Abstract
Three-dimensional (3D) customized scaffolds are anticipated to provide new frontiers in cell manipulation and advanced therapy methods. Here, we demonstrate the application of hybrid 3D porous scaffolds, representing networks of highly aligned self-assembled ceramic nanofibers, for culturing four types of cancer cells. Ultrahigh aspect ratio (∼107) of graphene augmented fibers of tailored nanotopology is shown as an alternative tool to substantially affect cancerous gene expression, eventually due to differences in local biomechanical features of the cell–matrix interactions. Here, we report a clear selective up- and down-regulation of groups of markers for breast cancer (MDA-MB231), colorectal cancer (CaCO2), melanoma (WM239A), and neuroblastoma (Kelly) depending on only fiber orientation and morphology without application of any other stimulus. Changes in gene expression are also revealed for Mitomycin C treatment of MDA-MB231, making the scaffold a suitable platform for testing of anticancer agents. This allows an opportunity for selective “clean” guidance to a deep understanding of mechanisms of cancer cells progressive growth and tumor formation without possible side effects by manipulation with the specific markers.
Keywords: scaffold, nanofibers, alumina, graphene, anisotropy, cancer, gene expression
Introduction
Understanding the interactions between cells and their microenvironment is of fundamental importance for tissue engineering and regenerative medicine. Tissue engineering methods and 3D cell cultures are a vital part of modern oncology and possess a great potential in searching for new cancer therapy.1 Traditional development of cancer treatment is facing serious limitations, as many compounds that cure diseases in rodent model systems fail to provide meaningful clinical benefit for humans.2 The role of proper in vitro protocols is vital for cost-effective and scientifically valid treatments, which must be comprised of reconsideration of the relevance of animal study known as refinement, reduction, replacement or the 3R approach and further minimization of the long and expensive clinical trials. Despite the advances of modern high-throughput screening systems aimed at the computational discovery of new molecule design, about 85% of novel cancer drugs fail in the phase II clinical trials because of lack of evident efficacy.2,3
There are clear changes in the microenvironment of the natural tissue architecture during the development of cancer. It has been suggested that tumor cells respond inappropriately to cues from the surrounding normal tissue. Microenvironmental signals are an important part of the whole dynamic interplay between the growing tumor and the adjacent tissues.2,4 Changes in gene expression can be modulated by cell plasticity, resulting in generation of so-called cancer stem cells with high mobility and differentiation potential.4 In addition, the complex and less understood role of inflammation and infections, which always present in the clinical cases, should be inevitably taken into consideration.5 It was obviously shown that tissue architecture is a crucial component of any cellular function.6 The extracellular matrix (ECM) and its functionality, composition, and dynamics are of a paramount importance for understanding tumor triggering formation, growth, expanded invasion, and metastasis.7 With respect to the tissue structure, the diseased and aging tissues are often found to be more fibrotic or inappropriately organized, compromising their mechanical compliance.8 For example, breast cancer development and metastasis were reported to be associated with an increased collagen density, its altered organization, changes in tumor solid stresses, and a stromal fluid flow.9,10 Earlier cancer and cell culture studies were mostly based on 2D substrates (such as polystyrene and glass); however, the significant drawbacks were demonstrated due to inappropriate translation to in vivo situations and, therefore, to clinical practice. The bioactivity of usually employed materials in 2D culture should be additionally modified; nevertheless, currently it is possible only to a limited extent. The structure, organization, and biomechanical properties of such systems are usually outside of the physiologic range.11
The physical 3D tissues are subjected to different gradients of oxygen tension and nutrient concentration. The relative hypoxia in central areas of the tissue bulk alters oncogene expression and upregulates the expression of ECM remodelling enzymes and angiogenic growth factors.7,12 Novel 3D cell cultures are usually based on natural, synthetic, or semisynthetic (often modified) matrixes,13 including a wide variety of hydrogels.14 Matrigel (one of the most used), harvested from the rodent sarcoma cell line, is known to have significant variations in the composition,15 making exact reproduction of the results difficult.2,7 Various synthetic and semisynthetic matrixes can be designed to avoid such drawbacks of naturally derived materials. Materials providing a better platform for independent management of matrix cues or specific biochemical signaling are currently under close attention in the cancer research community.7
