic stability when compared to the linear counterpart. Over the past 20 years, peptide stapling has benefited from the development of numerous chemical reactivities,5 including olefin metathesis,7 lactamization,8 and cycloadditions,9 but also cysteine-based reactions for disulfide bond formation,10 alkylation,11 and arylation.4 We recently developed a perfluoroaryl-cysteine SNAr chemistry for the rapid and selective synthesis under mild conditions of peptide macrocycles with enhanced biological properties.4a Nevertheless, the S−C(aryl) bond formed during this reaction can be eliminated to produce dehydroalanine under basic conditions and can be subject to oxidation.12 In addition, for S-arylation-mediated peptide macrocyclization, disulfide bond formation is a competing off-pathway reaction.4d Such limitations can be overcome by the discovery of new arylation chemistries for residues other than cysteine.
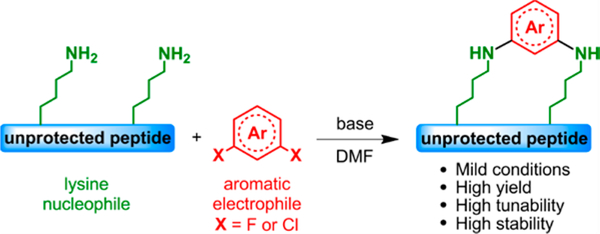
In this Communication, we report a macrocyclization methodology based on the discovery of lysine N-arylation of unprotected peptides (Figure 1). This synthetic approach is high yielding and mild, and it works over a range of macrocyclic loop sizes. Importantly, the N−C(aryl) bond overcomes the chemical stability issues sometimes encountered with cysteine S-arylation. Finally, to illustrate its potential, we apply our strategy to the macrocyclization of a known p53 peptide inhibitor against MDM2 protein.13
Figure 1.
Peptide macrocyclization via SNAr at lysine.
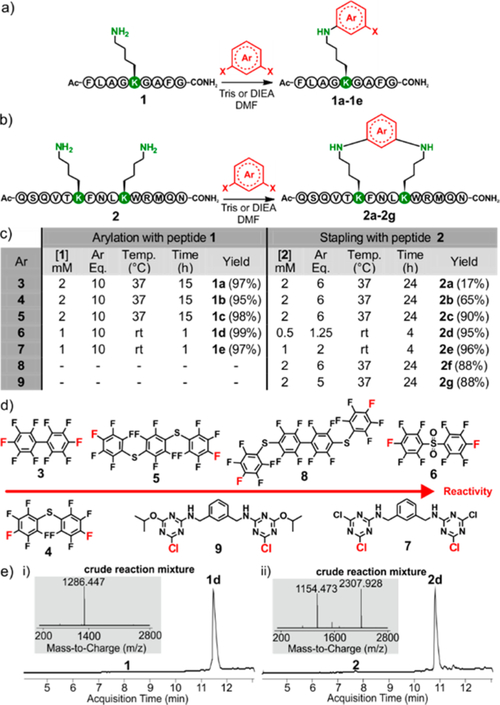
Our investigation commenced with the evaluation of lysine reactivity with perfluoroaryl compounds, since these electro-philes have been shown to be good candidates for SNAr cysteine arylation.4a Model peptide 1 was used to study monoarylation (Figure 2a), while peptide 2 was used to study i,i+4 macrocyclization. Both peptides were designed to possess most of the reactive side chains commonly found within bioactive peptides and proteins (Figure 2b). Our sights were aimed at developing the mildest reaction conditions while obtaining the highest conversion in the shortest time (Figure 1). Optimized conditions and electrophiles are summarized in Figure 2c,d. We found that dimethylformamide (DMF) was the best solvent and Tris base or DIEA was the best base for this perfluoro-lysine SNAr reaction. We were pleased to find that electrophiles 3, 4, and 5 reacted with peptide 1 at 37 °C after 15 h to give the corresponding mono-arylated products 1a, 1b, and 1c with high conversion. The decreased reaction rate of these compounds matches the lower nucleophilicity of lysine compared to cysteine. The reaction of peptide 2 with decafluorobiphenyl 3 resulted in low yields, owing to the deactivation of this electrophile after the first lysine SNAr.
