Summary
In the auditory system, Type I spiral ganglion neurons (SGNs) convey complex acoustic information from inner hair cells (IHCs) to the brainstem. Although SGNs exhibit variation in physiological and anatomical properties, it is unclear which features are endogenous and which reflect input from synaptic partners. Using single-cell RNA sequencing, we derived a molecular classification of mouse Type I SGNs into three subtypes that express unique combinations of Ca2+ binding proteins, ion channel regulators, guidance molecules, and transcription factors. Based on connectivity and susceptibility to age-related loss, these subtypes correspond to those defined physiologically. Additional intrinsic differences among subtypes and across the tonotopic axis highlight an unexpectedly active role for SGNs in auditory processing. SGN identities emerge postnatally and are disrupted in a mouse model of deafness that lacks IHC-driven activity. These results elucidate the range, nature, and origins of SGN diversity, with implications for treatment of congenital deafness.
Summary
Single cell analyses of mouse Type l spiral ganglion neurons characterize three functionally distinct subtype, reveling insights into auditory processing with implication for treating congenital deafness.
eTOC
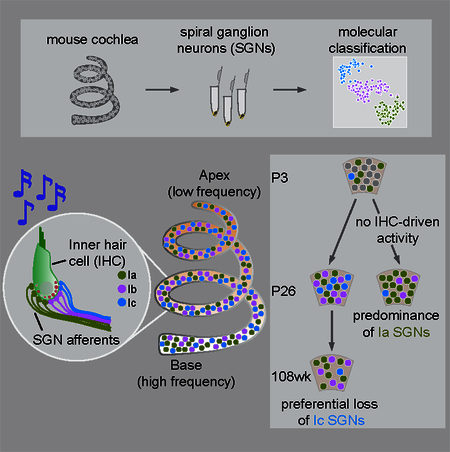
Using single cell RNA-sequencing, Shrestha and colleagues define three subtypes of inner ear neurons with differential expression of physiologically relevant molecules across subtypes and tonotopically. SGN diversification begins postnatally and requires IHC-driven activity, with implications for age-related and congenital deafness.
Introduction
Heterogeneity is a hallmark of the nervous system, with distinct types of neurons forming networks that subserve specific functions. Defining their cellular components is a necessary step toward understanding such networks. Across circuits with clearly different functional outputs, assignment of cell identity is relatively straightforward. For instance, in the somatosensory system, neurons that encode pain and light touch are readily distinguished by the identities of their peripheral end organs and morphologies of their endings (Abraira and Ginty, 2013). However, classification of neurons that share the same position in a circuit, perform similar function, and underlie a single modality can be challenging. For instance, the primary sensory neurons of the auditory system, the Type I spiral ganglion neurons (SGNs), are all bipolar and make connections with the same sensory cell type, consistent with their shared ability to encode sound. Their key differences are physiological, which could be imposed by other cells in the circuit. Whether Type I SGNs also exhibit endogenous differences that might contribute to auditory function has remained elusive.
Type I SGNs differ in sensitivity to sound and spontaneous firing rate (SR), as revealed over 50 years ago via single-fiber recordings in the cat auditory nerve (Kiang et al., 1965). Based on the relation between threshold and SR, they were classified into 3 groups: low-SR, medium-SR and high-SR (Liberman, 1978). These three subtypes are present in all regions of the cochlea and in many species (Barbary, 1991; Borg et al., 1988; Schmiedt, 1989; Taberner and Liberman, 2005; Winter et al., 1990). SGNs with different SRs form synapses at different positions along the basolateral surface of inner hair cells (IHCs) (Liberman, 1982) and project to different cell types in the brainstem. Such diversity enables the wide dynamic range of sound intensities encoded in the cochlea and helps maintain hearing in noisy environments (Costalupes et al., 1984; Liberman, 2017; Winter et al., 1990). In addition, immature SGNs display different firing properties in the base versus apex of the cochlea, which encode high and low sound frequencies, respectively (Mann and Kelley, 2011). Whether mature SGNs retain these differences and how they vary among SR subtypes is not known.
Because Type I SGNs have been classified physiologically, the nature and origin of their underlying heterogeneity is unclear. At one extreme, there may be one basic type of SGN that exhibits different firing properties because of the input received from its pre-synaptic partners. Indeed, both heterogeneity in presynaptic vesicle release probability in IHCs (Frank et al., 2009) and regulation by olivocochlear efferents (Guinan, 2017; Liberman, 1980) have been suggested to contribute to SGN physiological diversity. Alternatively, SGNs may express different collections of receptors and ion channel regulators that shape their sensitivity and SR. Indeed, hints of molecular heterogeneity have been noted (Adamson et al., 2002; Chen et al., 2011; Liu and Davis, 2014; Liu et al., 2014), although it was not possible to link any one marker with known electrophysiological signatures. Without a comprehensive understanding of SGN molecular profiles, the ways SGNs actively influence auditory coding remain to be defined.
The lack of knowledge of subtype-specific molecular signatures represents a major barrier in understanding SGN biology and function, with direct implications for human health. Low-SR SGNs are lost after noise damage (Furman et al., 2013) and as a consequence of aging (Schmiedt et al., 1996), which may explain the difficulties understanding speech amid background noise in older individuals. However, in the absence of subtype-specific molecular tools, efforts to assign functions to individual subtypes and to understand the mechanisms underlying selective vulnerability have been limited. Similarly, since SGN subtypes do not exhibit mature physiological properties until the end of the first month in mice (Liberman and Liberman, 2016), the factors that give rise to their diverse properties during development remain elusive, despite the potential for SGN pathology in congenital deafness.
Single cell transcriptome-based approaches offer a powerful way to investigate heterogeneity in neuronal populations. To characterize neuronal diversity in the cochlea, we generated high-resolution transcriptomic profiles of individual SGNs. We found three molecularly distinct subtypes (Ia, Ib, Ic) that match the anatomical features of high-, medium- and low-SR SGNs, with additional variation along the tonotopic axis. SGN identities emerge gradually in development and this diversification requires IHC-driven activity. This comprehensive view of SGN heterogeneity uncovers endogenous differences that could directly affect auditory coding and highlights the importance of activity for establishing these differences during development, with important implications for the effects of age-related and congenital hearing loss on SGN composition.
Results
Type I and Type II SGNs exhibit clear molecular differences
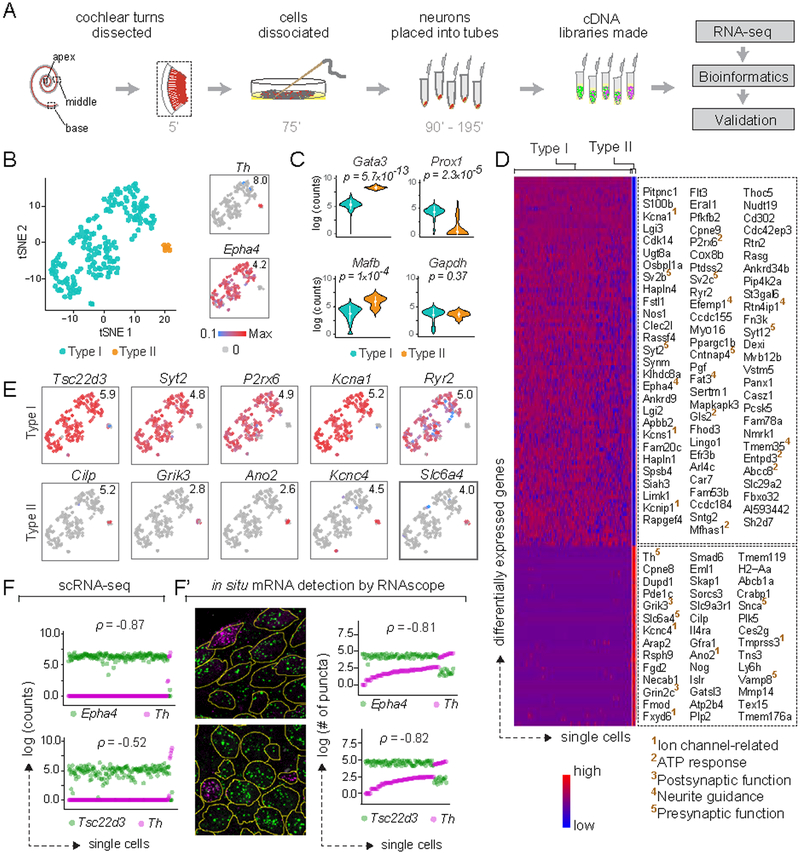
To survey SGN heterogeneity in an unbiased manner, we manually collected individual SGNs from the basal, middle and apical turns of cochlea of young adult mice (P25-P27, 11 animals) and analyzed them by single-cell RNA sequencing (scRNA-seq) (Fig. 1A; see Methods). A total of 186 wildtype SGN single-cell libraries passed all quality control criteria (Fig. S1A). In a two dimensional representation generated using t-stochastic neighbor embedding (tSNE), SGN libraries segregated into several distinct groups (Fig. 1B). Type I and II SGNs, which exhibit well-characterized differences in anatomy, glutamate-responsiveness, myelination, and synapse number (Berglund and Ryugo, 1987; Glowatzki and Fuchs, 2002; Spoendlin, 1979; Weisz et al., 2009), fell into distinct groups enriched for known markers such as Epha4 and Th, respectively. Presumed Type I and Type II SGNs also express different levels of Gata3 (p = 5.7×10−13), Mafb (p = 1×10−4) and Prox1 (p = 2.3×10−5), as previously reported (Fig. 1C) (Nishimura et al., 2017). Housekeeping genes such as Gapdh (p = 0.37) (Fig. 1C) and Eif2a (p = 0.43, not shown) are expressed at comparable levels in all SGNs. Overall, Type I and II SGNs comprised 96% and 4% of the samples, respectively, which is close to histological estimates of Type I/II proportions (Spoendlin, 1969).
Figure 1: Type I and II SGNs can be detected as molecularly distinct cell populations using scRNA-seq.
(A) Workflow for single cell RNA-seq (scRNA-seq) of spiral ganglion neurons (SGNs). Numbers indicate time elapsed (in minutes) since animal euthanization. (B) t-stochastic neighbor embedding (tSNE) of neuronal profiles (n = 186, 11 P25–P27 animals) revealed several clusters. Clusters of Type I (blue) and II (orange) SGNs were identified by expression of Epha4 and Th, respectively (insets). In this and all subsequent plots, numbers in the upper right corner indicate highest expression (Max) observed for each gene. (C) Violin plots comparing gene expression among Type I and II SGNs illustrate increased expression of Gata3 and Mafb in Type II SGNs (p = 5.7×10−13 and 1×10−4, respectively), and of Prox1 in Type I SGNs (p = 2.3×10−5), with no difference in expression of the housekeeping gene Gapdh (p = 0.37). White dot and bar indicate mean and standard deviation, respectively. (D) Heat map showing genes expressed differentially between Type I and II SGNs, with examples of Type I-enriched (top panel) and Type II-enriched (bottom panel) genes listed on the right. Superscripted numbers indicate gene functional groups annotated manually. (E-F) Several genes exhibit binary ON/OFF expression between the two subtypes (E), with clear correspondence between scRNA-seq (F) and RNAscope (F’) quantification in P25–P27 tissue sections for Epha4 and Th, as well as the novel Type I marker Tsc22d3. In images showing RNAscope puncta (F’), SGN cell bodies are outlined in yellow as visualized by immunostaining for parvalbumin (not shown). In scatterplots (F, F’), the two dots in each column indicate counts for two different genes in the same neuron, and neurons are sorted along the X-axis by the level of the gene in magenta. p represents Pearson’s correlation coefficient. See also Fig. S1.
Type I and II SGNs exhibit profound differences in connectivity and function that predict similar distinctions at the molecular level. The myelinated Type I SGNs encode all the signals we think of as “hearing” (Kiang et al., 1967; Sachs and Abbas, 1974), whereas unmyelinated Type II SGNs may mediate auditory pain and nociception (Flores et al., 2015). Like other neurons representing different modalities, Type I and Type II SGNs exhibit broad differences at the molecular level, with >1700 differentially expressed genes (p<0.01 or lower, Table S1A). Of these, 335 genes show a binary ON/OFF expression between the two cell classes (defined operationally as expressed in >70% and <20% of cells for ON and OFF, respectively) (examples in Fig. 1E). Gene ontology (GO) analysis of differential expression revealed overrepresentation of molecules involved in neuronal function and innervation (Fig. S1H). Additional manual categorization of the top 50 genes highlighted a stark difference in the presynaptic vesicle-associated protein profiles between Type I and II SGNs, with highly specific expression of Sv2b, Sv2c, Syt2, Syt12 in Type I SGNs and of Snca, Vamp8 in Type II SGNs (Fig. 1D). In addition, whereas both subtypes co-express several glutamate receptor subunits (e.g., Grina, Gria2, Gria4, data not shown), Type II SGNs show unique expression of Grin2c and Grik3 (Fig. 1D). Type I and Type II SGNs also express different sets of potassium channel subunits, with Kcns1 and Kcnip1 in Type I and Kcnc4 in Type II SGNs (Fig. 1D). These results suggest distinctions in glutamate responsiveness, presynaptic release properties, and neuronal excitability that are consistent with previous reports of different electrophysiological signatures between these SGN subtypes (Glowatzki and Fuchs, 2002; Weisz et al., 2014).
Validation using RNAscope, an in situ approach for semi-quantitative RNA detection (ACD Bio), confirmed that the variation detected by scRNA-seq represents bona fide differences. In tissue sections of P25–P27 mouse cochleas, levels of Th, which mark the small Type II subpopulation in our scRNA-seq data (Fig. 1B, inset), were highest in cells with the lowest levels of Epha4, a Type I marker (Fig. 1F’). Tsc22d3 transcripts were detected in neurons negative for the Type II marker Th (Fig. 1F’), and overlapped with expression of the Type I marker Epha4 (data not shown), indicating that it may be a reliable Type I marker as predicted by scRNA-seq (Fig. 1F). Thus, our approach allowed sampling and detection of the Type II subtype comprising only a small minority of SGNs (Spoendlin, 1969). Despite the small size and number of Type II SGNs, an average of 6320 genes/cell and >11000 genes across all cells could be detected, including cell type-specific molecules that influence neuron physiology and connectivity. These findings confirm that transcriptomic SGN profiles can be used to establish a molecular framework for subtype classification.
Three molecularly-defined subtypes of Type I SGNs
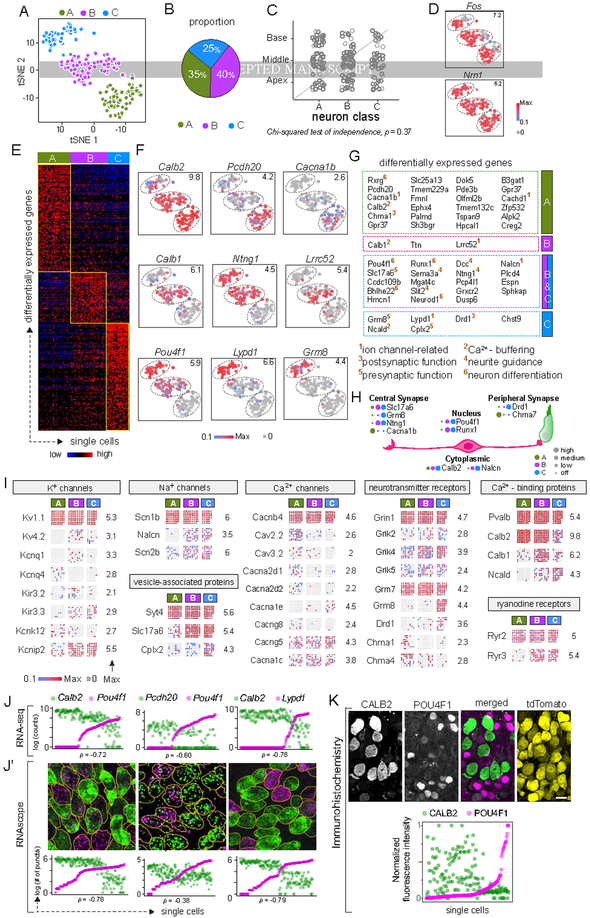
We next focused our analysis on Type I SGNs, which were identified as Epha4+ Th− cells in the scRNA-seq libraries (Fig. 1B, insets). In a tSNE plot, Type I SGNs further segregated into three distinct groups (Fig. 2A, S1B,C). Independent classification using an unsupervised machine-learning strategy (i.e., graph-based clustering, see Methods) categorized Type I SGNs into three distinct subtypes (Ia, Ib, Ic) that constitute 35%, 40%, and 25%, respectively, of the total population (Fig. 2B). Both projection of SGN transcriptomes onto principal component analysis (PCA) space (Fig. S1D) and hierarchical clustering by Ward’s method (Fig. S1E) also revealed three broad subtypes and close correspondence with the cell identities that were independently predicted by the graph-based clustering approach (Fig. 2A). Thus, multiple statistical methods indicate that there are three molecularly distinct Type I SGN subtypes.
