Abstract
BACKGROUND
Primary microcephaly (MCPH) is a rare developmental defect characterized by impaired cognitive functions, retarded neurodevelopment and reduced brain size. It is genetically heterogeneous and more than 17 genes so far have been identified that are associated with this disease.
OBJECTIVE
To study the genetic defect in a consanguineous Saudi family with primary microcephaly.
DESIGN
Cross-sectional clinical genetics study of a Saudi family.
SETTING
Medical genomics research center.
PATIENTS AND METHODS
Blood samples collected from six members of a family of healthy consanguineous parents were analyzed by whole exome sequencing to identify the underlying pathogenic mutations in two members of the family (23-year-old female and 7-year-old male) who presented with primary microcephaly, intellectual disability, delayed psychomotor development and walking difficulty, speech impediments and seizures.
MAIN OUTCOME MEASURE(S)
Detection of mutation in the WD repeat domain 62 (WDR62) gene in a family segregating autosomal recessive primary microcephaly.
RESULTS
The exome variant analysis identified a novel missense mutation (c.3878C>A) in WDR62 gene in exon 30 resulting in amino acid change from alanine to aspartate (p.Ala1293Asp). Further validation in the affected patients and healthy members of family and 100 unrelated healthy persons as controls confirmed it to be pathogenic.
CONCLUSIONS
Functional impairment of the WDR62 gene can lead to severe neurodevelopmental defects, brain malformations and reduced head size. A missense mutation of exon 30 changed alanine to aspartate in the WDR62 protein leading to the typical MCPH phenotype.
LIMITATIONS
Mutation was identified in a single family.
Microcephaly (MCPH) is an abnormally small head size. There are two sub categories: primary microcephaly is present at birth and is a static developmental anomaly; secondary microcephaly develops later in life (postnatally) and is a progressive neurodegenerative condition. Primary microcephaly is a congenital condition associated with incomplete brain development, where a baby’s head is smaller than expected when compared to babies of the same sex and age. It serves as an important neurological indication or warning sign, but no uniformity exists in its definition. It is usually defined as a head circumference (HC) more than four standard deviations (SD) below the mean for age and sex.1
Primary microcephaly (MCPH) is very rare; the rate is estimated to be 1 in 30 000 in Japan2 and 1 in 250 000 in Holland.3 This rate increases to 1 in 10 000 in regions where interfamily and cousin marriages are common.4 The birth incidence varies from 1.3 to 150 per 100 000 persons, depending on various factors, mainly population type and consanguinity.5
So far, 17 genes have been identified that underlie autosomal recessive primary microcephaly. These include microcephalin at MCPH1,6 WDR62 at MCPH2,7–9 CDK5RAP2 at MCPH3,10 CASC5 at MCPH4,11 ASPM at MCPH5,12 CENPJ at MCPH6,12 STIL at MCPH7,13 CEP135 at MCPH8,14 CEP152 at MCPH9,11,15 ZNF335 at MCPH10,16 PHC1 at MCPH11,17 CDK6 at MCPH 12,18 CENPR at MCPH13,19 SASS6 at MCPH14,20 MFSD2A at MCPH15,21 ANKLE2 at MCPH16,22 CIT at MCPH17.23–25 Furthermore, recently identified syndromic microcephaly AGMO, RTTN and PGAP2 genes have been reported in the Saudi population.26–28
However, the majority of mutations have been identified in two genes: ASPM, accounting for more than half of all mutations and WDR62, which accounts for around 10% of all cases.29 All of the WDR62 mutation cases presented up to now have shown the presence of mental retardation and also the presence of prominent microcephaly on physical examination, while some of the patients also suffered from seizures. The examination of the brain of the patients under high field strength (3 Tesla) magnetic resonance imaging (MRI) have identified hallmarks of a wide range of severe cortical malformations.8
In our study, we have ascertained the presence of a novel mutation in a consanguineous family from Saudi origin. There were two affected siblings born to a consanguineous union in this family. Whole exome sequencing was performed to identify the underlying genetic cause because of the potential of this technology in the molecular diagnostics of similar genetic disorders.30 We identified a novel mutation in the WDR62 gene in these Saudi patients.