One of the features of such substrates, which have not yet received sufficient attention, is their mechano-topology on a micro- and nanoscale. It is identified that the cancer cells and tumors have rather distinct biomechanical properties as compared to the normal cells and tissues.16 An increased interstitial pressure in tumors due to increased angiogenesis enhances the overall fluid flux (and, presumably, shear stresses) through the stroma.17,18 Growing tumors themselves also generate solid stress, which is reported to reach up to 10 kPa in some in vitro models.7,19 These stresses together with an altered ECM secretion and remodelling make the tumor stroma substantially stiffer than the normal ECM. The standard breast tissue, for example, was reported to have Young’s modulus of ∼0.15 kPa, which increases up to ∼4 kPa in advanced malignancies.20 However, due to the viscoelastic nature of the tissues and cells, their elastic properties cannot routinely be expressed in some simple numbers: they are functions of many other parameters depending also on the analysis method and on the characteristics of the substrate where located cells are being measured.21 Synthetic gel systems are often exploited to study the substrate stiffness-mediated cell behavior, spreading, and proliferation, yet with some controversial results. Some reports found that in glioma cell line, they were enhanced on stiffer substrates.22,23 Other studies24 indicate that these cancer cells “prefer” softer substrates. Yet another study reported the rigidity dependence of the substrate in only some cell lines, being insignificant for other cells’ proliferation, apoptosis, and spreading.25 Such differences are due to many interrelated factors, which cannot be easily separated and independently evaluated. Because of the specific bioactivity of many natural and hybrid matrixes, scaffold engineering for study of molecular signals presents a strong challenge.2,7 The traditional polymer scaffolds often require additional processing steps to create a matrix open to 3D cell seeding.26 The nanofiber 3D scaffolds such as poly-ε-caprolactone (PCL) made by electrospinning technique have potential to improve the situation. Several successful studies using the PCL nanofibers for Ewing’s sarcoma cells showed an advantage over the 2D monolayer scaffold.27 However, whereas these nanofibers might have some of the features closer to the natural matrixes, such polymers cannot fully represent necessary interactions with cancer cells. Many properties of 3D scaffolds (yet not very well quantified) are essential, even when the scaffold materials might be considered as bioinert ones (not causing immunological or other reactions to the cells). Whereas substrate stiffness and topology are known to modulate cell shape and morphology,22−25,28 their explicit interactions with cells are too complex to allow separation of contributions. Mechanical forces generated by the cells themselves (without any explicit external stimulus) were reported to affect cell proliferation, differentiation, and cell signaling pathways and up/downregulate inflammatory markers.29−32 The effect of substrate stiffness was a subject of many studies,22−25 but the mechanical anisotropy acting simultaneously at nano-, micro-, and macroscales has received much less attention: only homogeneous substrate stiffness (with or without proper porosity) response to cancer cells was studied.7,16,19,20,22,23,25 Anisotropy of the most nanofiber-based scaffolds is difficult to assess as nanofibers are usually randomly oriented.26,27 There are not many 3D scaffolds with aligned nanofibers, and practically none with these nanofibers made as self-aligned, consistently grown anisotropic properties. We recently demonstrated a method and the properties of such self-aligned nanofiber scaffolds made of aluminum oxide capable of guiding various cells reactions.30,33 Ceramics have substantially higher stiffness than any of known polymers or hydrogels and are not commonly considered to be a material mimicking cells natural environment (perhaps with the exception of bone tissue). Bare ceramics substrates however do not possess high versatility in nanotopology due to presence of covalent bonding and more rigid crystal lattice, even in the case of amorphous materials. For this study, we applied additional modification of the nanofibers surface using graphene deposition.34
Graphene is an important material in tissue engineering studies and regenerative medicine.35−39 Graphene-coated surfaces with different stiffness and roughness have been shown to accelerate cell adhesion and proliferation of human mesenchymal stem cells into osteocytes and adipocytes, and committed, for instance, toward cardiomyogenic lineage by culturing them on graphene by regulated expression of ECM and signaling molecules.40 However, nonfunctionalized graphene and graphene oxide have been reported to increase cellular toxicity,41−44 and thus, many studies utilize substantial surface modification of graphene (oxy- and hydroxyl groups, grafting of organic molecules, biofunctionalization, etc.) to minimize cell toxicity.35−37 We recently reported that one of the possible pathways of graphene toxicity could be associated with its role in oxygen reduction reactions and consequently reactive oxygen species activity.45 Hence, it is beneficial to assess which effects unmodified graphene on ceramic nanofiber scaffolds would present to cancer cells and how these reactions might be quantified and exploited in understanding and developing cancer therapy.