Figure 2: Three molecular subtypes of Type I SGNs exist in the mouse inner ear.
(A-B) tSNE embedding of Type I SGN transcriptomes (A) depicting three clusters — A, B, C — predicted by graph-based clustering, which are indicated by dot color. Overall proportions are illustrated in B. (C-D) SGN subtypes are present in all regions of the cochlea (C) and show expression of the activity-induced genes Fos and Nrn1 in all clusters (D). (E-G) The clusters exhibit broad differences in their transcriptomes, illustrated in a heat map for the top 100 differentially expressed genes (E) and in tSNE plots for individual genes that show subtype-specific expression patterns (F). Numbers in the upper right corner indicate highest expression (Max) observed for each gene. (G) Genes enriched across the three subtypes encode proteins associated with many aspects of neuronal differentiation and function. Superscripted numbers indicate gene functional groups annotated manually. (H) Examples of differentially expressed genes that encode proteins localized to different neuronal compartments, indicating that input and output properties vary among SGNs. For each gene, expression level among SGN subtypes is indicated by the size of each colored dot. (I) Expression of select genes relevant to neuronal physiology is illustrated in dot matrix plots of individual libraries, which are grouped by subtype. Some genes are expressed uniformly across all libraries (top row for each group), whereas others vary across subtypes (all other rows). Numbers on the right indicate the highest expression (Max) observed for each gene. (J-K) Differentially expressed genes identified by scRNA-seq (J) showed the same patterns of expression in individual SGNs analyzed by RNAscope of P25–P27 tissue sections (J’). SGN cell bodies are outlined in yellow as visualized by immunostaining for parvalbumin (not shown). Similarly, immunostaining (K) for CALB2 (calretinin) (green), POU4F1 (magenta) and tdTomato (yellow) in tissue sections of P25–P27 bhlhb5Cre/+: Ai14/+ mice revealed inverse gradients of CALB2 and POU4F1 expression, quantified below. In scatterplots (J, J’, K), the two dots in each column indicate expression levels of two different markers in the same neuron, and neurons are sorted along the X-axis by the level of the gene in magenta. Scale bars: 10 μm (K). See also Fig. S1, S2, S3.
This classification scheme was not influenced by experimental and biological variables. First, the clusters do not correspond to where the neurons originated in the cochlea (Chi-squared test, p = 0.37) (Fig. 2C, S1D,E). Second, clustering is not driven by the acoustic exposure history of the animal, as activity-induced genes such as Fos and Nrn1 were expressed in comparable proportions of cells (Fig. 2D). Furthermore, no significant differences were found across the clusters in experimental factors such as animal identity, cDNA processing batch, sequencing batch, genes detected, or proportions of spike-in and mitochondrial transcripts (Fig. S1F,G). Lastly, none of the single-cell libraries were positive for glial markers (Fig. S1H).
Differential expression analysis among Ia, Ib, and Ic SGNs revealed 425 genes (all at p < 0.01) with subtype-enriched expression (Table S1B, top 100 or all for each subtype depicted in Fig. 2E). The segregation into three subtypes is associated with striking differences in expression of multiple genes, including Calb2, Pou4f1, Ntng1, and Lypd1. Of these, Calb2 and Pou4f1 are expressed in complementary gradients: Calb2 is highest in Ia and lowest in Ic, with Pou4f1 highest in Ic and lowest in Ia (p = −0.73, Fig. 2F). Ib and Ic SGNs both express high levels of Ntng1, but can be further distinguished by expression of Lypd1, which overlaps partially with Pou4f1 and Ntng1 (p = 0.71, 0.49, respectively), and is expressed at higher levels in Ic SGNs than in Ib SGNs (Fig. 2F). Pcdh20, Cacna1b, Lrrc52, Grm8 were also expressed in subtype-specific patterns (Fig. 2F). In contrast, pan-neuronal genes, such as Tubb3, Gria2, and Atp1b1, were expressed broadly (100%, 99%, 99% of cells, respectively) (Fig. S1I).
Genes that are expressed differentially across subtypes are mostly involved in regulating neuronal differentiation, connectivity and physiology. In a GO analysis, ‘potassium channel activity’, ‘calcium ion binding’, ‘signal transducer activity’, ‘neuron differentiation’, ‘regulation of ion transport’, and ‘axon development’ were among the 54 biological processes enriched significantly (p <0.01) (Fig. S1K). Genes encoding proteins that localize to ‘synapse’, ‘cation channel complex’, and ‘plasma membrane’ were also overrepresented. Manual annotation revealed combinatorial expression of several transcription factors, further indicating that SGN subtypes express distinct cohorts of genes that might influence their differentiation: Rxrg is expressed mostly in Ia, Runx1 is primarily in Ib and Ic, and Pou4f1 is in a decreasing gradient from Ic to Ia. Similarly, the subtypes express unique combinations of genes encoding Ca2+-binding proteins, with Calb2, Calb1 and Ncald enriched in Ia, Ib and Ic SGNs, respectively. Ib and Ic SGNs also show enriched expression of molecules related to neurite guidance (e.g., Ntng1, Sema3a, Dcc, Slit2) and presynaptic release (e.g., Slc17a6, Cplx2) (Fig. 2G).
The molecular signatures of SGN subtypes suggest meaningful physiological differences. Several proteins related to synaptic transmission or neuronal physiology were among those differentially expressed, and a comprehensive assessment of their gene families revealed both broad and subtype-specific expression (Fig. 2I, S2, S3). Among glutamate receptors, several AMPA-type subunits (e.g., Gria2, Gria3, Gria4) were uniformly expressed, but Grik4 was expressed in an increasing gradient from Ia to Ic while the metabotropic receptor subunit Grm8 was expressed specifically in Ic SGNs (Fig. 2I, S3). Whereas the cholinergic receptor subunits Chrna1 and Chrna4 were enriched in Ia SGNs, Ic SGNs showed enriched expression of Drd1, which encodes a dopamine receptor subunit (Fig. 2I, S3). Among genes encoding K+ channel subunits, Kcnq4 and Kcnd2 were detected mostly in Ia and Ib, respectively, with enriched expression of Kcnip2 and Kcnj9 in Ib and Ic SGNs (Fig. 2I, S2). The voltage-gated Ca2+ channels Cacna1b, Cacna1h and Cacna2d1 were all enriched in Ia SGNs (Fig. 2I, S2). Expression of the leak Na+ channel Nalcn was restricted to Ib and Ic neurons, whereas the voltage-gated Na+ channel subunit Scn2b was expressed in a decreasing gradient from Ic to Ia (Fig. 2I, S2). Taken together, these findings suggest that the three molecularly defined Type I SGN subtypes constitute physiologically distinct subpopulations.
RNAscope validation confirmed that the same genes that drive SGN segregation in the scRNA-seq data can be used to identify Ia, Ib, and Ic subtypes in cochlear sections of P25–P27 mice. As seen by scRNA-seq (Fig. 2J), Calb2 and Pou4f1 are expressed in opposing gradients among SGNs (p = −0.78) (Fig. 2J’). Pcdh20 levels are highest in cells negative for Pou4f1 transcripts and lower in all other cells, indicating a weak negative correlation (p = −0.38). Lypd1 expression is restricted to cells largely off or low for Calb2 (p = −0.79). Thus, these results validated both individual gene-level variation as well as gene-gene relationships across SGN populations. Furthermore, a negative correlation was observed between CALB2 and POU4F1 protein levels in SGNs (p = −0.36) (Fig. 2K), suggesting that the observed transcript-level differences may be functionally significant.
Tonotopic differences
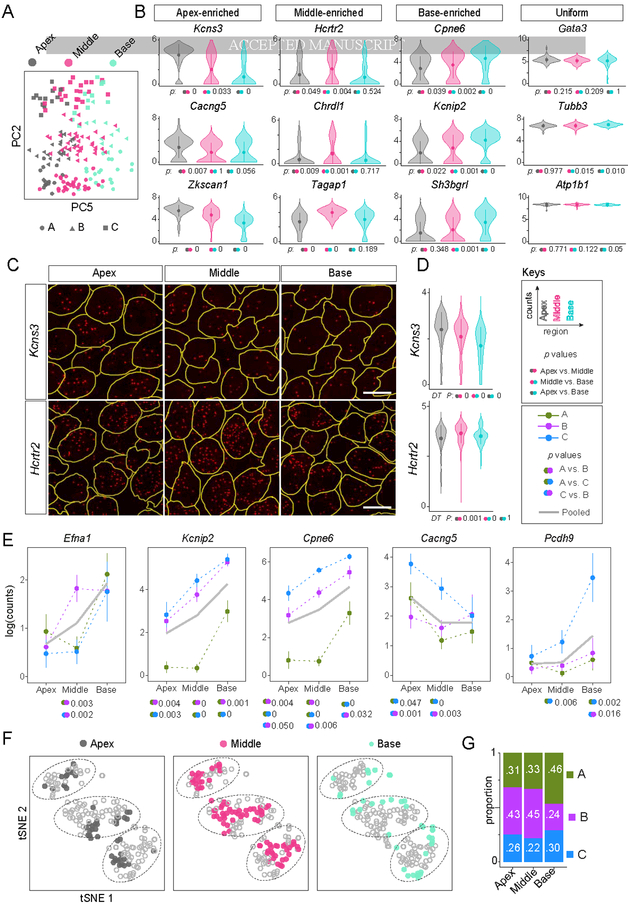
To gain further insights into Type I SGN heterogeneity, we examined other sources of variation in the data revealed by PCA. We found that although differences corresponding to the Ia, Ib, and Ic subtypes can be accounted for largely by PC2, additional heterogeneity represented by PC5 diversified SGNs within each subtype (Fig. 3A). This orthogonal axis of heterogeneity corresponded to the tonotopic origin of the SGNs, which was recorded for every cell during dissociation and manual collection. This result motivated further analysis of differences related to tonotopy both at the single-cell and tissue levels.
Figure 3: Type I SGNs exhibit both broad and subtype-specific tonotopic differences.
(A) Molecular heterogeneity exists along the tonotopic axis of the cochlea. Projection of single cell transcriptomes onto principal component analysis (PCA) space shows that PC2 reflects differences among the A, B, C subtypes while PC5 captures heterogeneity that corresponds to tonotopic origin. (B) Violin plots illustrate examples of genes that show either differential or uniform expression across the three tonotopic regions. Dunn’s test was used to assess significance for each possible comparison, as indicated by colored dots next to p values. See Keys. (C-D) Regional differences in expression of Kcns3 and Hcrtr2 were confirmed by RNAscope of P25 tissue sections (C), quantified in D. (E) Further analyses of scRNA-seq data revealed that some genes exhibit regional variation in a subtype-specific manner. Trends for all SGNs are shown in grey solid lines and for Ia (green), Ib (purple) and Ic (blue) SGNs in dashed lines. Error bars represent SEM. Pairs of dots indicate p values for comparisons across tonotopic regions by Tukey’s HSD test if the data were normally distributed and Dunn’s test otherwise. P values are reported only for statistically significant differences. (F) SGN subtypes are present in all regions of the cochlea, as revealed by tSNE plots showing the anatomic origin of cells (apex, middle, base) in each cluster. However, the proportions differ in the basal turn of the cochlea compared to the apex and the middle (G). Scale bars: 10 μm (C). See also Fig. S4.
We grouped the single-cell transcriptomic profiles based on cochleotopic position instead of molecular identity and examined gene expression differences among such groups. As predicted by PCA results (Fig. 3A), this analysis revealed an additional but subtler variation, with 114 genes expressed differentially among neurons from the three cochlear regions, compared to 425 genes that differ across subtypes (p < 0.01) (Fig. S4A, Table S1C). Unlike the genes that drive the Ia, Ib, and Ic classification, region-enriched genes were expressed mostly in gradients (Fig. 3B). For example, genes for two K+ channel subunits (Kcns3, Kcnip4) and several guidance-related factors (Ntng2, Wnt3) were enriched in the apex (Fig. S4A). RNAscope of P25–P27 mouse cochlea validated the tonotopic variation in expression observed by scRNA-seq. Levels of Kcns3, which encodes the Kv9.3 K+ channel subunit, were highest in the apex, lower in the middle and even lower in the base of the cochlea (median number of puncta per cell: 11.5, 9, 6, respectively, Fig. 3C,D). Similarly, expression of Hcrtr2, which encodes the hypocretin receptor type 2, was highest in the middle of the cochlea and lower in both the apex and the base (median number of puncta per cell: 44, 34, 33, respectively, Fig. 3C,D), consistent with scRNA-seq results (Fig. 3B). By contrast, expression of the pan-neuronal marker Tubb3 showed minimal variation (median number of puncta per cell: 86, 85, 89 in apex, middle, base, respectively, Fig. S4B), indicating that the observed variability in Kcns3 and Hcrtr2 transcripts is unlikely to be due to technical artifacts.
Closer analysis revealed that some genes vary along the tonotopic axis in a subtype-specific manner. For example, Efna1, which encodes the axon guidance molecule Ephrin A1, is expressed at higher levels in the middle and base relative to the apex among Ib SGNs, but only enriched in the base among Ia and Ic SGNs (Fig. 3E). Both Kcnip2 and Cpne6 are expressed in an apical-basal ascending gradient among Ib and Ic SGNs, but selectively enriched in the base among Ia SGNs (Fig. 3E). Cacng5, which encodes a voltage-gated Ca2+ channel subunit, shows a particularly complex pattern, with an apical-basal descending gradient among Ic SGNs, an apical-middle descending gradient among Ia SGNs, and relatively uniform expression among Ib SGNs. Pcdh9 is expressed in apical-basal gradient but only among Ic SGNs. We did not observe subtype-specific variation in expression for known pan-neuronal genes, such as Tubb3 and Atp1b1, or other weakly-expressed genes that exhibit tonotopic variation, such as Ntng2 and Wnt3 (Fig. S4C), suggesting that such differences are not artifactual. Overall, analysis across the tonotopic axis for each individual class of SGNs revealed 184, 157 and 183 genes with regional differences in expression among Ia, Ib, and Ic SGNs, respectively (Table S1D,E,F). Of the 524 total genes, 494 were unique, which is 3 times higher than the genes identified in subtype-blind comparisons. Only 77 of the 524 genes overlapped with the gene list derived from subtype-blind comparisons. Thus, a subtype-aware analysis of scRNA-seq data unmasked a new layer of tonotopic variation among SGNs.