Figure 2.
Various electrophiles and peptides undergo N-arylation at lysine. (a) Lysine mono-arylation of peptide 1. (b) Macrocyclization of peptide 2 via lysine N-arylation at the i,i+4 position. (c) Table of optimized conditions. Yields were determined by HPLC-MS analysis. (d) Electrophiles used during this study, ranked according to their relative reactivity. (e) HPLC-MS analysis of crude reaction mixture using electrophile 6 with (i) peptide 1 and (ii) peptide 2. See SI for all HPLC-MS traces and experimental details.
On the other hand, perfluoroaryls 4 and 5 showed increased macrocyclization conversion, presumably due to the stabilization of the Meisenheimer complex from the pre-installed sulfur atom.14 However, the use of these electrophiles required long reaction times and resulted in low yields, prompting us to investigate more reactive electrophiles. Based on these considerations, we decided to promote SNAr by introducing an electron-withdrawing group at the para position of the arene, thereby delocalizing the negative charge in the Meisenheimer complex.14 Such a compound was obtained by oxidizing the perfluorosulfide 4 to give perfluorosulfone 6 (Figure 2d). While we were preparing this manuscript, an elegant approach was successfully applied by the Derda laboratory for cysteine arylation on phage using linker 6.4d We also synthesized dichlorotriazine-based electrophile 7 in order to benefit from the reported high reactivity of such activated aromatic rings.15 Electrophiles 6 and 7 were first tested against peptide 1. High conversions and clean HPLC-MS traces (see Figure 2e and Supporting Information (SI), page S10) were observed for both electrophiles after only 1 h of reaction at room temperature. Thus, for subsequent work we favored the use of linkers 6 and 7. Since these two aryl halides are highly reactive, we had to tune the reaction parameters for each of them (Figure 2c). Indeed, judicious choice of peptide and electrophile concentration is important for highly efficient conversion of 2 into macrocyclic product. In fact, the double-arylation side product was observed in each case when reactions were run at concentrations >1 mM. We were delighted to find that 2 cyclized to give products 2d and 2e in the presence of electrophiles 6 and 7, respectively. Importantly, after 4 h at room temperature, the reaction yielded the desired stapled products with remarkably clean HPLC-MS traces (see Figure 2e and SI, page S10).
To further demonstrate the versatility of our lysine N-arylation methodology, we investigated its selectivity toward other residues. Therefore, we designed two model peptides that contained all nucleophilic residues except for cysteine. We were pleased to find that only the lysine-containing peptide was converted to the N-arylated product under these conditions (see SI, Figures S1–S4).
To expand the scope of this methodology, we synthesized two new electrophiles. Compound 8, a tetracyclic perfluorinated analogue of 3 and 5, was found to display reactivity similar to that of electrophile 5.
On the other hand, electrophile 9, an analogue of the triazine-based electrophile 7 where one chlorine atom has been substituted by an isopropoxy chain on both rings, had decreased reactivity when compared to its homologue 7. Nevertheless, the macrocyclization reaction was still complete in 24 h. Importantly, this result shows the feasibility of functionalization of electrophile 7 at position 5 of the triazine electrophile.
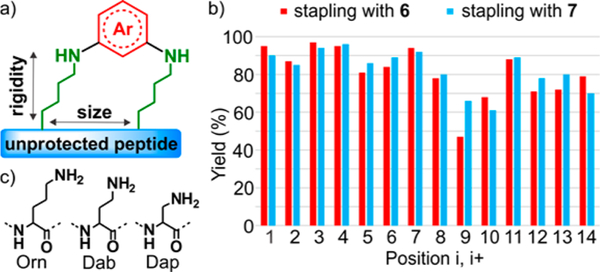
Encouraged by these results, we next aimed to expand the structural diversity of the peptide scaffold by tuning ring size and carbon side-chain length of lysine (Figure 3a). To modify the macrocyclic ring size, we performed a scan with peptide analogues of 2 by varying the site of lysine residues (i,i+1 to i,i +14) with two of the most reactive electrophiles, 6 and 7 (see SI, page S30). High conversions were observed (Figure 3b) for most of the peptides, except for two positions (i,i+9 and i,i +10). In these cases, ring strain is hypothesized to disfavor cyclization while favoring the undesired double-arylation products (see SI, pages S57–S60).