Results and Discussion
In this work, we demonstrate an alternative approach to conventional oncology studies using nanomaterials that do not aim to mimic natural hostile environment but rather to create “out-of-comfort” nanobiomechanical29−31 environment. The rationale of this research is that natural-like environments have a myriad of different signals and interactions (cell–cell, cell–substrate, cell–ECM), which result in extremely complex data sets incapable of manipulation and with connections between observed gene expression that cannot be understood without use of specific stimuli or transfection. We recently demonstrated that application of cells on graphene-augmented inorganic nanofiber (GAIN) scaffolds can trigger different cell reactions and mechanisms, leading to changes in their morphology, orientation, and gene expression.30,46 It has been shown that some cancer cells can change their behavior depending on the substrate usage. The current work studies this effect in more precise detail. We hypothesized that local and highly anisotropic mechano-transduction conditions combined with unique properties of subnanolayers of nonfunctionalized graphene on oriented ceramic fibers cause spectra of signals that might lead to a “soft touch” on cell activity, which would be reflected in one or another spectrum of gene expression; this result could be captured and correlated with time, scaffold type, and anticancer drug presence.
The main aim of the study was to demonstrate the appropriateness of the highly aligned graphene-augmented scaffolds for the biomedical cancer research, trying to gain a deeper understanding on whether different cancer cells provide different gene expression on the new anisotropic GAIN scaffolds vs traditional 2D control, and if yes, which ones, and whether these changes are sufficiently significant to be detected and deployed to up- and down-regulate specific markers without use of chemical stimuli. In addition, we attempted to analyze whether cancer cells seeded on the surface of GAIN scaffolds do react in a unique (and different form the standard 2D conditions) way on anticancer drug additions.
The substrate used as a scaffold for this work represents the network of highly aligned free-standing self-oriented alumina nanofibers with a single fiber diameter of 10 ± 3 or 40 ± 5 nm and aspect ratio (length to diameter) of order 106:1. Graphene layers were grown along the longitudinal axis of nanofibers using a hot-wall single-step process of catalyst-free chemical vapor deposition (CVD) at a treatment temperature of 1000 °C and in methane (CH4) gas flowing through a CVD chamber at a rate of 50 cm3 min–1 as detailed in refs (31) and (32). A number of the deposited layers is affected by process conditions such as gas flow kinetics, quantity of nucleation centers on the fiber surface, temperature, and time of exposition. Two types of structures were prepared for the current work, namely, GAIN-C3 consisting of the fibers covered by 2–5 graphene layers, and GAIN-C4 with at least 10 layers of graphene on the fiber surface, Figure 1. Depending on the process parameters, the nanostructures of different morphologies, starting from smooth coatings and ending by highly foliated 3D structures, can be produced. As soon as the weight gain of the carbon coating is reached of about ΔW = 150%, the formation of the graphene flakes/foliates along the longitudinal axis of the fiber was observed. The appearance of foliates is similar to the leaves of aquatic plant Leptodictyum riparium: nearly homogeneously distributed foliates grow perpendicular to the fiber surface, and their density is regulated by process parameters.
Figure 1.