Even some of the genes that vary among SGN subtypes exhibit additional variation along the cochlear axis. The most robust subtype markers, such as Calb2, Cacna1b, Ntng1, and Lypd1, were expressed at similar levels in the apex, middle and base (Fig. S4D). However, some functionally relevant genes (e.g., Lrrc52, Kcnip4, Anxa5, Ryr3) showed additional regional differences (Fig. S4E). For instance, the potassium channel subunit Kcnip4 was enriched in Ia SGNs in the apex and middle, but in Ic SGNs in the base. This indicated that, although the molecular identities of Ia, Ib, and Ic SGNs across the cochlea are largely similar, subtle variations exist in some regions, particularly in the base.
To further investigate how SGNs vary across the three major turns of the cochlea (apex, middle, base), we annotated the anatomic origin of every neuron in tSNE plots that depict SGN subtypes. We observed different proportions of SGN subtypes in each turn (Fig. 3F). In the middle turn, there were 33% Ia, 45% Ib, and 22% Ic SGNs. The proportions were similar in the apex (31% Ia, 43% Ib, and 26% Ic) (Fig. 3G). However, the base contained a significantly higher proportion of Ia SGNs (46%), largely at the expense of Ib SGNs (24%). Thus, tissue-level variation in SGN subtype proportions is a tonotopic feature that is superimposed on gene expression gradients across the mouse cochlea.
Type Ia, Ib, and Ic SGNs exhibit stereotyped anatomical and morphological differences
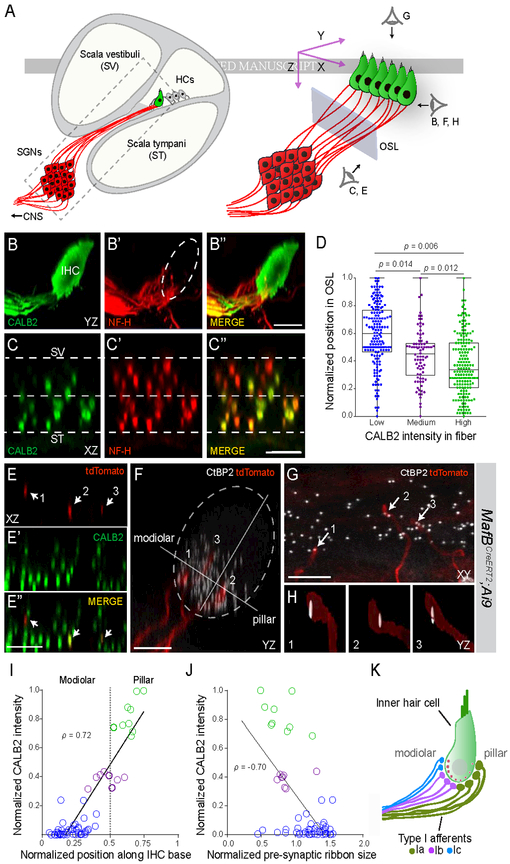
Previous studies showed synapse size differences and spatial segregation of the peripheral projections of the three SR groups (Kawase and Liberman, 1992; Liberman, 1982; Liberman et al., 2011). To map our molecular classification onto the known anatomy, we took advantage of sparse labeling of SGNs in the MafbCreERT2;Ai9 transgenic line to trace neuronal projections from the cell body to the peripheral synapse. Consistent with scRNA-seq and RNAscope results, levels of the calcium-binding protein CALB2 (calretinin) varied among SGN fibers (Fig. 4B-B”). Double staining for CALB2 revealed that levels in peripheral processes correlated strongly (p = 0.86) with those in the cell body (S5A, B). Thus, CALB2 levels in individual fibers can be used to identify SGN molecular subtypes.
Figure 4: Type I SGN peripheral processes and synapses are anatomically segregated by subtype.
(A) Schematic depicting a cross-section of the cochlea (left) with a magnified view of the boxed area on the right. The three perspectives corresponding to the cochlear wholemount images in BH are indicated (right). Blue rectangle represents the plane of section through confocal image stacks of afferent fibers (red) extending through the osseous spiral lamina (OSL) to terminate along the basolateral surface of the hair cell (HC) (green). (B-C) Side (B) and cross-sectional (C) views of a wholemount cochlea stained for CALB2 (green, B,C) and NF-H (neurofilament heavy chain) (red, B’,C’), with merged images (B”,C”). CALB2+ fibers preferentially project towards the pillar side of the inner hair cell (IHC) compared to the total population of all NF-H+ SGN processes and are segregated along the scala vestibuli (SV)-scala tympani (ST) axis in the OSL (C-C”). CALB2 antibody also labels IHCs. (D) Quantification of afferent fiber distribution in the OSL. CALB2 fluorescent intensity levels were measured for all NF-H+ fibers in the OSL cross-section (n = 5 animals). Fibers were split into three groups based on CALB2 levels: ‘low CALB2’ (n = 165 fibers), ‘medium CALB2’ (n = 82 fibers), and ‘high CALB2’ (n = 174 fibers). Distance from the median center of each nerve bundle was calculated for individual fibers from each cluster. P values indicate results of Tukey’s HSD test following one-way ANOVA. (E-H) Individual tdTomato-labeled fibers (red) (E, E”) were traced in cochlear wholemounts from MafbCreERT2;Ai9 animals that were also stained for CALB2 (green, E’, E”) to assign subtype identity. Presynaptic ribbons were defined by co-staining for CtBP2 (white, F-H). In this example, three individual tdTomato-labeled SGN fibers (arrows) express ‘high’ (2), ‘medium’ (3), and ‘low’ (1) levels of CALB2 as they project through the OSL (E, E”). The same three fibers segregate along the modiolar-pillar axis of the IHC, shown in side view in F. Each tdTomato-labeled fiber terminates opposite a single presynaptic ribbon, shown in high resolution reconstructions (H). (I-J) Quantification of all analyzed fibers (n = 61, 5 animals) revealed that both fiber position (I; p = 0.72) and ribbon size (J; p = −0.70) correlate with CALB2 intensity. (K) Type Ia (green), Ib (purple), and Ic (blue) SGNs extend peripheral processes that are segregated in the OSL and along the modiolar-pillar axis of the IHC where they are apposed by presynaptic ribbons that decrease in size along the same axis. These features match those described for high, medium, and low SR SGNs. Scale bars: 10 μm (B, C, E, F); 5 μm (G). See also Fig. S5.
We observed a striking organization of Ia, Ib, and Ic peripheral processes coursing through the osseous spiral lamina (OSL), which is flanked by scala vestibuli (SV) and scala tympani (ST) (Fig. 4A). In optical cross sections of image stacks capturing CALB2- and neurofilament (NF)-labeled afferent processes in the OSL in the P25–P27 cochlea (Fig. 4C-C”), fibers with the highest levels of CALB2 (CALB2high NF+) were closer to ST, while those with the lowest level of CALB2 (CALB2lo NF+) were closer to SV. The mid-OSL region was occupied by fibers with intermediate CALB2 levels (CALB2mid NF+) (Fig. 4D). This organization matches the projections of low-, medium-, and high-SR SGNs along the SV-ST axis (Kawase and Liberman, 1992). Examination of tdTomato-labeled olivocochlear efferent axons in ChatCre;Ai14 mice indicated comparatively even distribution across the OSL (Fig. S5E-G) in contrast with tdTomato− NF+ SGN processes. Thus, the presence of CALB2lo NF+ fibers closer to SV is not conflated by olivocochlear axon organization.
The position and morphology of synapses also matched known differences among SR subtypes. By following sparsely labeled SGN processes from the OSL to their IHC terminals (n = 61 SGNs, 5 P25–P27 MafbCreERT2;Ai9 animals; Fig. 4E-H), we found that peripheral axons with low CALB2 expression (Ic subtype) ran closer to SV and could be traced to the modiolar aspect of IHCs (Fig. 4I), where they formed synapses with large presynaptic ribbons (Fig. 4J). Conversely, fibers with high CALB2 levels (Ia subtype) were closer to ST, projected to the pillar face (Fig. 4I), and formed synapses with small ribbons (Fig. 4J). Processes from neurons with medium CALB2 levels (Ib subtype) were flanked by high and low CALB2 processes (Ia and Ic, respectively) along the modiolar-pillar axis and were apposed to medium-sized synaptic ribbons. Overall, there was a strong positive correlation between CALB2 fiber intensity and synapse position (p = 0.72) (Fig. 4I), and a strong negative correlation (p = −0.70) between CALB2 fiber intensity and the size of the associated ribbon (Fig. 4J). Thus, Ia, Ib, and Ic fibers are topographically organized in the OSL and this order is preserved along the modiolar-pillar axis of the IHC, accompanied by a gradient of presynaptic ribbon size in IHCs (Fig. 4K). Overall, based on fiber position, synapse location, and synapse morphology, the Ia, Ib, and Ic subtypes match the features of high-, medium- and low-SR subgroups, respectively.
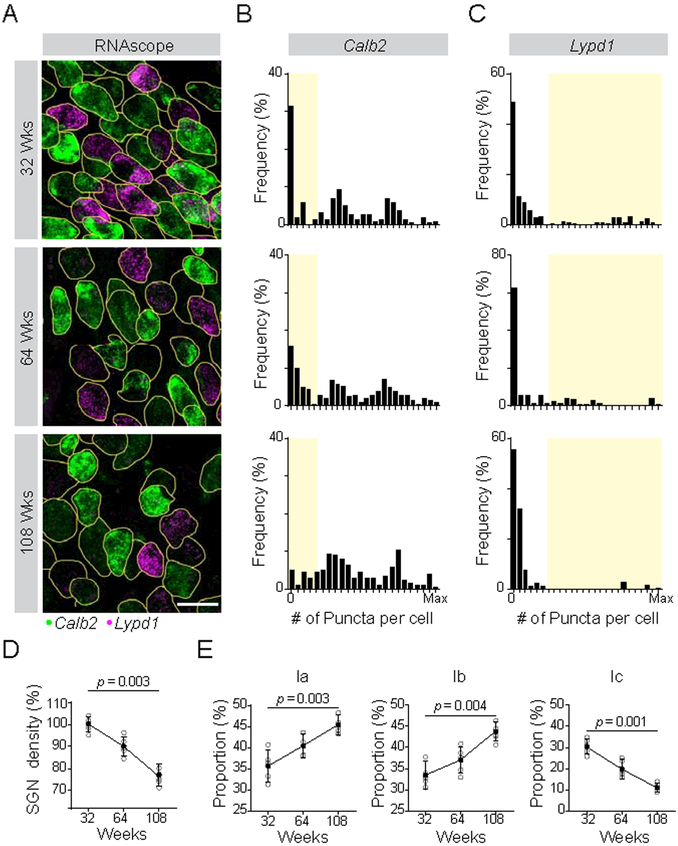
Type Ic SGNs are selectively vulnerable to age-related loss
In aging animals, SGNs are progressively lost, and low-SR fibers are the most affected (Schmiedt et al., 1996; Sergeyenko et al., 2013). Thus, we assessed Type Ia, Ib, and Ic SGNs in 32, 64, and 108 week old mice, as determined by expression of Calb2 and Lypd1. SGN loss in aging animals was characterized by a gradual loss of cells expressing low levels of Calb2 and high levels of Lypd1 (Fig. 5A), as evident in population histograms (Fig. 5B,C). To assess the composition of SGNs at each stage, we defined Calb2hi Lypd1off, Calb2mid Lypd1off, Calb2lo Lypd1hi neurons as Ia, Ib, Ic subtypes, respectively. As expected, overall SGN density declined with age (Fig. 5D). Only Type Ic SGNs showed a similar decrease, dropping from 30% at 32 weeks (wks) to 20% at 64 wks and still further to 11% at 108 wks (Fig. 5E). The proportion of Ia and Ib SGNs increased. Thus, Ic SGNs are selectively vulnerable to age-related hearing loss.
Figure 5: Type I SGN subtypes show differential vulnerability to age-related hearing loss.
(A) SGN subtype identity was assessed using RNAscope to quantify levels of Calb2 and Lypd1 transcripts at 32, 64 and 108 weeks, shown in representative tissue sections. (B-C) Histograms show the frequency distribution of Calb2 (B) and Lypd1 (C) mRNA levels for all analyzed SGNs at each age (n = 212 at 32 weeks (top), 175 at 64 weeks (middle), 155 at 108 weeks (bottom)). The Type Ic population, defined by low levels of Calb2 and high levels of Lypd1, is shaded in yellow. (D-E) SGN density (% relative to the 32 wk time point) decreases over time (D) and this loss is matched by a decrease in the proportion of Ic SGNs (E). Type Ia and Ib SGNs increase in proportion over the same time frame, indicating that loss of Ic SGNs likely accounts for the overall decrease in density. P values indicate results of Dunn’s test following one-way ANOVA for 5 animals. Scale bar: 10 μm (A). See also Fig. S6.
Signatures of SGN subtypes are present at birth and refined over the first postnatal week
The discovery of molecularly distinct SGN subtypes raises the important question of how this heterogeneity is established in development. In many systems, identities are apparent as soon as neurons become post-mitotic, with additional activity-dependent modification as the neurons are integrated into functional circuits. In the mouse cochlea, Type I and Type II SGNs exhibit distinct anatomical features by E16.5 (Koundakjian et al., 2007). By contrast, variation among Type I SGNs has not been detected until postnatal stages, after the peripheral processes have reached their IHC targets, and IHC-SGN synapse morphology is not fully mature until the 4th postnatal week (Liberman and Liberman, 2016). Since the nature of the synapses linking IHCs to Type I SGNs likely influences neuronal thresholds (Liberman et al., 2011), this relatively slow and late emergence of functional heterogeneity suggests that SGN subtype identity may not be fully established until the end of the first month of life. However, in the absence of a molecular toolkit, analysis of SGN development has been limited to young adult and adult stages, obscuring early developmental events that shape SGN identities.