PATIENTS AND METHODS
Sample collections
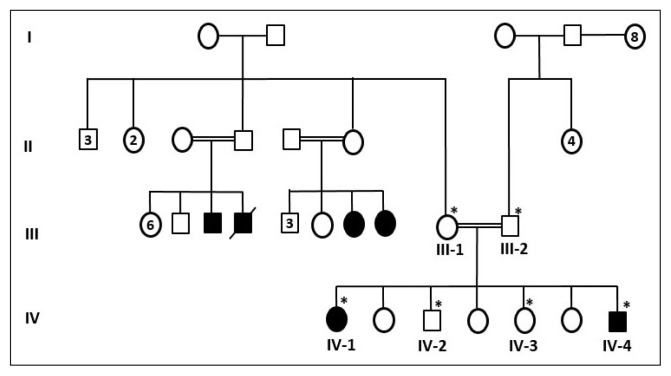
The pedigree (family chart) of the family was drawn by obtaining information from elders of the family (Figure 1). Pedigree construction suggested an autosomal recessive pattern of inheritance (Figure 1). Before the study was initiated, written informed consent was taken from all participants of the study. The study was also approved from ethical committee of the Center of Excellence in Genomic Medicine Research, King Abdulaziz University, Jeddah (013-CEGMR-2-ETH). The blood samples were collected from six members of the family (two affected and four normal individuals) and one hundred unrelated healthy people of Saudi origin as controls. Both the affected individuals underwent medical examination at King Abdulaziz University Hospital, Jeddah. We ruled out possible environmental factors and infections such as rubella, toxoplasma, cytomegalovirus, and Zika virus that may lead to microcephaly.
Figure 1.
A pedigree of a consanguineous family from Saudi Arabia showing the disease phenotype segregating in an autosomal recessive manner. The samples available for genetic testing are marked with asterisks.
Patient 1
Proband (IV-1) was a 23-year-old female who presented with MCPH and suffered from mental retardation. She was the first child of consanguineous healthy parents (Figure 1). She had a normal weight and height, but could not walk properly, had speech problems, had brain atrophy and reported seizures. Her head circumference was less than 5 standard deviations below the mean.
Patient 2
Proband (IV-4) was a 7-year-old male who presented with MCPH and suffered from mental retardation. This individual was born after five normal offspring as shown in the pedigree (Figure 1). He was not able to move due to muscular dystrophy, could not speak properly, and had a history of seizures. The head circumference was less than 5 standard deviations below the mean.
Whole exome sequencing
To identify the underlying pathogenic mutation behind this disease phenotype we planned whole exome sequencing (HiSeq 2500 System, Illumina, San Diego, CA, United States). Extraction of genomic DNA (gDNA) was performed using standard procedures. Briefly, the blood sample was used for direct gDNA isolation using the QIAamp DNA Blood Mini Kit, Cat. Nr. 51106 (Qiagen, Hilden, Germany) following manufacturer’s instructions and modified where necessary. The gDNA was analyzed by the Bioanalyzer system (Agilent Technologies, Santa Clara, CA, United States) and quantified by a NanoDrop Spectrophotometer and the genomic DNA was stored under appropriate conditions for future analysis.
The samples were prepared according to an Agilent SureSelect Target Enrichment Kit preparation guide (Capture kit, SureSelect_v6) by using genomic DNA directly. The genomic DNA libraries were sequenced using the Illumina HiSeq 2000/2500 sequencer. The resulting VCF (variant call format) file contained 89064 variants. These variants were filtered based on quality, frequency, genomic position, protein effect, pathogenicity and previous associations with the phenotype.