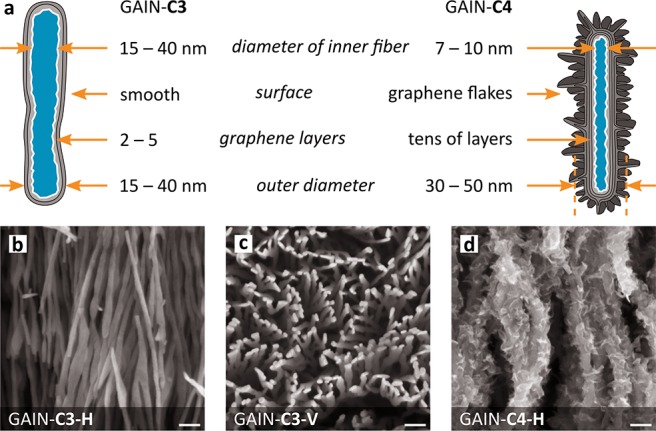
GAIN scaffolds and their differences for C3 and C4 type (a). Scaffold of C3 type were also tested in their horizontal (C3–H; b) and vertical (C3–V; c) alignment for cells seeding; horizontal scaffold for C4–H (d). Scale bars (b–d) = 100 nm.
Scaffolds of GAIN-C3 type consisting of relatively large fibers with average diameter 40 nm were used in two geometries, either with horizontal or vertical fiber orientation toward the seeding cells. Covering by graphene (estimated thickness 2–5 nm) turned highly hydrophilic properties into hydrophobic adding specific biocompatibility to the substrate.30,46 The GAIN-C4 type scaffold is based on fibers with 10 nm diameters, which forms the substrate of high stiffness being mechanically unreliable to be used as a self-standing scaffold. Carbon deposition increases the fiber diameters up to 30–50 nm, making them safer and easier to handle and increasing the stiffness compatible to stiffness of GAIN-C3 type. Presence of the graphene flakes (estimated layer thickness 20–40 nm) appeared perpendicularly to the carrier fiber in GAIN-C4, which makes the essential difference from the sleek fiber surface of GAIN-C3 (Figure 1). Highly porous and oriented GAIN scaffolds were designed to enable more natural reproduction of in vivo cell function within an in vitro model system.
To understand the effects of various scaffold types on cancer cell growth, behavior, and expression profile, we chose four different cancer cell lines: colorectal cancer (CaCO2), breast cancer (MDA-MB231), melanoma (WM239A), and neuroblastoma (Kelly). These cells were cultured on different GAIN scaffold types (marked as C3–H, C3–V, and C4–H, respectively) and the control (culture plates without scaffold material) and analyzed. To show the structure of the cytoskeleton on the surface of scaffolds, cellular actin was stained with FITC-phalloidin. The images of cancer cells on the GAIN horizontal fibers (C3–H, C4–H) demonstrate cells adherence to the substrate and more natural spatial cell shape and morphology as compared to 2D culture model (Figure 2), where cells flattened against surface of the culture plates.
Figure 2.
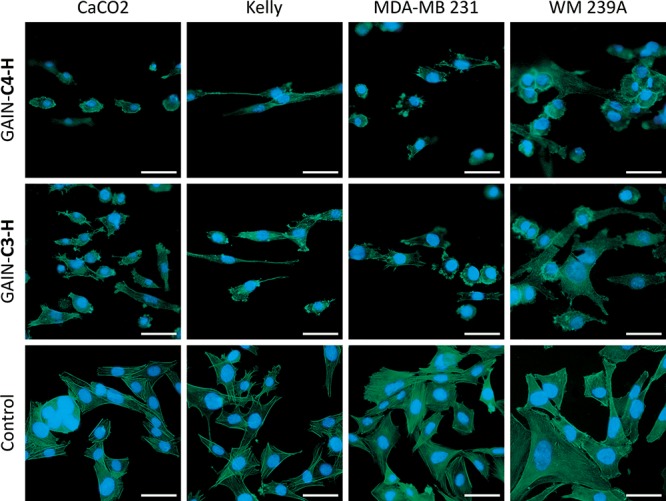
Fluorescence images of cancer cells on C3–H and C4–H scaffolds for horizontal orientation. Magnification bar = 50 μm.