Figure 3.
N-Arylation enables a macrocyclization scan and chemistry at other nitrogen-containing amino acids. (a) Tuning size and rigidity of the macrocycles. (b) Bar graph reaction yield summary of the macrocyclization scan. The positions of the two lysines were varied from i,i+1 to i,i+14. Yield of desired product was determined by HPLC-MS. (c) Amino acids Orn, Dab, and Dap used as lysine surrogates.
Finally, to modulate the carbon side-chain length of lysine, we installed non-natural amino acids of variable hydrocarbon chain lengths (ornithine (Orn), diaminobutyric acid (Dab), and diaminopropionic acid (Dap), see Figure 3c) at i,i+7 spacing for each residue. Excellent conversions (72−97%) for all variant peptides were found with aryl halides 6 and 7 (see SI, pages S72–S77). Taken together, this macrocyclic scan may enable the design of highly tunable and diverse scaffolds.
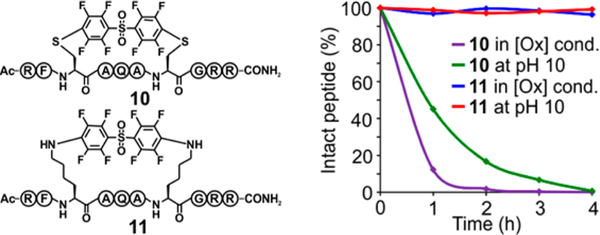
We then aimed to investigate the stability of the lysine-aryl stapled peptides compared to their cysteine-aryl homologues. We synthesized two macrocyclic peptides, 10 and 11, where cysteine and lysine were arylated with electrophile 6, respectively (Figure 4). Stability of these aryl conjugates was first studied under oxidative conditions and analyzed by HPLCMS (Figure 4 and SI, Figures S5 and S6). Peptide 10 degraded rapidly and produced multiple unidentified products with no intact peptide after 4 h. Strikingly, no detectable degradation was observed for 11 when it was treated with these oxidative conditions. We next focused on the stability of these peptides under basic conditions (Figure 4). 10 underwent elimination to give the double dehydroalanine-containing peptide after 4 h, whereas 11 showed complete stability to base (Figure 4 and SI, Figures S7 and S8). These results confirmed our initial hypothesis that activated S−C(aryl) bonds may undergo degradation under certain chemical conditions,4d,12 emphasizing the need for N-arylation to produce very stable macrocyclic peptides.
Figure 4.
N-Arylation enables generation of chemically stable macrocycles. Chemical stability of stapled peptides 10 and 11 under oxidative conditions (80 mM H2O2 in a buffered solution at pH 8.0 at 37 °C) and basic conditions (pH 10.0 at 37 °C).