Sanger sequencing
To confirm the mutation in patients and family, we did Sanger sequencing (ABI 3700). To confirm the mutation as pathogenic, we also sequenced this DNA variant in 100 unrelated control people.
RESULTS
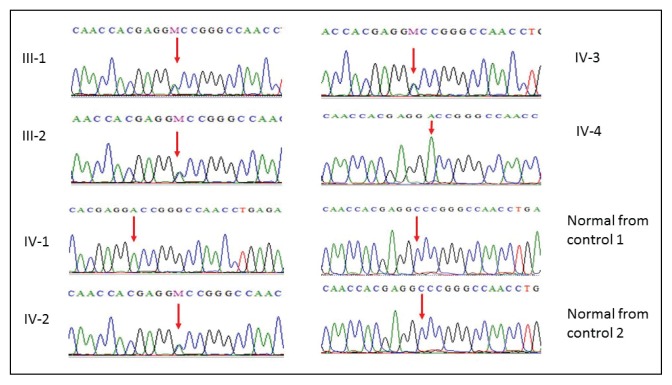
Candidate variants were first searched in a broad panel of genes which had been previously associated with microcephaly, intellectual disability, and one of the reported features (in human and model organisms). The exome variant analysis yielded a plausible candidate variant in the WDR62 gene where cytosine (C) at position 3878 is replaced by adenine (A), resulting in conversion of amino acid alanine at position 1293 to aspartate, thus showing a novel missense mutation 3878C>A, at exon 30 in affected members of the family as shown in Figure 2. Furthermore, in the Greater Middle East variome, the minor allele frequency was 0.0005 and there was single heterozygous and no homozygous individuals found in the database. Moreover, PolyPhen, MutationTaster and SIFT predicted this disease-causing mutation. This mutation was absent in the Human Gene Mutation database (HGMD, www.hgmd.cf.ac.uk/) and MIM. 1000 genome (http://www.internationalgenome.org/) and Exome Aggregation Consortium (http://exac.broadinstitute.org/) database.
Figure 2.
Sanger sequence analysis: mother III-1, father III-2, IV-2, IV-3 are normal parents showing C and A in heterozygous state, while IV-1 and IV-4 are affected children showing only homozygous A in exon 30 of WDR62 gene.
Other mutations in this gene are known to cause primary autosomal recessive microcephaly type 2, with or without cortical malformations (MCPH2). This disorder shows phenotypic overlap with the symptoms reported for this individual. Hence, this variant was considered as a plausible candidate, but needed further investigation to validate its clinical significance.
To check the other mutations elsewhere in the whole exomes, we also broadened the analysis to all genes applying all inheritance modes. However, no additional potential candidate variant that might be of relevance for the reported phenotype could be identified. To confirm this novel mutation in WDR62 gene, further validation was carried out in all available affected patients and healthy members of family and 100 unrelated healthy persons as controls. This mutation was not detected in any healthy individual, which confirmed it to be pathogenic. The parents of the affected members were heterozygous, which also confirms the autosomal recessive mode of inheritance.
DISCUSSION
To date about 25 mutations have been identified in WDR62 gene (Table 1). In our family, a rare, homozygous missense variant was detected in the WDR62 gene in patients in a homozygous state. This gene encodes a protein which is required for proper neurogenesis and cerebral cortical development.31 It is proposed to play a role in neuronal proliferation and migration.7,8 The expression of WDR62 was found to be widespread in the developing mouse brain, with highest expression in the forebrain.7 Mechanistically, WDR62 associates and genetically interacts with Aurora A to regulate spindle formation, mitotic progression and brain size. It is also reported that WDR62 interacts with Aurora A to control mitotic progression, and loss of these interactions leads to mitotic delay and cell death of neural progenitor cells (NPCs) which could be a potential cause of human microcephaly.32 Mutations in this gene have been associated with microcephaly 2, primary, autosomal recessive, with or without cortical malformations (MCPH2).7–9,33–36 This is a disease characterized by microcephaly associated with other manifestations and shows wide phenotypic variability.37 Associated features include moderate to severe mental retardation, and various types of cortical malformations in most patients. Cortical malformations may include pachygyria with cortical thickening, microgyria, lissencephaly, hypoplasia of the corpus callosum, and schizencephaly. All affected individuals have delayed psychomotor development. Some patients have seizures.