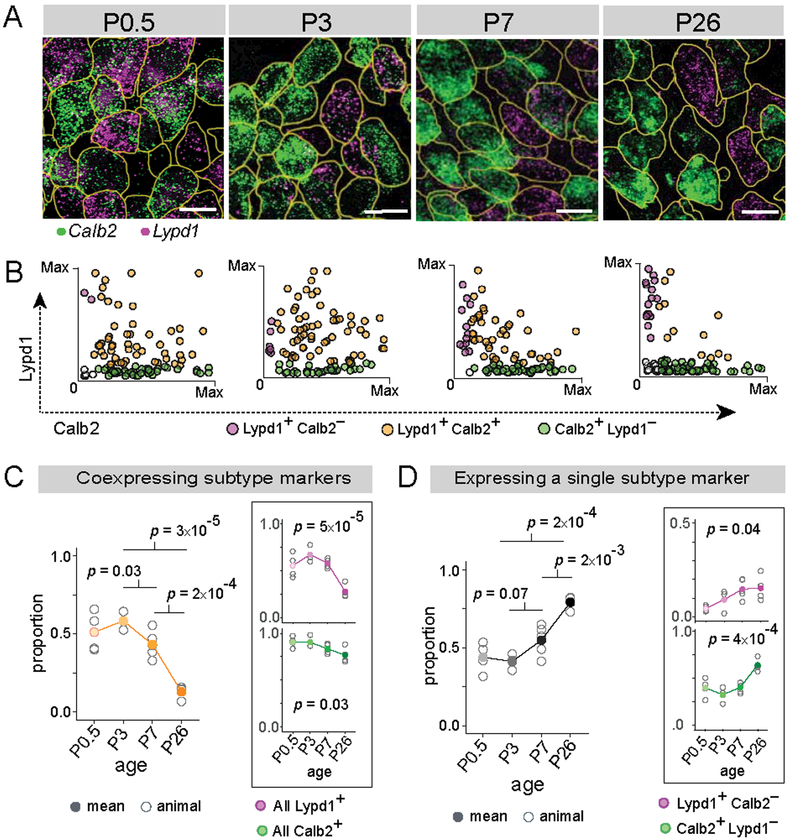
Using subtype-specific markers, we found that SGNs begin to diversify perinatally, followed by a period of refinement over the first postnatal week (Fig. 6). The Ia marker Calb2 and Ic marker Lypd1 are already expressed in SGNs by late embryonic stages (E18.5) and at P0.5, but differ sharply in breadth of expression: whereas Calb2 is expressed in almost all neurons (>90%), Lypd1 expression is more restricted (60–70%) at both time points (Fig. 6A, Fig. S7A-C). Calb2 levels are relatively uniform at these early stages, unlike the strong variability observed in the mature cochlea (Fig. 2J). Given the extensive co-expression and lack of a strong Calb2 gradient, SGN identities cannot be assigned confidently. By P3, Calb2 levels are more heterogeneous, and a small fraction of cells begin to show mutually exclusive expression of Calb2 and Lypd1, with significantly more Lypd1+Calb2− neurons compared to P0.5 (Fig. 6B, magenta dots). The gradient of Calb2 expression is even sharper at P7, and more low-Calb2 or Calb2− neurons express high levels of Lypd1. At P26, Calb2 and Lypd1 show mutually exclusive expression in ~90% of SGNs (Fig. 6D). These observations suggest that identities take shape through the gradual downregulation of Calb2, followed by maintenance of Lypd1 in select SGNs. Consistently, most SGNs initially express Calb2, but this proportion drops significantly after P3 (p = 0.03, Fig. 6C inset, green line). Expression of Lypd1, in comparison, first ramps up, possibly reflecting a slight delay in the onset of Lypd1 relative to Calb2, and then decreases over time (p = 5×10−5, Fig. 6C inset, magenta line). These trends coincide with a sharp decline in the proportion of cells co-expressing subtype-specific markers both within the first postnatal week (P3 to P7: p = 0.03) and over the first 3 weeks of postnatal development (P3 to P26, p= 3×10−5) (Fig. 6C, yellow line). At the same time, the proportion of cells expressing only one of the two markers increases (P3 to P26: p = 2×10−4; P3 to P7: p = 0.07) (Fig. 6D). Thus, SGN molecular identities begin to take shape around birth, with broad and overlapping expression of markers that becomes progressively restricted and signs of adult-like mutually exclusive expression appearing by the end of the first postnatal week.
Figure 6: Type I SGN subtypes emerge gradually over the first postnatal week.
(A-B) Representative images of Calb2 (green) and Lypd1 (magenta) mRNA detected using RNAscope in tissue sections of cochlea at various developmental stages (A). Scatterplots (B) show expression levels of Calb2 alone (green), Lypd1 alone (magenta) or both markers (orange) in individual SGNs (n = 100 randomly selected cells at each time point). (C-D) Over time, the proportion of cells expressing both Calb2 and Lypd1 decreases (orange, B, C), shown also for expression of each gene individually (magenta and green, insets). In parallel, there is an increase in the proportion that express a single subtype marker (D), shown for the whole population (black), as well as separately for Lypd1+ only SGNs (magenta, inset) and Calb2+ only SGNs (green, inset). Means are shown in solid dots, with raw data from each individual animal in open circles. P values indicate results of Tukey’s HSD test (left) and one-way ANOVA (right, inset). Scale bars: 10 μm (A).
Synaptic transmission from IHCs is necessary for SGN diversification
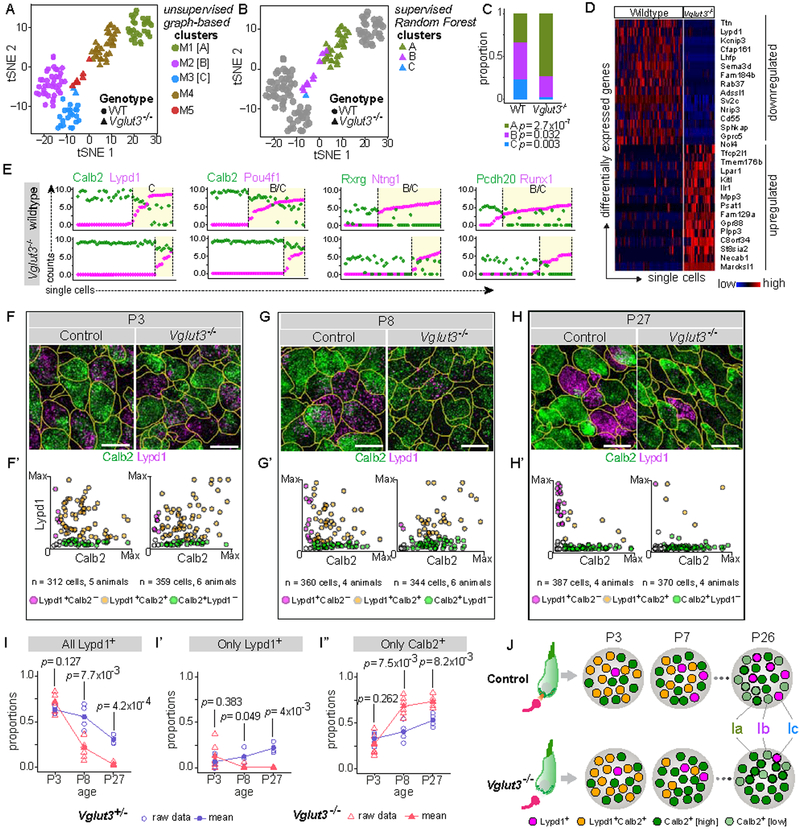
Mice begin to respond to airborne sound around P12 and cochlear function continues to mature into the 3rd postnatal week (Shnerson and Pujol, 1981). However, our data indicate that molecularly distinct SGNs emerge just after peripheral synapses begin to form at birth. To investigate whether synaptic communication influences SGN diversification, we performed scRNA-seq in P25–P27 Vglut3−/− mice, in which glutamatergic transmission from IHCs to SGNs is abolished. Transcriptional profiles of 40 SGNs from Vglut3−/− animals and 88 origin-matched wildtype SGNs were analyzed as described above. In a tSNE plot, wildtype (WT) middle-turn SGNs again segregated into three distinct groups (Fig. 7A). SGNs from Vglut3−/− animals, on the other hand, split into only two highly asymmetric groups. Unsupervised graph-based clustering sorted WT SGNs into 3 clusters (M1, M2, M3), containing 33%, 43% and 24% of the cells. In contrast, 80% of Vglut3−/− SGNs formed a single cluster (M4), with the remaining 20% mostly in a small fifth cluster (M5). Based on expression of subtype-specific markers, WT cells in the M1, M2, M3 clusters were identified as Ia, Ib, and Ic SGNs. The mutant cells in cluster M4 expressed Ia markers (such as Calb2) (Fig. S7D), while those in M5 expressed mostly Ib markers (Ntng1, Pou4f1) and very low or zero levels of Ic markers (Lypd1, Grm8) (Fig. S7D). Thus, the vast majority of SGNs (80%) from Vglut3−/− animals are Ia-like and the rest are Ib/Ic-like (20%).
Figure 7: SGN heterogeneity is altered in a mouse model of congenital deafness.
(A) tSNE embedding of single cell transcriptomic profiles from wildtype (WT, circles) and Vglut3−/− (triangles) animals, with 5 distinct clusters (M1–M5) predicted by graph-based unsupervised clustering indicated by color. M1, M2 and M3 correspond to WT Ia, Ib, and Ic SGNs, respectively, whereas M4 and M5 consist of SGNs from Vglut3−/− animals. (B) Subtype identities of neurons from Vglut3−/− animals were assigned using supervised clustering by the Random Forest method. WT cells are shown in grey, and cells from Vglut3−/− animals are shown in colors corresponding to their predicted subtype identities. Cluster M4 (brown, A) consists of Type Ia-like SGNs (green, B), whereas the remaining neurons in cluster M5 (red, A) are either more like Type Ib (purple, B) or Type Ic (blue, C) SGNs. (C) Subtype proportions are significantly altered in Vglut3−/− animals compared to WT, with a dramatic loss of Ic SGNs. P values indicate results of Test of Equal Proportions between Ia, Ib, and Ic subtypes in the two genetic backgrounds. (D-E) SGNs from WT and Vglut3−/− animals show broad differences in gene expression reflective of a shift from Ic to Ia identity, shown both in a heatmap (D) and in scatterplots (E) of the level of expression of Ia (Calb2, Rxrg, Pcdh20) and Ib/Ic (Lypd1, Pou4f1, Ntng1, Runx1) subtype markers (WT, top; Vglut3−/−, bottom). Yellow shading marks cells belonging to B/C or C clusters. (F-H) RNAscope of tissue sections from P3 (F), P8 (G), and P27 (H) WT (left) and Vglut3−/− (right) animals shows that expression of Calb2 (green) and Lypd1 (magenta) initiates normally at P3, quantified in F’. However, the proportion of Lypd1+ SGNs is decreased at P8 (G’), and there are almost no Lypd1+ SGNs remaining at P27 (H’). Scatterplots (F’-H’) show individual cells expressing Calb2 alone (green), Lypd1 alone (magenta) or co-expressing both markers (orange). (I-I”) Proportions of SGNs in control (blue circles) and Vglut3−/− (red triangles) animals that express any Lypd1 (I), that express only Lypd1 (I’), or that express only Calb2 (I”). Means shown in solid symbols, with raw data from individual animals in open symbols. P values refer to results of independent samples t-test between the two genetic backgrounds at each time point. (J) Schematic showing developmental emergence of mutually exclusive expression between Calb2 and Lypd1. In Vglut3−/− animals, in which glutamate release from IHCs is abolished, Lypd1+ SGNs are severely underrepresented compared to control animals, beginning after P3, resulting in overabundance of Ia SGNs by the fourth postnatal week. See also Fig. S7. Scale bars: 10 μm (F-H).
To corroborate these results, we utilized Random Forest, an ensemble machine learning approach, for supervised classification of Vglut3−/− cells based on subtype definitions derived from WT cells (Fig. 7B). Importantly, this approach is independent of sample size. After verifying that the classifier performed robustly (Fig. S7E-G, see Methods), predictions for SGNs from Vglut3−/− animals were generated (colored triangles in Fig. 7B). We found that 73% of the mutant cells were transcriptionally similar to Ia SGNs, 25% were Ib-like, and 3% were Ic-like (Fig. 7C). Consistent with the results of unsupervised classification, this indicated an overabundance of Ia-like neurons at the expense of Ib and Ic subtypes. Indeed, pooled comparison of scRNA-seq profiles of mutant and WT SGNs revealed a broad transcriptional shift towards the Ia phenotype, with downregulation of multiple Ib and Ic markers, such as Ttn and Lypd1, respectively (Fig. 7D).
Variability in Calb2 expression further supported this interpretation: Calb2 levels ranged from moderate to high (standard deviation, s = 0.07) across all Vglut3−/− cells, as expected for a population comprised mostly of Ia SGNs. This contrasts with the steep gradient from zero to high expression observed among WT cells (standard deviation, s = 0.78) (Fig. 7E). In addition, Lypd1 expression was limited to ~18% of cells, all of which lie near the WT Ib/Ic clusters in the tSNE plot (Fig. S7D). Changes in proportions of neurons expressing other Ib/Ic-specific or Ia-enriched transcription factors and axon guidance molecules (Fig. 7E) are consistent with the interpretation that SGN subtypes are significantly altered in Vglut3−/− animals.
Defective consolidation of Ic identity in Vglut3−/− animals
Depolarization of IHCs triggers action potential in SGNs, both in pre-hearing animals (<P12), when waves of spontaneous activity propagate from IHCs to SGNs and up the auditory axis, and in post-hearing animals, when IHC-SGN transmission is essential for mEPSCs and sound-evoked action potentials. The change in SGN proportions seen in Vglut3−/− animals could arise from disruption of activity at either stage. To determine when SGN diversification goes awry, we examined expression of subtype-specific markers at earlier developmental stages. At P3, both Calb2 and Lypd1 are expressed in the expected proportion of SGNs in Vglut3−/− animals compared to WT (all Lypd1+ SGNs: 0.63 and 0.72, p = 0.13; only Lypd1+ SGNs: 0.07 and 0.13, p = 0.38; only Calb2+ SGNs: 0.33 and 0.26, p = 0.26 in control and Vglut3−/−, respectively) (Fig. 7F-F’, II”). Thus, the onset of subtype specification does not require IHC-driven activity. However, by P8, the proportion of cells expressing the Ic marker Lypd1 is decreased significantly in Vglut3−/− animals (All Lypd1+ SGNs: 0.55 and 0.22, p = 7.7×10−3; only Lypd1+ SGNs: 0.13 and 0.01, p = 0.049 in control and Vglut3−/−, respectively) (Fig. 7G-G’, I-I’). This is accompanied by an increased proportion of Calb2+ SGNs (only Calb2+ SGNs: 0.40 and 0.68, p = 7.5×10−3 in control and Vglut3−/−, respectively) (Fig 7I”). These changes persist into the fourth postnatal week (Fig. 7H-H’), with significantly lower Lypd1+ SGN proportion at P27 in Vglut3−/− animals (All Lypd1+ SGNs: 0.30 and 0.03, p = 4.2×10−4; only Lypd1+ SGNs: 0.22 and 0.007, p = 4×10−3 in control and Vglut3−/−, respectively) (Fig. 7I-I’), consistent with scRNA-seq results (Fig. 7A-E).
Comparison of marker expression over time suggests that Vglut3−/− animals fail to produce Ib/c subtypes from among a pool of SGNs with mixed identity, which instead assume Ia identities. In control animals, the proportion of Lypd1+ SGNs (−13%) decreases modestly between P3 and P8 (Fig. 7I, blue line), consistent with the expected gradual refinement of SGN identities (Fig. 6). However, in Vglut3−/− animals, the proportion of Lypd1+ SGNs drops drastically (−70%) (Fig. 7I, red line). This is unlikely to reflect accelerated maturation because the proportions continue to decrease to near-absence of Lypd1+ SGNs by P27 (−96% vs. P3, in contrast to −52% in control). At the same time, the proportion of SGNs that express only Lypd1+ (and not Calb2) does not increase over time, in contrast to controls (Fig. 7I’). Instead, the proportion of Calb2+-only SGNs increases between P3 and P8 to a much greater extent in Vglut3−/− animals (+161%) than in controls (+21%) (Fig. 7I”). Thus, in Vglut3−/− animals, the Calb2 gradient does not sharpen and there is a dramatic loss in Lypd1 expression, as if most remaining SGNs retained an Ia identity rather than taking on a Ib/c identity in the first postnatal week and beyond (Fig. 7J).
Thus, our data indicate that IHC-driven activity is required for the developmental refinement of SGN identities from an early phase of broad and overlapping marker expression to subtype-restricted expression, with impairment of this process causing a drastic shift in SGN subtype proportions within the first postnatal week, which persists in the mature cochlea (Fig. 7J).
Discussion
SGNs enable the wide dynamic range needed to sense sounds from the rustle of a leaf to the roar of a jet engine. Although SGNs show well-characterized differences in morphological and functional properties that subserve dynamic range expansion (Kawase and Liberman, 1992; Liberman, 1982, 1991; Liberman et al., 2011; Taberner and Liberman, 2005), the molecular correlates of such heterogeneity have been a mystery. Using single-cell transcriptomics in mice, we define three subtypes of Type I SGNs that match the anatomical features of those originally defined by physiology, including the subtype that is preferentially affected by age-related hearing loss. These SGN subtypes express distinct cohorts of ion channels, receptors, synaptic proteins, and adhesion molecules that vary both locally and along the tonotopic axis, indicating that a complex reservoir of SGNs is available for coding auditory information. Unexpectedly, SGNs with distinct identities emerge during the first postnatal week in an activity-dependent manner. Together, these results establish a new molecular logic for cochlear circuitry, reveal the potential for many endogenous differences among SGNs, and suggest a new model for when and how SGN diversification is achieved, with important implications for the treatment of congenital deafness.