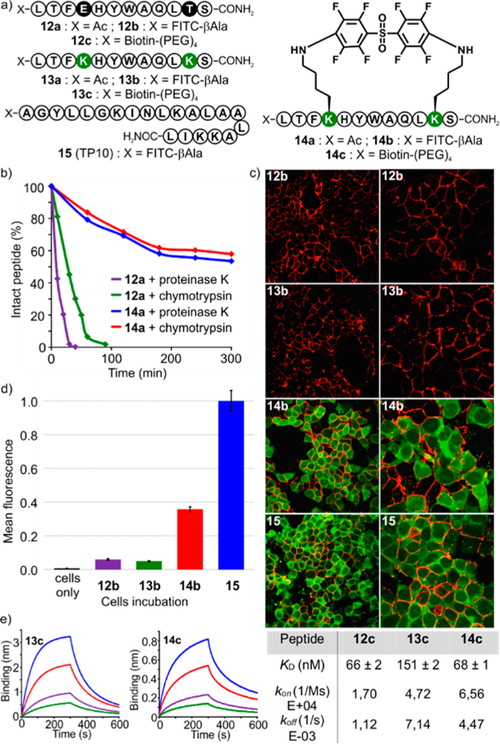
Peptide macrocyclization can improve pharmacodynamic and pharmacokinetic profiles of peptides by enhancing binding affinity to protein targets, cell permeability, and proteolytic stability.6 To further illustrate the usefulness of our macrocyclic peptides in the biological context, we decided to synthesize a macrocyclic analogue of a known p53 peptide inhibitor of MDM2 protein13 (pDI, 12a) and evaluate its properties with N-aryl linkages. Drawing from a recent study,6e peptide 12a (Figure 5a), featuring 12 amino acid residues, was modified with two lysines positioned in an i,i+7 fashion to give peptide 13a. To investigate the cell-penetrating properties of perfluoroaryl macrocycles, 13a was further reacted with 6 on 10 mg scale, affording the desired 14a in 71% yield after RPHPLC purification. Stability was assayed via incubation of 12a and 14a with chymotrypsin and proteinase K. Macrocyclic peptide 14a showed improved proteolytic stability compared to linear peptide 12a. For example, in 20 min of incubation with proteinase K, only 20% of 12a remained undigested, while 14a remained intact (Figure 5b and SI, Figures S9 and S10).
Figure 5.
N-Aryl p53 macrocyclic peptide retains binding capacity, is cell permeable, and is stable to proteolysis. (a) Derivatives of peptides 12, 13, 14, and 15. (b) Proteolytic stability assay. (c) Confocal microscopy imaging (cell membrane, red (WGA staining); FITC-labeled peptides, green) of HEK293T cells treated with peptides 12b, 13b, 14b, and 15 (10 μM). Left row, 63×, and right row, 126× magnification. (d) Flow cytometry analysis (cells were incubated with 10 μM for each peptide). Mean fluorescence was normalized against positive control 15. (e) Bilayer interferometry binding sensograms of immobilized 13c and 14c with 25 nM (green), 50 nM (purple), 100 nM (red), and 200 nM (blue) MDM2.
In order to evaluate cell uptake, we resorted to the synthesis of FITC-conjugated 12b, 13b, 14b and used the transportan cell-penetrating peptide 15 as a positive control (Figure 5a and SI, page S91). Cell uptake was determined by flow cytometry and confocal microscopy. Incubation of HEK-293T cells with the constructs followed by confocal imaging showed significant cell uptake for stapled 14b and positive control 15, whereas no intracellular signal was detected for unstapled 12b and 13b (Figure 5c and SI, Figures S11–S15). This trend was confirmed for concentrations ranging from 1 to 20 μM, while flow cytometry provided further support for cell uptake, with construct 14b showing a 5-fold increase compared to unstapled 12b and 13b (Figure 5d and SI, Figures S16–S18).
Previous reports demonstrated that rigidifying peptidic scaffolds may lead to improved binding affinities.6a,e,11a Therefore, to assess the effect of N-arylation on the binding to a protein target, we compared affinities of biotinylated 13c and 14c for MDM2 using an Octet RED96 bio-layer interferometry system (Figure 5e and SI, Figures S20 and S21). In our hands, stapled 14c displayed binding improvement (KD = 68 ± 1 nM) compared to linear 13c (KD = 151 ± 2 nM), while the biotinylated pDI 12c, which serves as a reference, falls in the same range as 14c (KD = 66 ± 2 nM).