Table 1.
Mutation spectrum of WDR62 gene mutations.
| S. No | Nucleotide variation | Amino acid variation | Location | Reference |
|---|---|---|---|---|
|
| ||||
| 1 | c.28G>T | p.Ala10Ser | Exon 1 | 41 |
| 2 | c.189G>T | p.Glu63Asp | Exon 2 | 41 |
| 3 | c.193 G>A | p.Val65Met | Exon 2 | 7, 9 |
| 4 | c.332G>C | p.Arg111Thr | Exon 3 | 19, 29 |
| 5 | c.363delT | p.Asp112MetfsX5 | Exon 4 | 7 |
| 6 | c.535_536insA | p.Met179fsX21 | Exon 5 | 33 |
| 7 | c.671 G>C | p.Trp224Ser | Exon 6 | 8 |
| 8 | c.900C>A | p.Cys300X | Exon 8 | 33 |
| 9 | c.1043+1 G>A | p.Ser348RfsX63 | Intron 8 | 7 |
| 10 | c.1143delA | p.H381PfsX48 | Exon 9 | 42 |
| 11 | c.1194G>A | p.Trp398 | Exon 9 | 18, 29 |
| 12 | c.1313 G>A | p.Arg438His | Exon 10 | 9 |
| 13 | c.1408C>T | p.Gln470X | Exon 11 | 8 |
| 14 | c.1531 G>A | p.Asp511Asn | Exon 11 | 9 |
| 15 | c.1576 G>T | p.Glu526X | Exon 12 | 8 |
| 16 | c.1576 G>A | p.Glu526Lys | Exon 12 | 8 |
| 17 | c.1942 C>T | p.Gln648X | Exon 15 | 43 |
| 18 | c.2867+4_c2867+7delGGTG | p.Ser956CysfsX38 | Intron 23 | 7 |
| 19 | c.3232 G>A | p.Ala1078Thr | Exon 27 | 9 |
| 20 | c.3361delG | p.Ala1121Glnfs*6 | Exon 28 | 18, 29 |
| 21 | c.3503G>A | p.Trp1168* | Exon 29 | 18, 29 |
| 22 | c.3839_3855delGCCAAGAGCCTGCCCTG | p.Gly1280AlafsX21 | Exon 30 | 7, 8 |
| 23 | c.3878C>A | p.Ala1293Asp | Exon 30 | This study |
| 24 | c.3936dupC/3936_3937incC | p.Val1314ArgfsX18/Val1314GlyfsX17 | Exon 30 | 7, 9 |
| 25 | c.4205delTGCC | p.Val1402GlyfsX12 | Exon 31 | 8 |
| 26 | c.4241dupT | p.Leu1414LeufsX41 | Exon 31 | 9 |
The variant detected here is a substitution that affects a highly conserved alanine residue that is the last amino acid of the last WD domain (WD stands for tryptophan-aspartic acid dipeptide). The underlying common function of all WD-repeat proteins is coordinating multi-protein complex assemblies, where the repeating units serve as a rigid scaffold for protein interactions. This mutation was not previously reported. Previously reported pathogenic mutations include missense (e.g. E526K; W224S; R438H) and truncating mutations (e.g. Val1402GlyfsTer12; Q470X; Gly1280AlafsTer21; 2083delA; 2472_2473delAG; c.390G>A; c.2527dupG; p.R438H; p.D955Afs*112).8,9,34,35,38–40 The frameshift mutations reported by Murdock et al were reported to cause nonsense-mediated mRNA decay and loss of function.34 However, the effect of the missense variant detected here remains to be elucidated. Interestingly, this variant is predicted to be deleterious by the majority of in silico prediction tools. This variant is absent from population databases such as 1000 Genomes Project and the Exome Aggregation Consortium. The variant was tested for likely pathogenic effect by in silico tools like MutationTaster,44 SIFT and Polyphen, and it was predicted as “disease causing” with high pathogenicity scores (Table 2). Further functional analysis is needed to determine the functional effect of this variant and its possible contribution to the reported phenotype.