It was not a given that SGNs could be parsed molecularly, since the response properties that differentiate them could be imposed entirely by other cells in the circuit, e.g. functional presynaptic differences within IHCs (Wichmann and Moser, 2015) or differential innervation by olivocochlear efferents (Yin et al., 2014). To the contrary, we find that individual SGNs are intrinsically different in ways that could fundamentally alter their input and output properties. For instance, each subtype expresses unique cohorts of K+ channel subunits, Na+ channel subunits, and Ca2+-binding proteins that could yield distinct responses to input from IHCs and efferent neurons, such that both endogenous and circuit-level differences underlie SR-group heterogeneity. Notably, one of the most robust markers for Ic SGNs is Lypd1, which encodes a transmembrane receptor implicated in cholinergic signaling (Tekinay et al., 2009). Thus, Type Ic SGNs may exhibit unique responses to acetylcholine released by olivocochlear efferents. However, Lypd1 is only one of many class-specific genes that could create other, as yet uncharacterized, diversity in SGN electrical properties beyond SR and threshold. For instance, Type Ib and Ic SGNs are similar at the molecular level, suggesting they might be difficult to distinguish physiologically and highlighting the need for other means of classification. Similarly, whole-cell recordings from immature Type I somata reveal a range of electrical properties (Crozier and Davis, 2014; Liu et al., 2014), some of which depend on voltage-gated K+ channels (Liu et al., 2014) or correlate with levels of Ca2+-binding proteins (Liu and Davis, 2014). Given this broad range of possible physiological differences, SR differences are unlikely to be explained by single genes, and other variable features may also contribute to auditory coding in ways we do not yet understand.
SGNs may also differ in how they transmit signals to their brainstem targets in the cochlear nucleus complex (CNC). Type Ia, Ib, and Ic SGNs express distinct complements of axonal molecules, including proteins associated with presynaptic terminals such as the metabotropic glutamate receptor Grm8 and the exocytosis regulator Cplx2. Such molecular differences could generate distinct modes of plasticity and presynaptic release centrally, consistent with suggestions that presynaptic release is not uniform in the CNC (Cao and Oertel, 2010). Given the differences in expression of presynaptic molecules, as well as cell-adhesion molecules such as the Netrin family gene Ntng1, SGN subtypes may stimulate separate networks of CNC targets by virtue of their unique synaptic properties and connectivity. Indeed, previous studies have revealed both mixed and selective innervation of central targets by different SR groups. Multipolar cells in one part of the anteroventral cochlear nucleus (AVCN) receive inputs from all SR groups, whereas multipolar cells in another part of the AVCN, and in the small-cell cap region, are almost exclusively innervated by low- and medium-SR fibers (Liberman, 1991; Ryugo, 2008).
With the discovery of molecular differences among Type I SGNs comes the opportunity to not only test long-standing hypotheses but also pursue fresh questions regarding their functional properties and significance. Much has been learned about the physiology of Type I SGNs in adult ears via single-fiber recordings from the VIIIth nerve, work that showed correlations between SR and numerous other physiological features, as well as vulnerability to aging and noise damage. Our results make it possible to further elucidate the features and functions of individual subtypes, for instance by assessing the circuit-level contributions of each subtype to auditory behavior by using genetic approaches to perform targeted ablations. Some caution is warranted before embarking on such studies, however, due to the broad distribution of subtype-specific markers early in development. Careful consideration of expression dynamics will be critical for obtaining interpretable results. In this regard, combining subtype-specific markers with approaches that permit precise spatiotemporal control of gene expression, for instance via intersectional targeting strategies, may prove useful. Our data also stimulate new hypotheses regarding the mechanistic basis of SGN physiology, including but not limited to SR. Due to the technical difficulties of recording from individual adult SGNs both in vivo and in vitro, it will take time to work out how individual molecules contribute to SGN subtype function and ultimately hearing. Indeed, the breadth of molecular differences uncovered here identifies new questions beyond the origins of SR heterogeneity and opens many doors that will fuel the field for years to come.
Multiple layers of tonotopic variation among Type I SGNs
Tonotopy is a fundamental organizing principle in the auditory system that originates with the orderly gradient of frequency tuning along the cochlear spiral. Although emphasis is placed on this frequency axis and the gradient in hair cell mechanics that underlies it, tonotopic differences in SGN physiology have also been described, such as differences in action potential kinetics at neonatal stages (Adamson et al., 2002). However, with only a few genes identified (Adamson et al., 2002; Flores-Otero and Davis, 2011), neither the origin nor the extent of such variation is clear. Our results reveal differences in molecules spanning diverse gene families across the tonotopic axis. Many are involved in neuronal connectivity, such as the Ephrin gene Efna1, and might therefore help maintain topographic organization within the VIIIth nerve and in the CNC. Others could influence SGN physiology, such as the K+ channel subunit Kcns3, as well as Cacng5, Cpne6, and Vamp2. Thus, rather than serving as identical conduits within a topographically ordered circuit, SGNs may exhibit endogenous differences in their response properties that affect encoding of low versus high sound frequencies. Intriguingly, some tonotopic variation was limited to individual SGN subtypes, such that there could be three overlapping maps along the cochlear spiral. Nonetheless, the difference across subtypes, rather than tonotopic variation, was the dominant form of molecular heterogeneity in the adult cochlea. This suggests that the ear’s dynamic range is extended by the development of molecularly distinct cohorts of SGNs that are further specialized based on their tonotopic position.
Detection of complex sounds can depend not only on the properties and connectivity of SGN subtypes, but also on the proportions of these subtypes along the tonotopic axis. Interestingly, the distribution of SR rates is uniform in some species (Liberman, 1978; Tsuji and Liberman, 1997), whereas others show striking differences across the tonotopic axis (Schmiedt, 1989). The situation in mice is ambiguous, since it is harder to record from large populations of SGNs in such small animals (Taberner and Liberman, 2005). Here, we find a larger proportion of Ia neurons and smaller proportion of Ib neurons in the base compared to the rest of the cochlea. Such regional variation could enable heightened responses to certain, ethologically relevant sound frequencies, much as the presence of a high cone:rod photoreceptor ratio in the fovea of the retina enables strong color perception (Curcio and Hendrickson, 1991). Thus, SGNs could contribute to stimulus coding at multiple levels, from cell-based differences within and across SGN classes to tissue-level differences in their pattern of distribution along the cochlear spiral.
SGN subtypes emerge early via an activity-dependent process
The presence of molecular SGN subtypes presents a challenge for wiring the developing cochlea, where one IHC connects to 10–30 SGNs that share the same basic patterns of connectivity, but also exhibit diverse anatomical and functional phenotypes (Liberman, 1982; Liberman et al., 2011). Heterogeneity in the size of postsynaptic glutamate receptor patches is detectable by P14 in mice, but other features such as synapse number, efferent innervation density, ribbon size gradient, and ion channel localization reach maturity only between the third and fourth postnatal weeks (Kim and Rutherford, 2016; Liberman and Liberman, 2016). It was therefore surprising that Ib/Ic markers appear in subsets of SGNs shortly after their processes reach hair cells around E15.5 (Koundakjian et al., 2007), and that well-defined subtypes emerge coincident with a period of synaptogenesis and pruning during the first postnatal week (Huang et al., 2012; Meyer et al., 2009). Thus, SGN diversity is seeded soon after they form connections, pointing to a major role for signaling between IHCs and SGNs in the acquisition of mature identities.
Although mature Type I SGNs express class-specific genes, these differences emerge after several days of co-expression (Fig. 6). Transition from such a mixed state is defined by a singular rule: those that maintain Ib/Ic-specific genes lose Ia identity and those that do not continue as Ia neurons. Although transient periods of co-expression are a recurrent theme in the developing nervous system, conflicting identities are typically resolved intrinsically, for instance by mutual repression of class-specific transcription factors within individual progenitors (Jukam and Desplan, 2010). SGN identity, on the other hand, appears to be shaped by neuronal activity. When IHCs can no longer release glutamate to activate SGNs, as occurs in Vglut3−/− animals (Seal et al., 2008), the proportion of Ib/Ic SGNs is drastically decreased. Since markers for Ib/Ic SGNs appear normally at P3 in these animals, activity seems to bias how the mixed state is ultimately resolved, rather than directly specifying Ib/Ic identity. Furthermore, additional unbiased transcriptional profiling in Vglut3−/− animals confirmed that most SGNs assume a nearly normal Ia identity. Thus, IHC-driven activity appears to select a subset of SGNs to take on Ib and Ic fates from a pool of SGNs that would otherwise become Ia.
Although we have focused on IHC-driven events, any change in cochlear activity may also contribute to the final mix of SGNs. Indeed, SGN activity in the neonatal cochlea depends not only on IHCs but also on input from the olivocochlear efferent system. For example, medial efferents form transient cholinergic synapses with IHCs during the first postnatal week (Glowatzki and Fuchs, 2000) and are thus poised to influence IHC-evoked SGN activity during this period of SGN diversification. Lateral efferents could also influence SGN identity, perhaps in a maintenance role as suggested previously (Yin et al., 2014). Sun*, Babola*, Pregernig* et al. also report in this issue of Cell that Type I SGNs can be classified into three broad molecular subtypes that match those we describe here. Consistent with our findings, they demonstrate that subtype-specific marker expression changes upon disruption of mechanotransduction (Tmie−/−) or glutamate release from IHCs (Vglut3−/−), both of which significantly alter SGN spontaneous activity patterns. Regardless of which factors pattern SGN activity during this critical developmental window, their effects may materialize as changes in expression or function of subtype-specific transcription factors, such as Pou4f1 and Runx1. In fact, both Pou4f1 and Runx1 contribute to diversification of dorsal root ganglion neurons (Chen et al., 2006; Qi et al., 2017; Yoshikawa et al., 2007; Zou et al., 2012) and could play similar roles in SGNs, acting downstream of a generic differentiation program headed by the master regulator Gata3 (Appler et al., 2013; Yu et al., 2013). Interestingly, a similar activity-dependent mechanism diversifies cortical interneurons with distinct firing properties, in this case via the transcription factor Er81 (Dehorter et al., 2015). Such a mechanism of neuronal diversification may be particularly effective in the cochlea, where patterns of IHC-driven activity during an early plastic phase, achieved for instance via efferent modulation, could yield mature subtype proportions across the cochlea from a finite pool of generic postmitotic neurons. Under this model, SGN heterogeneity is established by both intrinsic and extrinsic mechanisms: presynaptically patterned activity informs identity during development, yet the SGNs also express transcription factors that induce and maintain subtype-specific cohorts of genes to further shape each neuron’s mature functional properties. An important question for the future is whether SGN identities remain sensitive to activity into adulthood or whether these early developmental events lock them in place.
Implication for disease and therapy
Loss of Vglut3−/− causes total deafness in mice (Seal et al., 2008), while mutation in Vglut3 underlies the deafness DFNA25 in humans (Ruel et al., 2008). Therefore, a major implication of the influence of IHC-driven activity on SGN identity is that some forms of congenital deafness—specifically those that disrupt SGN activity before the onset of hearing—may have an associated SGN pathology. This underscores the need to look beyond the primary molecular lesion in assessments of cellular phenotypes associated with deafness-causing mutations. An outstanding question is whether restoring expression of a defective gene in IHCs is sufficient to correct the sequelae of SGN phenotypes associated with a congenital lesion. The answer lies in part in whether plasticity of SGN identity, particularly its susceptibility to extrinsic influence, persists into adulthood. Should it be necessary to directly target SGNs to re-establish correct proportions, valuable insights may be gained from studies focusing on molecular genetic regulation of SGN subtype identity during development and beyond.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lisa Goodrich (Lisa_Goodrich@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Harvard Medical School and the Massachusetts Eye and Ear Infirmary as appropriate, and conducted following ethical guidelines described in the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. The following mouse lines were used:
Mice harboring the transgenes Bhlhb5Cre/+; Ai14/+ and Bhlhb5Cre/+; Ai14/+; Vglut3−/−were used for single cell RNA-seq experiments. Both males and females aged P25 to P27 were used and all mice were of mixed C57BL/6J and CD1 backgrounds. The Bhlhb5Cre line, in which the coding sequence at the Bhlhe22 locus is replaced with that for Cre recombinase, was kindly provided by Dr. Michael Greenberg (Harvard Medical School, Boston, USA) (Ross et al., 2010). The Vglut3−/− line was obtained from The Jackson Laboratory (B6;129S2-Slc17a8tm1Edw/J; Stock No: 016931) and contains an EGFP sequence knocked into exon 2 of the Slc17a8 locus, which abolishes Vglut3 expression in these mice (Seal et al., 2008). The Ai14 line was obtained from The Jackson Laboratory (Madisen et al., 2010). In addition, the following lines were used to study the developmental emergence of subtype identities by RNAscope: 1) Bhlhb5Cre/+; Ai14/+, 2) Vglut3+/− and 3) Vglut3−/− .All mice used for data collection were born and reared in a barrier animal facility at Harvard Medical School, Boston, USA.
Mice harboring the transgenes MafbCreERT2/+; Ai9/+ were used for immunohistochemistry and neuroanatomical tracing. MafbCreERT2, a bacterial artificial chromosome (BAC) transgenic line (MGI: 5474190), was kindly provided by Dr. Filippo Rijli (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) (Di Meglio et al., 2013). The Ai9 line was obtained from The Jackson Laboratory (Madisen et al., 2010). All mice used for data collection were of mixed C57BL/6J and CD1 background and born and reared in a barrier animal facility at Harvard Medical School, Boston, USA.
Cochleae of ChatCre/+; Ai14/+ mice were used for immunohistochemistry. Both the ChatCre (Rossi et al., 2011) and Ai14 (Madisen et al., 2010) lines were obtained from The Jackson Laboratory. All mice used for data collection were of mixed C57BL/6J and CD1 background and born and reared in a barrier animal facility at Harvard Medical School, Boston, USA.
Animals used for age-related loss of spiral ganglion neurons were of CBA/CaJ background and born and reared in a barrier animal facility at the Massachusetts Eye and Ear Infirmary, Boston, USA. The acoustic environment of the facility has been described previously (Sergeyenko et al., 2013). Mice of both sexes were used at all time points.
CD1 mice (Charles River, Stock No. 022) of both sexes and aged P25–P27 were used for validation of gene expression by RNAscope. Mice were housed 0 to 7 days after purchase in a barrier animal facility at Harvard Medical School, Boston, USA before tissue collection.
METHOD DETAILS
Single cell RNA-sequencing
High throughput scRNA-seq approaches such as Dropseq and Indrop offer ease of profiling and classifying cells but at the cost of low information content per cell. Since our goal was to both classify neurons and gain a deep understanding of the transcriptomic basis of neuronal identities, we chose to combine manual neuron collection with the Smart-seq2 approach that offers a high rate of transcript sampling per cell and sequencing of full-length transcripts. Our workflow is described in detail below.
Collection of single neurons.