In summary, we present the first lysine N-arylation of unprotected peptides and its application to macrocyclization. This methodology enables efficient access to a large variety of macrocyclized scaffolds under mild conditions using numerous electrophiles. DMF was used as the primary reaction solvent to solubilize both the unprotected peptide and electrophile. The use of organic solvent will hinder efforts to modify proteins in one step, and our future efforts are to design soluble electrophiles for N-arylation of proteins. Since there is much need for new macrocyclization methodologies that produce shelf- and solution-stable constructs, our approach expands this chemical toolkit and addresses the possible chemical stability issues with cysteine arylation. Building on these findings, we evolved a known MDM2 inhibitor into a perfluoroaromatic N-arylated macrocyclic peptide that displayed many desirable characteristics that may prompt additional cancer studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Stephen L. Buchwald for encouragement and support. This work was supported by an MIT start-up fund, the National Institutes of Health (NIH R01GM110535), and the Sontag Foundation Distinguished Scientist Award (to B.L.P.). This work was partially funded by Human Frontier Science Program (HFSP, F.T.). The authors acknowledge the Biological Instrument Facility of MIT for providing the Octet BioLayer Interferometry System (NIH S10OD016326) and the Koch Institute for providing the flow cytometer. We thank Dr. Amy Rabideau for MDM2 protein expression, Colin Fadzen for help using the FACS instrument, and Chi Zhang and Ethan Evans for technical assistance and fruitful discussions throughout the course of the work.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b03757.
Full experimental and characterization details (PDF)
REFERENCES
- (1).(a) Hili R; Yudin AK Nat. Chem. Biol. 2006, 2, 284. [DOI] [PubMed] [Google Scholar]; (b) Fischer C; Koenig B Beilstein J. Org. Chem. 2011, 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bariwal J; Van der Eycken EV Chem. Soc. Rev. 2013, 42, 9283. [DOI] [PubMed] [Google Scholar]
- (2).(a) Cano R; Ramon DJ; Yus M J. Org. Chem. 2011, 76, 654. [DOI] [PubMed] [Google Scholar]; (b) Diness F; Fairlie DP Angew. Chem., Int. Ed. 2012, 51, 8012. [DOI] [PubMed] [Google Scholar]
- (3).(a) Yang BH; Buchwald SL J. Organomet. Chem. 1999, 576, 125. [Google Scholar]; (b) Beletskaya I; Cheprakov A Coord. Chem. Rev. 2004, 248, 2337. [Google Scholar]
- (4).(a) Spokoyny AM; Zou Y; Ling JJ; Yu H; Lin YS; Pentelute BL J. Am. Chem. Soc. 2013, 135, 5946. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brown S; Smith AJ Am. Chem. Soc. 2015, 137, 4034. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Vinogradova EV; Zhang C; Spokoyny AM; Pentelute BL; Buchwald SL Nature 2015, 526, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kalhor-Monfared S; Jafari MR; Patterson JT; Kitov PI; Dwyer JJ; Nuss JM; Derda R Chem. Sci. 2016, 7, 3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lau YH; de Andrade P; Wu Y; Spring DR Chem. Soc. Rev. 2015, 44, 91. [DOI] [PubMed] [Google Scholar]