Table 2.
In silico tools used for prediction of pathogenicity of missense variants.
| S. No | Online tools | Pathogenicity score |
|---|---|---|
|
| ||
| 1 | Ensembl (http://www.ensembl.org, ENST00000401500) | 0.0 |
| 2 | SIFT (http://sift.jcvi.org/) | 1.0 |
| 3 | 1000 Genomes (http://www.internationalgenome.org/) |
0.0 |
| 4 | Exome Aggregation Consortium (http://exac.broadinstitute.org/) |
0.0 |
| 5 | Polyphen -2 (http://genetics.bwh.harvard.edu/pph2/) | 0.99 |
| 6 | MutationTaster (http://www.mutationtaster.org/) | 1.1 |
| 7 | MutationAssessor | 0.93 |
| 8 | PhyloP (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702902/) | 0.76 |
| 9 | GERP++ (http://mendel.stanford.edu/SidowLab/downloads/gerp/) | 0.97 |
Footnotes
Conflict of interest
The authors declare no conflict of interest.
REFERENCES
- 1.Faheem M, Naseer MI, Rasool M, Chaudhary AG, Kumosani TA, Ilyas AM, et al. Molecular genetics of human primary microcephaly: An overview. BMC Med Genomics. 2015;8(1):54. doi: 10.1186/1755-8794-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komai T, Kishimoto K, Ozaki Y. Genetic study of microcephaly based on Japanese material. Am J Hum Gen. 1955;7:51–65. [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch JVD. Microcephaly in the Netherlands: a clinical and genetical study. Ann Hum Gen. 1959;23:91–116. doi: 10.1111/j.1469-1809.1958.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 4.Cox J, Jakson AP, Bond J, Woods CG. What primary microcephaly can tell us about brain growth. Trends Mol Med. 2006;12:358–366. doi: 10.1016/j.molmed.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Kaindl AM, Passemard S, Kumar P, Kraemer N, Issa L, Zwirner A, et al. Many roads lead to primary autosomal recessive microcephaly. Prog Neurobiol. 2010;90:363–383. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, et al. Mutations in WDR62, encoding a centrosome associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture, Nat. Genet. 2014;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilgüvar K, Öztürk AK, Louvi A, Kwan KY, Choi M, Tatl B, et al. PamirWhole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–10. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholas AK, Khurshid M, Désir J, Carvalho OP, Cox JJ, Thornton G. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bond J, Robertsm E, Springell K, Lizarraga SB, Scott S, Higgins J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 11.Genin A, Desir J, Lambert N, Biervliet M, Van DA, Pierquin G, et al. Inetochore KMN network gene CASC5 mutated in primary microcephaly. Hum Mol Genet. 2012;21:5306–5317. doi: 10.1093/hmg/dds386. [DOI] [PubMed] [Google Scholar]
- 12.Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2001;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 13.Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–290. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain MS, Baig SM, Neumann S, Nurnberg G, Farooq M, Ahmed A, et al. A truncating mutation of CEP135 is associated with primary microcephaly and disturbed centrosomal function. Am J Hum Genet. 2012;90:871–878. doi: 10.1016/j.ajhg.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang YJ, Baltus AE, Mathew RS, Murphy EA, Evrony GD, Gonzalez DM, et al. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell. 2012;15:11097–1112. doi: 10.1016/j.cell.2012.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Awad S, Al-Dosari MS, Al-Yacoub N, Colak D, Salih MA, Alkuraya FS, Poizat C. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum Mol Genet. 2013;22:2200–2213. doi: 10.1093/hmg/ddt072. [DOI] [PubMed] [Google Scholar]