Cochleae of P25 - P27 mice (Genotypes: Bhlhb5Cre/+; Ai14/+ and Bhlhb5Cre/+; Ai14/+; Vglut3−/−) were dissected out of the inner ear. Each cochlea was further dissected to isolate either the most apical, middle, or basal turn and subsequent processing of each turn was done separately to permit tracking of each neuron’s origin along the tonotopic axis. Pieces of the cochlea were then digested first with collagenase (25 min at 37 °C) then 40 U/ml papain (25 min at 37 °C) (Worthington, LK003150) before passing through a discontinuous density gradient of ovomucoid protease inhibitor (Worthington, LK003182). The crude dissociation extract was passed through a 40 mm cell strainer (Corning, Inc., 352340) and placed in a petri dish with a glass bottom microwell (Cellvis, D60-14-1N) for manual collection. Neurons expressing tdTomato were identified under a dissecting microscope (Olympus, MVX10) and a small number of them (typically <100) were gently transferred to a clean droplet of transfer buffer (10% ovomucoid protease inhibitor in 1x EBSS (Invitrogen, 14155–063)) using a glass micropipette attached to suction tubing. To get rid of debris and non-neuronal cells that accompany tdTomato+ neurons during the transfer, cells were rinsed five times by aspirating and expelling them sequentially into a series of clean transfer buffer droplets on the same petri dish, taking care to aspirate only tdTomato+ cells each time. After the final rinse, a neuron-enriched pool with no visible tdTomato-negative cells or floating debris was obtained. Neurons were then placed one-by-one at the bottom of individual 0.2 mL tubes (Axygen, 321-10-061) using a fine glass micropipette (typically 40 μm internal diameter at the tip) attached to suction tubing. To avoid contamination resulting from cells that lyse during transfer, glass micropipettes and tubes were never reused after failed transfers. A typical transfer resulted in 1–2 μl of transfer buffer in the tube, most of which was aspirated out of the tube, leaving ~0.3 μL behind. To ensure collection of a healthy single neuron per tube, maintenance of strong red fluorescence, lack of cell shrinkage, and absence of other neurons in the same tube were visually verified for every sample under high magnification. Any sample that did not meet all three of these criteria were discarded. Cells were immediately frozen after transfer and stored at −80 °C for further processing. All cells were collected between 1 PM and 8 PM. Collection of single cells started ~75 minutes after sacrificing the animal and ended after 2 hrs. Time elapsed between euthanization of the animal and freezing of cells varied between 90 and 195 minutes. Both ears of every mouse and mice of both sexes were used.
cDNA and library preparation.
cDNA libraries were made by the Smart-seq2 method, essentially as described previously (PMID 24385147) but with the following modifications: 1) ERCC spike-in controls (1 μl at 1:50000 dilution, Ambion, 4456740) were added at the cell lysis step; 2) first-strand synthesis was done using Primescript Reverse Transcriptase (Takara, 2680A); 3) 19 cycles of PCR amplification was done; 4) the following primers were used:
Oligo-dT30VN: 5’-AATGATACGGCGACCACCGATCGT30VN-3’
Template-switching oligo: 5’-AGACGTGTGCTCTTCCGATCTNNNNNrGrGrG-3’
ISPCR: 5’-AGACGTGTGCTCTTCCGATCT-3’
Sample quality and yield were analyzed using the high-sensitivity DNA kit in a bioanalyzer (Agilent).
qPCR screening.
SGN cell bodies in mice are naturally ensheathed by satellite glia after neonatal stages. We found that a small proportion of SGNs dissociated using the protocol described above lost their associated ensheathing glia, but most retained it. This posed a challenge for obtaining neuron-specific transcriptome due to possible contamination from transcripts derived from satellite glia. Given the low proportion of cells that visibly lacked ensheathing glia under high magnification, we chose to pick tdTomato-positive SGNs without regard to their ensheathment status, and instead enriched for neuron-specific libraries by screening out those containing glia-derived transcripts by qPCR. Specifically, presence of Mpz (F: 5’-GTCAAGTCCCCCAGTAGAA-3’, R: 5’-AGGAGCAAGAGGAAAGCAC-3’) and Plp (F: 5’-AGCAAAGTCAGCCGCAAAAC-3’, R: 5’-CCAGGGAAGCAAAGGGGG −3’) were probed and only libraries that were negative for both transcripts were included in the library enrichment step of the Smart-seq2 protocol.
Tissue processing for mRNA/protein detection
Cochlear whole mounts and sections were processed as described previously. The following primary antibodies were used: anti-Calb2 (1:600), anti-Pou4f1 (anti-Brn3a) (1:300), anti-Parvalbumin (1:500), anti-GluA2 (1:500), anti-CtBP2 (1:500), anti-Myo7a (1:1000), anti-dsRed (1:1000), anti-NF-H (1:1000). For immunostaining of Pou4f1, antigen retrieval in 10 mM citrate buffer (pH 6.0) was done for 20 minutes before commencing with the staining protocol.
For mRNA detection by RNAscope (Advanced Cell Diagnostics), the manufacturer’s protocol was used with the exception that at the end of the protocol, tissues were stained overnight with anti-Parvalbumin, followed by incubation in the appropriate secondary for 1 hr the next day. The following probes were used: Mm-Calb2 (Cat#313641-C3), Mm-Pou4f1 (Cat#414671-C2), Mm-Th (Cat#317621), Mm-Tsc22d3 (Cat#448341), Mm-Epha4 (Cat#419081), Mm-Lypd1 (Cat#318361), Mm-Hcrtr2 (Cat#460881), Mm-Kcns3 (Cat#467371).
QUANTIFICATION AND STATISTICAL ANALYSIS
Sequencing and bioinformatic analysis
Sequencing was done in a NextSeq platform (Illumina) over 2 runs, with similar numbers of samples corresponding to different genetic backgrounds, ages, and cochlear origin mixed across the runs. De-multiplexed raw reads were processed using custom-written UNIX scripts in a shared high-performance compute cluster running CentOS Linux. The following steps and settings were used: 1) low-quality and short reads were trimmed using Trimmomatic (0.33) (PE-phred33 SLIDINGWINDOW:4:20 MINLEN:22 PE-phred33 SLIDINGWINDOW:4:20 MINLEN:22); 2) reads were aligned using STAR to the mouse genome (mm10) to which sequences corresponding to ERCC spike-ins and tdTomato had been appended (--sjdbOverhang 37 \ --outFilterScoreMinOverLread 0.4 \ --outFilterMatchNminOverLread 0.4); 3) reads mapped to the mouse transcriptome were counted using the featureCounts program of the Subread package (v1.4.6). Transcriptome-wide counts for all samples were merged into a table, which was then imported into R (v3.3.2). Analyses and visualization of results within the R environment were done using a custom-written script that utilized Seurat (v 1.4.0.12) for dimension reduction, tSNE, unsupervised clustering and differential expression analyses, edgeR for normalization, and randomForest for supervised classification.
Data normalization.
Type II SGNs have smaller cell bodies than Type I neurons (Brown, 1987). In addition, heterogeneities in cell size both locally and across the tonotopic axis have been noted (personal observations and (Echteler and Nofsinger, 2000)). To account for differences in RNA content arising from cell size heterogeneity, we performed trimmed mean of M-values (TMM) normalization on raw read counts using the edgeR package before importing the expression matrix into Seurat. Cell classification was also conducted without TMM normalization with nearly identical results. No other normalization or transformation was done beside log normalization of counts performed within Seurat. All log-normalized maximum count values for individual genes reported in tSNE plots were normalized further to account for gene length differences. However, these values were used strictly for visual reporting in such plots and not for any other aspect of our analyses.
Cell filter.
Only libraries meeting the following criteria were analyzed: 1) free of glial-markers (i.e., Mpz, Gjc3, Mbp all fl1); 2) total reads >750000; 3) proportion of ERCC spike-in transcripts < 20%; 4) proportion of transcripts from mitochondrial genome < 10%; 4) genes detected Ł 4000.
Dimension reduction.
Principal component analysis (PCA) was done using variable genes defined as described in the Seurat manual (Satija, 2018). Genes expressed (>1 count) in fewer cells than a set cutoff (i.e., cubed root of no. of cells in the quality-filtered dataset) were deemed to be ultra-sparse and hence omitted from the variable gene list used for PCA. In addition, all exogenous transcripts (ERCC spike-in and tdTomato) were removed. Between 8 and 20 top PCs were then selected to generate 2-dimensional embeddings for data visualization using tSNE for various subsets. The set of PCs used always included those contained within the first elbow in a scree plot as well as those deemed statistically significant in Jack Straw plots and explained 60–70% variance in the dataset. Because tSNE outcome can vary slightly from run to run even under the same settings, tSNE coordinates generated in initial runs were saved and re-used in subsequent runs for consistency in visualization.
Unsupervised clustering.
scRNA-seq profiles were classified via an unsupervised graph-based clustering algorithm implemented in Seurat (v1.4.0.12) that embeds cells in a K-nearest neighbor (KNN) graph and determines clusters by iteratively grouping cells while optimizing the standard modularity function (Butler et al., 2018; Satija et al., 2015). In this approach, granularity of the clustering is dependent on a user-specified ‘resolution’ parameter within the FindCluster function in Seurat. To ensure unbiased selection of the appropriate number of clusters for classifying Type I SGNs, clustering was conducted using 46 different values of this parameter, ranging from 0.4 to 1.3 with increments of 0.02. Predictions of 3 clusters were obtained across 42 out of those 46 runs (91%) (Fig. S1B), with 97% of cells being assigned the same identity across such runs (Fig. S1C). This led to the conclusion that the major structure in the single-cell transcriptomic data was captured by classification into 3 clusters, and by extension, reflected the existence of 3 distinct neuronal subtypes. Additionally, hierarchical clustering based on Ward’s method and Euclidean distances was conducted using the hclust package in R. In both approaches, only highly variable genes (as described above) were used.
Training an ensemble learning-based classifier.
Consistent with previous reports, we observed that deriving consistent cell type classification became progressively harder with decreasing cell number. To rule out that identifying subtypes among neurons from Vglut3−/− mice was not affected by sample size, we implemented an ensemble learning method to derive an independent classification. The same 3 subtypes identified across all Type I SGNs (n = 179) in our data could be identified even when we analyzed SGNs from the middle turn alone (n = 88). Therefore, we employed ensemble learning to generate a classifier trained on just the middle turn neurons using the randomForest package in R. The training set consisted of 44 randomly selected neurons (15, 18, 11 of A, B, C subtypes, respectively). We reasoned that differentially expressed genes must be detectable broadly within a cluster for it to be informative for classifying unknown cells, so only genes expressed in >50% of cells in the cluster they are markers for and <50% in the rest were selected. This resulted in a 44 × 233 cell-by-gene matrix, which was used as the training data set. To assess the classifier’s performance, we used it to predict classes for all wildtype cells not part of the training set, which comprised the test set and included 44, 50, and 41 cells from the middle, apex, and base, respectively. The results were compared against classification produced by graph-based clustering implemented in Seurat. Training parameters were tuned until the median out-ofbag error rate reached zero and performance in the test set peaked (99%) across >10000 combinations of the mtry, nodesize and ntree parameters. The following settings were picked: mtry = 3, nodesize = 1, ntree = 1000.
Image acquisition and analysis
All RNAscope images were acquired at Nyquist settings using a point-scanning confocal microscope (Leica SP8, 63x oil-immersion objective) equipped with hybrid detectors. Image stacks were then analyzed semi-automatically using Imaris (Bitplane, UK). Briefly, cell volumes were segmented using either anti-Parvalbumin stain or anti-dsRed stain (in animals expressing the tdTomato transgene in SGNs). Any cell clipped along the XYZ axes was removed before automatic detection of RNAscope puncta. Parameters for both cell segmentation and puncta detection were set after visually guided search for optimal parameters. To account for variability in signal:noise ratio across experimental conditions, puncta counts for cells from each image were rescaled to fit max and min values across all cells within each biological group. Cells with puncta count above 10% of the maximum value were considered positive for the marker.
Confocal images of cochlear wholemounts were acquired using the same hardware as above. Images were taken as a 1024 × 1024 pixel raster at 63x (2x digital zoom) and a Z-sampling rate of 0.308 μm. Image stacks were analyzed using Amira (FEI, USA). Volumes of individual pre-synaptic ribbons were computed using Amira’s 3D rendering and masking functions. Briefly, each pre-synaptic ribbon was reconstructed by a 3D ‘isosurface’ which surrounded all the pixels that defined the ribbon. The volume of the isosurface defined the volume of the pre-synaptic ribbon. Differences in sample preparations were normalized by dividing each isosurface by the median isosurface size in a given Z-stack. The spatial segregation of individually labelled Type I SGN fibers along the basolateral surface of the inner hair cell were analyzed by generating an isosurface around both the afferent fiber and the target pre-synaptic puncta. The YZ coordinates of the pre-synaptic ribbon and adjacent afferent terminal were remapped onto an XY plane and assessed. A line bisecting the inner hair cell nucleus and cuticular/basolateral axis defined the 0 position of the pre-synaptic ribbons along the modiolar/pillar axis.
Optical sections of the OSL bundle were taken 50 μm away from the base of the IHCs in Amira (FEI, USA) and all subsequent analyses involving fluorescence intensity measurements was done using Image J (1.49V). To analyze CALB2 levels in SGN fibers in the OSL, regions of interest (ROI) were drawn around SGN fibers positive for NF-H in optical cross-sections of confocal Z-stacks. Mean fluorescence intensity values for CALB2 were then calculated for all ROIs and normalized to the highest value within an image. Somatic CALB2 fluorescence intensity was quantified in MafbCreERT2;Ai9 animals, which exhibit sparse labeling of SGN cell bodies with the tdTomato reporter . ROIs were drawn around tdTomato+ SGN cell bodies and fluorescence intensity values for CALB2 across all ROIs within an image were normalized to the highest mean fluorescence value in the same image.
Clustering of RNAscope-based expression data.
An unsupervised machine learning algorithm (K-means clustering) was utilized to group the CALB2 OSL fluorescence intensity data (Fig. S5C, D) into a specified number of clusters. The number of clusters (k) was determined based on an elbow plot (sum of squared errors, SSE vs. k clusters) and chosen at the point where the slope approached zero. A total of 3 clusters were found to be appropriate for the CALB2 OSL dataset. Cluster boundaries defined by K-means clustering was used to set the fluorescence intensity cutoffs for ‘low’, ‘medium’, and ‘high’ CALB2 groups (Fig. S5D). K-means clustering was also performed on the RNAscope-based Lypd1/Calb2 expression data presented in Figure 6. Elbow plots utilized for each dataset are shown in Fig. S6C, D. All statistical analyses of in situ expression data and immunostaining data in Figs. 4 and 5 were done in GraphPad Prism 6.0. Analyses of such data in all other figures were done in R.
DATA AND SOFTWARE AVAILABILITY
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE114997.
Supplementary Material
Figure S1: Quality control metrics and Gene Ontology results. Related to Fig. 1 and 2. (A) Scatterplots showing technical and biological criteria used to filter cells before analysis. Red dotted lines indicate cutoff value and red arrow indicates direction of exclusion. (B) Setting the resolution parameter to values between 0.40 and 1.30 for graph-based clustering in Seurat yielded 2 to 5 clusters, with 42 out of 46 values (91%) predicting 3 clusters. (C) Agreement with regard to cell identity among all variations of the resolution parameter that yielded 3 clusters. For each cell, a consensus score was calculated as the percentage of times the same identity was assigned to it across clustering runs that varied only in terms of resolution. Consensus was 100% for 174 out of 179 cells (97%). (D) Projection of SGN transcriptomes onto PCA space (PC2 versus PC4) reveals groups that correspond closely with those identified by graph-based clustering. Each group in the PCA plot contains SGNs from all tonotopic regions. (E) Results of hierarchical clustering by Ward’s method depicted as a dendrogram. The three major branches correspond well with SGN subtype identities predicted by graph-based clustering but not with their tonotopic position. (F) Segregation of SGNs is not driven by technical variables or experimental factors such as sequencing batch. P values refer to Chi-squared test of independence. (G) There were no significant subtype-specific differences in the detection of spike-in transcripts, mitochondrial transcripts, or number of genes detected. P values are indicated next to pairs of dots that represent subtypes compared by Tukey’s HSD test if the data was normally distributed data and Dunn’s test otherwise. (H,I) Absence of glial gene expression (H) and uniform expression of known pan-neuronal genes (I) across the three Type I SGN clusters. Numbers in upper right corner indicate highest expression (Max) observed for each gene. (J, K) Graphical representation of results of Gene Ontology analysis of genes expressed differentially between Type I and II SGNs (J) or among Type I SGNs (K). A few examples of statistically significantly enriched categories are named.