- (6).(a) Walensky LD; Kung AL; Escher I; Malia TJ; Barbuto S; Wright RD; Wagner G; Verdine GL; Korsmeyer SJ Science 2004, 305, 1466. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Walensky LD; Pitter K; Morash J; Oh KJ; Barbuto S; Fisher J; Smith E; Verdine GL; Korsmeyer SJ Mol. Cell 2006, 24, 199. [DOI] [PubMed] [Google Scholar]; (c) Gavathiotis E; Reyna DE; Davis ML; Bird GH; Walensky LD Mol. Cell 2010, 40, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Grossmann TN; Yeh JTH; Bowman BR; Chu Q; Moellering RE; Verdine GL Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 17942. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Chang YS; Graves B; Guerlavais V; Tovar C; Packman K; To KH; Olson KA; Kesavan K; Gangurde P; Mukherjee A; Baker T; Darlak K; Elkin C; Filipovic Z; Qureshi FZ; Cai H; Berry P; Feyfant E; Shi XE; Horstick J; Annis DA; Manning AM; Fotouhi N; Nash H; Vassilev LT; Sawyer TK Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E3445. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Walensky LD; Bird GH J. Med. Chem. 2014, 57, 6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).(a) Blackwell HE; Grubbs RH Angew. Chem., Int. Ed. 1998, 37, 3281. [DOI] [PubMed] [Google Scholar]; (b) Schafmeister CE; Po J; Verdine GL J. Am. Chem. Soc. 2000, 122, 5891. [Google Scholar]; (c) Verdine G; Hilinski G Methods Enzymol. 2012, 503, 3. [DOI] [PubMed] [Google Scholar]
- (8).(a) Felix AM; Heimer EP; Wang GT; Lambros TJ; Fournier AJ; Mowles TF; Maines S; Campbell RM; Wegrzynski BB; Toome V; Fry D; Madison VS Int. J. Pept. Protein Res. 1988, 32, 441. [DOI] [PubMed] [Google Scholar]; (b) Taylor JW Biopolymers 2002, 66, 49. [DOI] [PubMed] [Google Scholar]; (c) de Araujo AD; Hoang HN; Kok WM; Diness F; Gupta P; Hill TA; Driver RW; Price DA; Liras S; Fairlie DP Angew. Chem., Int. Ed. 2014, 53, 6965. [DOI] [PubMed] [Google Scholar]
- (9).(a) Scrima M; Le Chevalier-Isaad A; Rovero P; Papini AM; Chorev M; D’Ursi AM Eur. J. Org. Chem. 2010, 2010, 446. [Google Scholar]; (b) Lau Y; de Andrade P; Quah S-T; Rossmann M; Laraia L; Skold N; Sum T; Rowling P; Joseph T; Verma C; Hyvonen M; Itzhaki L; Venkitaraman A; Brown C; Lane D; Spring D Chem. Sci. 2014, 5, 1804. [Google Scholar]
- (10).Jackson D; King D; Chmielewski J; Singh S; Schultz PJ Am. Chem. Soc. 1991, 113, 9391. [Google Scholar]
- (11).(a) Muppidi A; Doi K; Edwardraja S; Drake E; Gulick A; Wang H; Lin Q J. Am. Chem. Soc. 2012, 134, 14734. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jo H; Meinhardt N; Wu Y; Kulkarni S; Hu Z; Low KE; Davies PL; DeGrado WF; Greenbaum DC J. Am. Chem. Soc. 2012, 134, 17704. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wang Y; Chou D Angew. Chem., Int. Ed. 2015, 54, 10931. [DOI] [PubMed] [Google Scholar]; (d) Assem N; Ferreira D; Wolan D; Dawson P Angew. Chem., Int. Ed. 2015, 54, 8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).(a) Baertschi SW; Jansen PJ; Alsante KM In Pharmaceutical Stress Testing:Predicting Drug Degradation, 2nd ed.;, Alsante KM, Reed RA, Eds.; Informa Healthcare: London, 2011. [Google Scholar]; (b) Hovorka SW; Schoneich CJ Pharm. Sci. 2001, 90, 253. [DOI] [PubMed] [Google Scholar]; (c) Shen BQ; Xu K; Liu L; Raab H; Bhakta S; Kenrick M; Parsons-Reponte KL; Tien J; Yu SF; Mai E; Li D; Tibbitts J; Baudys J; Saad OM; Scales SJ; McDonald PJ; Hass PE; Eigenbrot C; Nguyen T; Solis WA; Fuji RN; Flagella KM; Patel D; Spencer SD; Khawli LA; Ebens A; Wong WL; Vandlen R; Kaur S; Sliwkowski MX; Scheller RH; Polakis P; Junutula JR Nat. Biotechnol. 2012, 30, 184. [DOI] [PubMed] [Google Scholar]; (d) Toda N; Asano S; Barbas CF Angew. Chem., Int. Ed. 2013, 52, 12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Phan J; Li Z; Kasprzak A; Li B; Sebti S; Guida W; Schonbrunn E; Chen JJ Biol. Chem. 2010, 285, 2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fahey DR; Ash CE Macromolecules 1991, 24, 4242. [Google Scholar]
- (15).(a) Blotny G Tetrahedron 2006, 62, 9507. [Google Scholar]; (b) Shannon DA; Banerjee R; Webster ER; Bak DW; Wang C; Weerapana EJ Am. Chem. Soc. 2014, 136, 3330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.