- 18.Hussain MS, Baig SM, Neumann S, Peche VS, Szczepanskim S, Nurnberg G, Tariq M, et al. CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly. Hum Mol Genet. 2013;22:5199–214. doi: 10.1093/hmg/ddt374. [DOI] [PubMed] [Google Scholar]
- 19.Mirzaa GM, Vitre B, Carpenter G, Abramowicz I, Gleeson JG, Paciorkowski AR, et al. Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum Genet. 2014;133:1023–1039. doi: 10.1007/s00439-014-1443-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan MA, Rupp VM, Orpinell M, Hussain MS, Altmuller J, Steinmetz MO, et al. A missense mutation in the PISA domain of HsSAS-6 causes autosomal recessive primary microcephaly in a large consanguineous Pakistani family. Hum Mol Genet. 2014;23:5940–5949. doi: 10.1093/hmg/ddu318. [DOI] [PubMed] [Google Scholar]
- 21.Alakbarzade V, Hameed A, Quek DQY, Chioza BA, Baple EL, Cazenave-Gassiot A, et al. A partially inactivating mutation in the sodium-dependent lysophosphatidylcholine transporter MFSD2A causes a non-lethal microcephaly syndrome. Nat Genet. 2015;47:814–817. doi: 10.1038/ng.3313. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaheen R, Hashem A, Abdel-Salam GMH, Al-Fadhli F, Ewida N, Alkuraya FS. Mutations in CIT, encoding citron rho-interacting serine/threonine kinase, cause severe primary microcephaly in humans. Hum Genet. 2016;135:1191–1197. doi: 10.1007/s00439-016-1722-2. [DOI] [PubMed] [Google Scholar]
- 24.Sulman B, Khalid M, Al-Harbi KM, Sabri AM, Alhijji SA, Alia M, et al. CIT, a gene involved in neurogenic cytokinesis, is mutated in human primary microcephaly. Hum Genet. 2016;135:1199–1207. doi: 10.1007/s00439-016-1724-0. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Bielas SL, Zaki MS, Ismail S, Farfara D, Um K, et al. Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am J Hum Genet. 2016;99:501–510. doi: 10.1016/j.ajhg.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alrayes N, Mohamoud HAS, Ahmed S, Almramhi MM, Shuaib TM, Wang J, et al. The alkylglycerol monooxygenase (AGMO) gene previously involved in autism also causes a novel syndromic form of primary microcephaly in a consanguineous Saudi family. J Neurol Sci. 2016;363:240–244. doi: 10.1016/j.jns.2016.02.063. [DOI] [PubMed] [Google Scholar]
- 27.Naseer MI, Rasool M, Jan MM, Chaudhary AG, Pushparaj PN, Abuzenadah AM, Al-Qahtani MH. A novel mutation in PGAP2 gene causes developmental delay, intellectual disability, epilepsy and microcephaly in consanguineous Saudi family. J Neurol Sci. 2016;371:121–125. doi: 10.1016/j.jns.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 28.Shamseldin H, Alazami AM, Manning M, Hashem A, Caluseiu O, Tabarki B, et al. RTTN Mutations Cause Primary Microcephaly and Primordial Dwarfism in Humans. Am J Hum Genet. 2015;97:862–868. doi: 10.1016/j.ajhg.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain MS, Bakhtiar MS, Farooq M, Anjum I, Janzend E, Toliat MR, et al. Genetic heterogeneity in Pakistani microcephaly families. Clin Genet. 2013;83:446–451. doi: 10.1111/j.1399-0004.2012.01932.x. [DOI] [PubMed] [Google Scholar]