Figure S2: SGN subtypes express unique cohorts of genes that influence neuronal physiology. Related to Fig. 2.
Dot-matrix plots showing expression of gene families relevant to neuronal physiology. Each dot is a neuron profiled by scRNA-seq and neurons are grouped based on subtype identity assigned by unsupervised clustering. Numbers on the right indicate the highest expression (Max) observed for each gene.
Figure S3: SGN subtypes express unique cohorts of genes that influence neurotransmission and neurite guidance. Related to Fig. 2.
Dot-matrix plots showing expression of gene families relevant to neurotransmission and neurite guidance. Each dot is a neuron profiled by scRNA-seq and neurons are grouped based on subtype identity assigned by unsupervised clustering. Numbers on the right indicate the highest expression (Max) observed for each gene.
Figure S4: Tonotopic differences in gene expression across all SGNs and within SGN subtypes. Related to Fig. 3.
(A) Heat map showing the top 100 most differentially expressed genes across the tonotopic axis. Each column represents a single cell and genes are in rows. Cells were grouped based on tonotopic origin. Panels on the right show examples of genes enriched in the apex (top), middle (middle) and base (bottom). (B) RNAscope of tissue sections (P25–P27) confirms even expression of the pan-neuronal gene Tubb3 in the apex, middle, and base of the cochlea, quantified below. (C) Plots showing expression (mean ± SEM) of genes that were invariant across the tonotopic axis for all pooled SGNs (dashed lines) and for each subtype (Ia, green; Ib, purple; Ic, blue). (D) Examples of genes that are robust markers for Ia, Ib, and Ic subtypes across all tonotopic regions. Mean expression levels (± SEM) are shown for Ia (A), Ib (B), and Ic (C) SGNs taken from the apex (gray), middle (red), or base (blue) of the cochlea. (E) Examples of genes whose cross-subtype variation in expression (± SEM) differs among SGNs from different cochlear regions (apex, gray; middle, red; base, blue). For example, there is significantly lower expression of Anxa5 in Ia SGNs from the base relative to those from the middle and the apex, but little variation among Ib or Ic SGNs. Pairs of dots indicate p values for comparisons across tonotopic regions by Tukey’s HSD test if the data was normally distributed data and Dunn’s test otherwise. P values are reported only for statistically significant differences. Scale bars: 10 μm (B).
Figure S5: Operational definition of SGN subtypes according to CALB2 intensity. Related to Fig. 4.
(A-B) Wholemount cochlea from MafbCreERT2; Ai9 animals were double-stained for CALB2 (green) and tdTomato (red). Individual tdTomato labeled SGNs could be traced from their cell bodies to the organ of Corti, shown in a top down (XY) view (top). Position in the OSL was assessed in cross-sectional (XZ) views through confocal image stacks (below). CALB2 intensities in cell bodies and processes were correlated for each neuron. Examples of one CALB2+ SGN (yellow arrow) and one CALB2low SGN (white arrow) are shown, with quantification for all analyzed SGNs in B. (C–D) SGN subtypes were operationally defined based on CALB2 levels. K-means clustering of all measured CALB2 fluorescence intensities in fibers revealed that the expression values are best split into three clusters (red dotted line, C). Fibers were divided into low (n=113), medium (n=78), and high (n=116) CALB2+ groups as defined by the K-means cluster boundaries, indicated by colored shading in a histogram of all analyzed fibers (n=214, 5 animals). (E) Cross-sectional views (XZ) through the OSL of ChatCre;Ai14 cochlear wholemount preparations immunostained for CALB2 (green) and tdTomato (red). ChatCre;Ai14 labels olivocochlear efferents. Dotted lines indicate the upper limit (towards SV), mid-point and lower limit (towards ST) of the afferent fiber bundle. (F-G) Histograms show even distribution of efferent fibers (red) but SV-biased distribution of low CALB2 (blue) fibers in the OSL (F). The average number of fibers per region is quantified in G. P values indicate results of Kolmogorov-Smirnov’s test (F) and results of Tukey’s HSD test following one-way ANOVA (G). Scale bars: 10 μm.
Figure S6: Analysis of Lypd1 and Calb2 mRNA levels in the aging mouse cochlea. Related to Fig. 5.
(A–B) In sections of the cochlea at 108 wks, autofluorescent aggregates (arrows) appear in all fluorescent channels (anti-parvalbumin (PARV), A; Calb2 RNAscope, A’; Lypd1 RNAscope, A”). In order to count mRNA puncta reliably, this signal was removed prior to cell segmentation and quantification by applying a Gaussian filter across the fluorescent channels (B-B”). (C) Scatterplots of Lypd1 and Calb2 levels in all SGNs analyzed at 32, 64, and 108 wks (n=5 animals, 212 cells at 32 weeks, n=5 animals, 175 cells at 64 weeks, n=5 animals, 151 cells at 108 weeks, respectively). (D–E) K-means clustering analysis showed that SGN populations could be sorted into three groups based on Calb2 levels at 32 wks (top) and 64 wks (middle), but only into 2 groups at 108 wks (bottom), consistent with a loss of Type Ic SGNs. Likewise, whereas two clusters of Lypd1+ SGNs can be defined at 32 and 64 wks, only one cluster remains at 108 wks (E). Scale bar: 10 μm (A,B).
Figure S7: Marker expression in embryonic and Vglut3-/- animals, and comparison of supervised and unsupervised clustering approaches. Related to Fig. 6 and 7.
(A–C) Detection of Lypd1 (magenta) and Calb2 (green) transcripts by RNAscope at E18.5 reveals early co-expression in individual SGNs, with Calb2 expressed broadly but Lypd1 expressed in a subset, quantified in B. Whereas the vast majority of SGNs express Calb2, a smaller proportion express Lypd1 (C). (D) The level of expression of select subtype-specific markers is overlaid in a tSNE plot containing both WT (M1–M3 clusters) and Vglut3−/− cells (M4–M5 clusters). Cluster identities in the key to the right are based on Fig. 7A. Numbers on the upper left indicate the highest expression (Max) observed for each gene. (E) tSNE plots with dot colors indicating subtype identity assigned by unsupervised graph-based clustering (left) and by Random Forest (right), a supervised clustering approach. (F) Scatterplot showing subtype identity of every cell predicted by unsupervised (X-axis, “Graph-based”) and supervised clustering (Y-axis, “Random Forest”) methods. Only cells that had not been used for training the Random Forest classifier are shown. Cells that fall on the diagonal dotted line are the ones predicted correctly by the supervised method. (G) Subtype proportions among SGNs from three tonotopic regions predicted by each clustering method. Scale bar: 10 μm (A).
Table S1, Related to Figure 1,2,3: Differentially expressed genes across all cell types/groups analyzed in the study. Each tab in the spreadsheet (A–F) contains data for a different set of comparisons as indicated. Top 100 or all genes (if <100) enriched in a subgroup are indicated in red. Statistical significance was assessed by conducting the likelihood-ratio test for gene expression in single cells in the Seurat R package.
Table S2, Related to Figures 1–7: Number of cells, animals and their age across all experiments reported in this study.
Highlights.
Three molecular subtypes of Type I spiral ganglion neurons (SGNs) identified
Additional molecular variations exist across subtypes and along the tonotopic axis
Subtype identities emerge around birth and are refined over postnatal stages
Disruption of hair cell-driven activity leads to aberrant SGN subtype diversity
Acknowledgments
We thank Dr. Sinisa Hrvatin (Harvard) for help with sequencing and helpful discussions, Dr. Aravind Parthasarathy and Karina Gaft (MEEI) for help with processing of aged cochleas, Jocelyn Curran and Laura Schell (Harvard) for technical help, Rory Kirchner (Harvard) for input on bioinformatics, the Image and Data Analysis Core (IDAC, Harvard) for guidance on image analysis, Dr. Filipo Rijli (FMI, Switzerland) for sharing MafbCreERT2;Ai9 mice, and Drs. David Corey and Michael Greenberg (Harvard) for sharing equipment. We also thank Drs. Matthew B. Kelley (NIH) and Ulrich Mueller (Johns Hopkins) for communicating their preliminary results and Drs. Matthew Kelley (NIDCD), David Ginty, Gord Fishell, and lab members (Harvard) for critical reading of the manuscript. Bioinformatic analysis was conducted partly on the Orchestra High Performance Compute Cluster (Harvard), a resource supported partially through grant NCRR 1S10RR028832–01. This work was supported by the Lefler Family Small Grant, Harvard Brain Initiative, NIH T32 AG 222–24 (CC) and 5 T32 NS007484–14 (BRS), an F32 Postdoctoral Individual National Research Service Award 5F32DC014371 (BRS), a Goldenson Postdoctoral Fellowship (BRS), and NIH R01 DC00188 (MCL) and DC009223 (LVG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Abraira VE, and Ginty DD (2013). The sensory neurons of touch. Neuron 79, 618–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson CL, Reid M. a., Mo ZL, Bowne-English J, and Davis RL (2002). Firing features and potassium channel content of murine spiral ganglion neurons vary with cochlear location. J. Comp. Neurol 447, 331–350. [DOI] [PubMed] [Google Scholar]
- Appler JM, Lu CC, Druckenbrod NR, Yu W-M, Koundakjian EJ, and Goodrich LV (2013). Gata3 Is a Critical Regulator of Cochlear Wiring. J. Neurosci 33, 3679–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Barbary A (1991). Auditory nerve of the normal and jaundiced rat. I. Spontaneous discharge rate and cochlear nerve histology. Hear. Res 54, 75–90. [DOI] [PubMed] [Google Scholar]
- Berglund AM, and Ryugo DK (1987). Hair cell innervation by spiral ganglion neurons in the mouse. J. Comp. Neurol 255, 560–570. [DOI] [PubMed] [Google Scholar]
- Borg E, Engström B, Linde G, and Marklund K (1988). Eighth nerve fiber firing features in normal-hearing rabbits. Hear. Res 36, 191–201. [DOI] [PubMed] [Google Scholar]
- Brown MC (1987). Morphology of labeled afferent fibers in the guinea pig cochlea. J. Comp. Neurol 260, 591–604. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X-J, and Oertel D (2010). Auditory Nerve Fibers Excite Targets Through Synapses That Vary in Convergence, Strength, and Short-Term Plasticity. J. Neurophysiol 104, 2308–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Broom DC, Liu Y, De Nooij JC, Li Z, Cen C, Samad OA, Jessell TM, Woolf CJ, and Ma Q (2006). Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron 49, 365–377. [DOI] [PubMed] [Google Scholar]
- Chen WC, Xue HZ, Hsu Y. (Lucy), Liu Q, Patel S, and Davis RL (2011). Complex distribution patterns of voltage-gated calcium channel Į-subunits in the spiral ganglion. Hear. Res 278, 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalupes JA, Young ED, and Gibson DJ (1984). Effects of continuous noise backgrounds on rate response of auditory nerve fibers in cat. J. Neurophysiol 51, 1326–1344. [DOI] [PubMed] [Google Scholar]
- Crozier RA, and Davis RL (2014). Unmasking of Spiral Ganglion Neuron Firing Dynamics by Membrane Potential and Neurotrophin-3. J. Neurosci 34, 9688–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio CA, and Hendrickson AE (1991). Organization and development of the primate photoreceptor mosaic. Prog. Retin. Res 10, 89–120. [Google Scholar]
- Dehorter N, Ciceri G, Bartolini G, Lim L, Del Pino I, and Marín O (2015). Tuning of fast-spiking interneuron properties by an activity-dependent transcriptional switch. Science (80-. ). 349, 1216–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio T, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, Hrycaj SM, Roska B, Peters AHFM, Eichmann A, et al. (2013). Ezh2 Orchestrates Topographic Migration and Connectivity of Mouse Precerebellar Neurons. Science (80-. ). 339, 204–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echteler SM, and Nofsinger YC (2000). Development of ganglion cell topography in the postnatal cochlea. J. Comp. Neurol 425, 436–446. [PubMed] [Google Scholar]
- Edgar R, Domrachev M, and Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores EN, Duggan A, Madathany T, Hogan AK, Marquez FG, Kumar G, Seal RP, Edwards RH, Liberman MC, and Garcia-Anoveros J (2015). A non-canonical pathway from cochlea to brain signals tissue-damaging noise. Curr. Biol 25, 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Otero J, and Davis RL (2011). Synaptic proteins are tonotopically graded in postnatal and adult type I and type II spiral ganglion neurons. J. Comp. Neurol 519, 1455–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank T, Khimich D, Neef A, and Moser T (2009). Mechanisms contributing to synaptic Ca2+ signals and their heterogeneity in hair cells. Proc. Natl. Acad. Sci. U. S. A 106, 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman AC, Kujawa SG, and Liberman MC (2013). Noise-induced cochlear neuropathy is selective for fibers with low spontaneous rates. J. Neurophysiol 110, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowatzki E, and Fuchs PA (2000). Cholinergic synaptic inhibition of inner hair cells in the neonatal mammalian cochlea. Science 288, 2366–2368. [DOI] [PubMed] [Google Scholar]
- Glowatzki E, and Fuchs PA (2002). Transmitter release at the hair cell ribbon synapse. Nat. Neurosci 5, 147–154. [DOI] [PubMed] [Google Scholar]
- Guinan JJ (2017). Olivocochlear efferents: Their action, effects, measurement and uses, and the impact of the new conception of cochlear mechanical responses. Hear. Res 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LC, Barclay M, Lee K, Peter S, Housley GD, Thorne PR, and Montgomery JM (2012). Synaptic profiles during neurite extension, refinement and retraction in the developing cochlea. Neural Dev. 7, 7–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, and Desplan C (2010). Binary fate decisions in differentiating neurons. Curr. Opin. Neurobiol 20, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase T, and Liberman MC (1992). Spatial organization of the auditory nerve according to spontaneous discharge rate. J. Comp. Neurol 319, 312–318. [DOI] [PubMed] [Google Scholar]
- Kiang NY, Watanabe T, Thomas EC, and Clark LF (1965). Discharge Patterns of Single Fibers in the Cat’s Auditory Nerve (Cambridge, MA: MIT Press; ). [Google Scholar]
- Kiang NYS, Sachs MB, and Peake WT (1967). Shapes of Tuning Curves for Single Auditory-Nerve Fibers. J. Acoust. Soc. Am 42, 1341–1342. [DOI] [PubMed] [Google Scholar]
- Kim KX, and Rutherford MA (2016). Maturation of NaV and KV Channel Topographies in the Auditory Nerve Spike Initiator before and after Developmental Onset of Hearing Function. J. Neurosci 36, 2111–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundakjian EJ, Appler JL, and Goodrich LV (2007). Auditory Neurons Make Stereotyped Wiring Decisions before Maturation of Their Targets. J. Neurosci 27, 14078–14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC (1978). Auditory-nerve response from cats raised in a low-noise chamber. J. Acoust. Soc. Am 63, 442–455. [DOI] [PubMed] [Google Scholar]
- Liberman MC (1980). Efferent synapses in the inner hair cell area of the cat cochlea: An electron microscopic study of serial sections. Hear. Res 3, 189–204. [DOI] [PubMed] [Google Scholar]
- Liberman MC (1982). Single-neuron labeling in the cat auditory nerve. Science 216, 1239–1241. [DOI] [PubMed] [Google Scholar]
- Liberman MC (1991). Central projections of auditory-nerve fibers of differing spontaneous rate. I. Anteroventral cochlear nucleus. J. Comp. Neurol 313, 240–258. [DOI] [PubMed] [Google Scholar]
- Liberman MC (2017). Noise-induced and age-related hearing loss: new perspectives and potential therapies. F1000Research 6, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LD, and Liberman MC (2016). Postnatal maturation of auditory-nerve heterogeneity, as seen in spatial gradients of synapse morphology in the inner hair cell area. Hear. Res 339, 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LD, Wang H, and Liberman MC (2011). Opposing Gradients of Ribbon Size and AMPA Receptor Expression Underlie Sensitivity Differences among Cochlear-Nerve/Hair-Cell Synapses. J. Neurosci 31, 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, and Davis RL (2014). Calretinin and calbindin distribution patterns specify subpopulations of type I and type II spiral ganglion neurons in postnatal murine cochlea. J. Comp. Neurol 522, 2299–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Lee E, and Davis RL (2014). Heterogeneous intrinsic excitability of murine spiral ganglion neurons is determined by Kv1 and HCN channels. Neuroscience 257, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AC, Frank T, Khimich D, Hoch G, Riedel D, Chapochnikov NM, Yarin YM, Harke B, Hell SW, Egner A, et al. (2009). Tuning of synapse number, structure and function in the cochlea. Nat. Neurosci 12, 444–453. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Noda T, and Dabdoub A (2017). Dynamic expression of Sox2, Gata3, and Prox1 during primary auditory neuron development in the mammalian cochlea. PLoS One 12, e0170568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, and Sandberg R (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098. [DOI] [PubMed] [Google Scholar]
- Qi L, Huang C, Wu X, Tao Y, Yan J, Shi T, Cao C, Han L, Qiu M, Ma Q, et al. (2017). Hierarchical Specification of Pruriceptors by Runt-Domain Transcription Factor Runx1. J. Neurosci 37, 5549–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, Jung C, Hu L, Mok SI, Shah A, Savner EM, et al. (2010). Loss of Inhibitory Interneurons in the Dorsal Spinal Cord and Elevated Itch in Bhlhb5 Mutant Mice. Neuron 65, 886–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J, Balthasar N, Olson D, Scott M, Berglund E, Lee CE, Choi MJ, Lauzon D, Lowell BB, and Elmquist JK (2011). Melanocortin-4 Receptors Expressed by Cholinergic Neurons Regulate Energy Balance and Glucose Homeostasis. Cell Metab. 13, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel J, Emery S, Nouvian R, Bersot T, Amilhon B, Van Rybroek JM, Rebillard G, Lenoir M, Eybalin M, Delprat B, et al. (2008). Impairment of SLC17A8 Encoding Vesicular Glutamate Transporter-3, VGLUT3, Underlies Nonsyndromic Deafness DFNA25 and Inner Hair Cell Dysfunction in Null Mice. Am. J. Hum. Genet 83, 278–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryugo DK (2008). Projections of low spontaneous rate, high threshold auditory nerve fibers to the small cell cap of the cochlear nucleus in cats. Neuroscience 154, 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MB, and Abbas PJ (1974). Rate versus level functions for auditory nerve fibers in cats: tone burst stimuli. J. Acoust. Soc. Am 56, 1835–1847. [DOI] [PubMed] [Google Scholar]
- Satija R, 2018. Seurat - Guided Clustering Tutorial [WWW Document]. URL http://satijalab.org/seurat/pbmc3k_tutorial.html (accessed 4.25.18).
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedt RA (1989). Spontaneous rates, thresholds and tuning of auditory-nerve fibers in the gerbil: Comparisons to cat data. Hear. Res 42, 23–35. [DOI] [PubMed] [Google Scholar]
- Schmiedt RA, Mills JH, and Boettcher FA (1996). Age-related loss of activity of auditory-nerve fibers. J. Neurophysiol 76, 2799–2803. [DOI] [PubMed] [Google Scholar]
- Seal RP, Akil O, Yi E, Weber CM, Grant L, Yoo J, Clause A, Kandler K, Noebels JL, Glowatzki E, et al. (2008). Sensorineural Deafness and Seizures in Mice Lacking Vesicular Glutamate Transporter 3. Neuron 57, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeyenko Y, Lall K, Liberman MC, and Kujawa SG (2013). Age-Related Cochlear Synaptopathy: An Early-Onset Contributor to Auditory Functional Decline. J. Neurosci 33, 13686–13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnerson A, and Pujol R (1981). Age-related changes in the C57BL/6J mouse cochlea. I. Physiological findings. Brain Res 254, 65–75. [DOI] [PubMed] [Google Scholar]
- Spoendlin H (1969). Innervation patterns in the organ of corti of the cat. Acta Otolaryngol. 67, 239–254. [DOI] [PubMed] [Google Scholar]
- Spoendlin H (1979). Sensory neural organization of the cochlea. J. Laryngol. Otol 93, 853–877. [DOI] [PubMed] [Google Scholar]
- Taberner AM, and Liberman MC (2005). Response Properties of Single Auditory Nerve Fibers in the Mouse. J. Neurophysiol 93, 557–569. [DOI] [PubMed] [Google Scholar]
- Tekinay ABA, Nong Y, Miwa JM, Lieberam I, Ibanez-Tallon I, Greengard P, and Heintz N (2009). A role for LYNX2 in anxiety-related behavior. Proc. Natl. Acad. Sci. U. S. A 106, 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji J, and Liberman MC (1997). Intracellular labeling of auditory nerve fibers in guinea pig: Central and peripheral projections. J. Comp. Neurol 381, 188–202. [PubMed] [Google Scholar]
- Weisz C, Glowatzki E, and Fuchs P (2009). The postsynaptic function of type II cochlear afferents. Nature 461, 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz CJC, Glowatzki E, and Fuchs PA (2014). Excitability of Type II Cochlear Afferents. J. Neurosci 34, 2365–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann C, and Moser T (2015). Relating structure and function of inner hair cell ribbon synapses. Cell Tissue Res. 361, 95–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter IM, Robertson D, and Yates GK (1990). Diversity of characteristic frequency rate-intensity functions in guinea pig auditory nerve fibres. Hear. Res 45, 191–202. [DOI] [PubMed] [Google Scholar]
- Yin Y, Liberman LD, Maison SF, and Liberman MC (2014). Olivocochlear innervation maintains the normal modiolar-pillar and habenular-cuticular gradients in cochlear synaptic morphology. JARO - J. Assoc. Res. Otolaryngol 15, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Senzaki K, Yokomizo T, Takahashi S, Ozaki S, and Shiga T (2007). Runx1 selectively regulates cell fate specification and axonal projections of dorsal root ganglion neurons. Dev. Biol 303, 663–674. [DOI] [PubMed] [Google Scholar]
- Yu WM, Appler JM, Kim YH, Nishitani AM, Holt JR, and Goodrich LV (2013). A Gata3-Mafb transcriptional network directs post-synaptic differentiation in synapses specialized for hearing. Elife e01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Li S, Klein WH, and Xiang M (2012). Brn3a/Pou4f1 regulates dorsal root ganglion sensory neuron specification and axonal projection into the spinal cord. Dev. Biol 364, 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Quality control metrics and Gene Ontology results. Related to Fig. 1 and 2. (A) Scatterplots showing technical and biological criteria used to filter cells before analysis. Red dotted lines indicate cutoff value and red arrow indicates direction of exclusion. (B) Setting the resolution parameter to values between 0.40 and 1.30 for graph-based clustering in Seurat yielded 2 to 5 clusters, with 42 out of 46 values (91%) predicting 3 clusters. (C) Agreement with regard to cell identity among all variations of the resolution parameter that yielded 3 clusters. For each cell, a consensus score was calculated as the percentage of times the same identity was assigned to it across clustering runs that varied only in terms of resolution. Consensus was 100% for 174 out of 179 cells (97%). (D) Projection of SGN transcriptomes onto PCA space (PC2 versus PC4) reveals groups that correspond closely with those identified by graph-based clustering. Each group in the PCA plot contains SGNs from all tonotopic regions. (E) Results of hierarchical clustering by Ward’s method depicted as a dendrogram. The three major branches correspond well with SGN subtype identities predicted by graph-based clustering but not with their tonotopic position. (F) Segregation of SGNs is not driven by technical variables or experimental factors such as sequencing batch. P values refer to Chi-squared test of independence. (G) There were no significant subtype-specific differences in the detection of spike-in transcripts, mitochondrial transcripts, or number of genes detected. P values are indicated next to pairs of dots that represent subtypes compared by Tukey’s HSD test if the data was normally distributed data and Dunn’s test otherwise. (H,I) Absence of glial gene expression (H) and uniform expression of known pan-neuronal genes (I) across the three Type I SGN clusters. Numbers in upper right corner indicate highest expression (Max) observed for each gene. (J, K) Graphical representation of results of Gene Ontology analysis of genes expressed differentially between Type I and II SGNs (J) or among Type I SGNs (K). A few examples of statistically significantly enriched categories are named.
Figure S2: SGN subtypes express unique cohorts of genes that influence neuronal physiology. Related to Fig. 2.
Dot-matrix plots showing expression of gene families relevant to neuronal physiology. Each dot is a neuron profiled by scRNA-seq and neurons are grouped based on subtype identity assigned by unsupervised clustering. Numbers on the right indicate the highest expression (Max) observed for each gene.
Figure S3: SGN subtypes express unique cohorts of genes that influence neurotransmission and neurite guidance. Related to Fig. 2.
Dot-matrix plots showing expression of gene families relevant to neurotransmission and neurite guidance. Each dot is a neuron profiled by scRNA-seq and neurons are grouped based on subtype identity assigned by unsupervised clustering. Numbers on the right indicate the highest expression (Max) observed for each gene.
Figure S4: Tonotopic differences in gene expression across all SGNs and within SGN subtypes. Related to Fig. 3.
(A) Heat map showing the top 100 most differentially expressed genes across the tonotopic axis. Each column represents a single cell and genes are in rows. Cells were grouped based on tonotopic origin. Panels on the right show examples of genes enriched in the apex (top), middle (middle) and base (bottom). (B) RNAscope of tissue sections (P25–P27) confirms even expression of the pan-neuronal gene Tubb3 in the apex, middle, and base of the cochlea, quantified below. (C) Plots showing expression (mean ± SEM) of genes that were invariant across the tonotopic axis for all pooled SGNs (dashed lines) and for each subtype (Ia, green; Ib, purple; Ic, blue). (D) Examples of genes that are robust markers for Ia, Ib, and Ic subtypes across all tonotopic regions. Mean expression levels (± SEM) are shown for Ia (A), Ib (B), and Ic (C) SGNs taken from the apex (gray), middle (red), or base (blue) of the cochlea. (E) Examples of genes whose cross-subtype variation in expression (± SEM) differs among SGNs from different cochlear regions (apex, gray; middle, red; base, blue). For example, there is significantly lower expression of Anxa5 in Ia SGNs from the base relative to those from the middle and the apex, but little variation among Ib or Ic SGNs. Pairs of dots indicate p values for comparisons across tonotopic regions by Tukey’s HSD test if the data was normally distributed data and Dunn’s test otherwise. P values are reported only for statistically significant differences. Scale bars: 10 μm (B).
Figure S5: Operational definition of SGN subtypes according to CALB2 intensity. Related to Fig. 4.
(A-B) Wholemount cochlea from MafbCreERT2; Ai9 animals were double-stained for CALB2 (green) and tdTomato (red). Individual tdTomato labeled SGNs could be traced from their cell bodies to the organ of Corti, shown in a top down (XY) view (top). Position in the OSL was assessed in cross-sectional (XZ) views through confocal image stacks (below). CALB2 intensities in cell bodies and processes were correlated for each neuron. Examples of one CALB2+ SGN (yellow arrow) and one CALB2low SGN (white arrow) are shown, with quantification for all analyzed SGNs in B. (C–D) SGN subtypes were operationally defined based on CALB2 levels. K-means clustering of all measured CALB2 fluorescence intensities in fibers revealed that the expression values are best split into three clusters (red dotted line, C). Fibers were divided into low (n=113), medium (n=78), and high (n=116) CALB2+ groups as defined by the K-means cluster boundaries, indicated by colored shading in a histogram of all analyzed fibers (n=214, 5 animals). (E) Cross-sectional views (XZ) through the OSL of ChatCre;Ai14 cochlear wholemount preparations immunostained for CALB2 (green) and tdTomato (red). ChatCre;Ai14 labels olivocochlear efferents. Dotted lines indicate the upper limit (towards SV), mid-point and lower limit (towards ST) of the afferent fiber bundle. (F-G) Histograms show even distribution of efferent fibers (red) but SV-biased distribution of low CALB2 (blue) fibers in the OSL (F). The average number of fibers per region is quantified in G. P values indicate results of Kolmogorov-Smirnov’s test (F) and results of Tukey’s HSD test following one-way ANOVA (G). Scale bars: 10 μm.
Figure S6: Analysis of Lypd1 and Calb2 mRNA levels in the aging mouse cochlea. Related to Fig. 5.
(A–B) In sections of the cochlea at 108 wks, autofluorescent aggregates (arrows) appear in all fluorescent channels (anti-parvalbumin (PARV), A; Calb2 RNAscope, A’; Lypd1 RNAscope, A”). In order to count mRNA puncta reliably, this signal was removed prior to cell segmentation and quantification by applying a Gaussian filter across the fluorescent channels (B-B”). (C) Scatterplots of Lypd1 and Calb2 levels in all SGNs analyzed at 32, 64, and 108 wks (n=5 animals, 212 cells at 32 weeks, n=5 animals, 175 cells at 64 weeks, n=5 animals, 151 cells at 108 weeks, respectively). (D–E) K-means clustering analysis showed that SGN populations could be sorted into three groups based on Calb2 levels at 32 wks (top) and 64 wks (middle), but only into 2 groups at 108 wks (bottom), consistent with a loss of Type Ic SGNs. Likewise, whereas two clusters of Lypd1+ SGNs can be defined at 32 and 64 wks, only one cluster remains at 108 wks (E). Scale bar: 10 μm (A,B).
Figure S7: Marker expression in embryonic and Vglut3-/- animals, and comparison of supervised and unsupervised clustering approaches. Related to Fig. 6 and 7.
(A–C) Detection of Lypd1 (magenta) and Calb2 (green) transcripts by RNAscope at E18.5 reveals early co-expression in individual SGNs, with Calb2 expressed broadly but Lypd1 expressed in a subset, quantified in B. Whereas the vast majority of SGNs express Calb2, a smaller proportion express Lypd1 (C). (D) The level of expression of select subtype-specific markers is overlaid in a tSNE plot containing both WT (M1–M3 clusters) and Vglut3−/− cells (M4–M5 clusters). Cluster identities in the key to the right are based on Fig. 7A. Numbers on the upper left indicate the highest expression (Max) observed for each gene. (E) tSNE plots with dot colors indicating subtype identity assigned by unsupervised graph-based clustering (left) and by Random Forest (right), a supervised clustering approach. (F) Scatterplot showing subtype identity of every cell predicted by unsupervised (X-axis, “Graph-based”) and supervised clustering (Y-axis, “Random Forest”) methods. Only cells that had not been used for training the Random Forest classifier are shown. Cells that fall on the diagonal dotted line are the ones predicted correctly by the supervised method. (G) Subtype proportions among SGNs from three tonotopic regions predicted by each clustering method. Scale bar: 10 μm (A).
Table S1, Related to Figure 1,2,3: Differentially expressed genes across all cell types/groups analyzed in the study. Each tab in the spreadsheet (A–F) contains data for a different set of comparisons as indicated. Top 100 or all genes (if <100) enriched in a subgroup are indicated in red. Statistical significance was assessed by conducting the likelihood-ratio test for gene expression in single cells in the Seurat R package.
Table S2, Related to Figures 1–7: Number of cells, animals and their age across all experiments reported in this study.