- 30.Ng SB, Buckingham KJ, Lee1 C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, Zhang F, Wang Y, Sun Y, Xu Z. Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Rep. 2014;6:104–116. doi: 10.1016/j.celrep.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Chen JF, Zhang Y, Wilde J, Hansen KC, Lai F, Niswander L. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat communicat. 2014;3885:1–13. doi: 10.1038/ncomms4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat V, Girimaji SC, Mohan G, Arvinda HR, Singhmar P, Duvvari MR, Kumar A. Mutations in WDR62, encoding a centrosomal and nuclear protein, in Indian primary microcephaly families with cortical malformations. Clin Genet. 2011;80:532–540. doi: 10.1111/j.1399-0004.2011.01686.x. [DOI] [PubMed] [Google Scholar]
- 34.Murdock DR, Clark GD, Bainbridge MN, Newsham I, Wu YQ, Muzny DM, et al. Whole-exome sequencing identifies compound heterozygous mutations in WDR62 in siblings with recurrent polymicrogyria. Am J Med Genet Part A 155A. 2011;155(9):2071–2077. doi: 10.1002/ajmg.a.34165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts E, Jackson AP, Carradice AC, Deeble VJ, Mannan J, Rashid Y, et al. The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1–13.2. Eur J Hum Genet. 1999;7:815–820. doi: 10.1038/sj.ejhg.5200385. [DOI] [PubMed] [Google Scholar]
- 36.Stark AE. The genetic epidemiology of the form of microcephaly ascribed to mutation at the WDR62 locus. Ann Trans Med. 2016;4:1–7. doi: 10.21037/atm.2016.07.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulton CJ, Schot R, Seufert K, Lequin MH, Accogli A, Annunzio GD, et al. Severe presentation of WDR62 mutation: is there a role for modifying genetic factors? Am J Med Genet Part A. 2014;164:2161–2171. doi: 10.1002/ajmg.a.36611. [DOI] [PubMed] [Google Scholar]
- 38.Bastaki F, Mohamed M, Nair P, Saif F, Tawfiq N, Aithala G, et al. Novel splice-site mutation in WDR62 revealed by whole-exome sequencing in a Sudanese family with primary microcephaly. Congenit Anom. 2016;56:135–137. doi: 10.1111/cga.12144. [DOI] [PubMed] [Google Scholar]
- 39.Rupp V, Rauf S, Naveed I, Windpassinger C, Mir A. A novel single base pair duplication in WDR62 causes primary microcephaly. BMC Med Genet. 2014;15:1–6. doi: 10.1186/s12881-014-0107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farag HG, Froehler S, Oexle K, Ravindran E, Schindler D, Staab T, et al. Abnormal centrosome and spindle morphology in a patient with autosomal recessive primary microcephaly type 2 due to compound heterozygous WDR62 gene mutation. Orphanet J Rare Dis. 2013;8:1–14. doi: 10.1186/1750-1172-8-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerjee S, Chen H, Huang H, Wu J, Yang Z, Deng W, et al. Novel mutations c.28G>T (p.Ala10Ser) and c.189G>T (p.Glu63Asp) in WDR62 associated with early onset acanthosis and hyperkeratosis in a patient with autosomal recessive microcephaly type 2. Oncotarget. 2016:7–48. doi: 10.18632/oncotarget.13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Memon MM, Raza SI, Basit S, Kousar R, Ahmad W, Ansar M. A novel WDR62 mutation causes primary microcephaly in a Pakistani family. Mol Biol Rep. 2015;40:591–595. doi: 10.1007/s11033-012-2097-7. [DOI] [PubMed] [Google Scholar]
- 43.Kousar R, Hassan MJ, Khan B, Basit S, Mahmood S, Mir A, et al. Mutations in WDR62 gene in Pakistani families with autosomal recessive primary microcephaly. BMC Neurol. 2011;11:1–6. doi: 10.1186/1471-2377-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]