All four types of cancer cells were well attached to the scaffold surfaces. Extended pseudopodia of cells were observed already after 24 h of culture, and the increase of these attachments and cell spreading were observed over time (data not shown). After 72 h, all cells were spread completely and attached tightly on the scaffold surface. Changes in cells morphology were more distinct when cells were allowed to grow both on the C3 scaffolds with vertical (C3–V) and horizontal (C3–H) orientation (Figure 3). Cancer cells grown on the vertical GAIN formed disorganized networks with variable size and shapes of cells, while horizontal scaffolds promoted cells orientation, especially for Kelly neuroblastoma cell line. Such stretching of cells is not typical for cancer cell lines, and this interesting phenomenon should be analyzed in the future in more detail.
Figure 3.
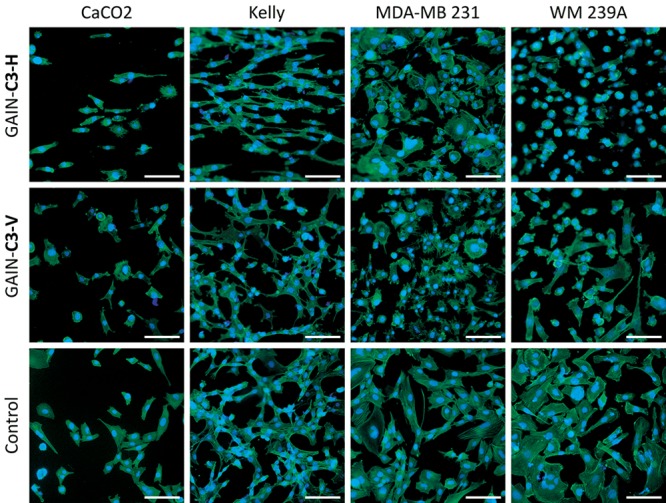
Fluorescence images of cancer cells on vertical (C3–V) and horizontal (C3–H) fiber orientations. Magnification bar = 100 μm.
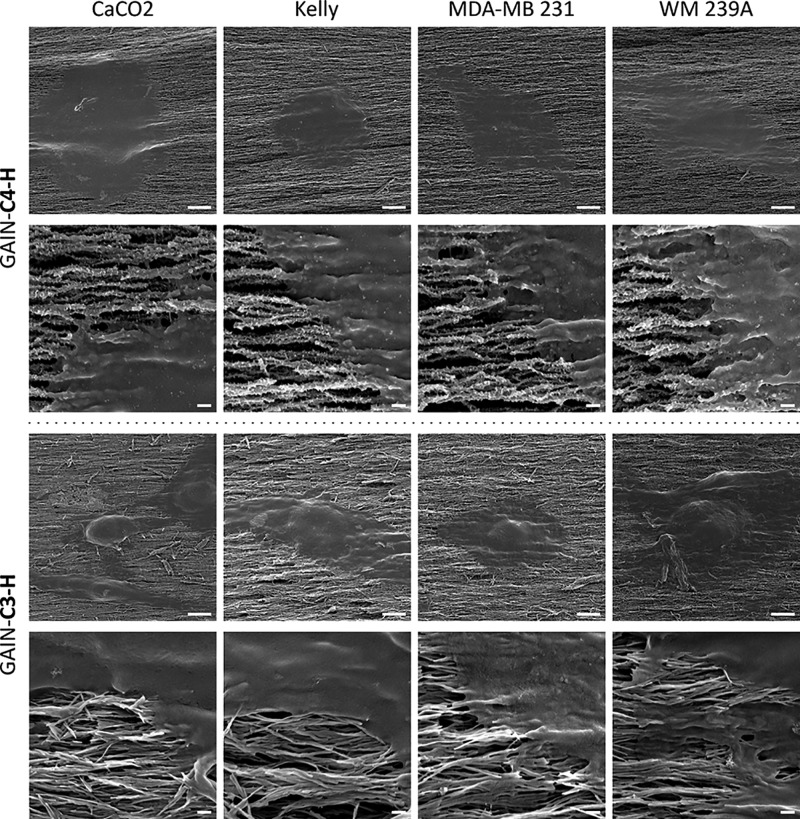
The spreading and infiltration potentials of cancer cells on the GAIN scaffolds were evaluated 24 h after seeding (scanning electron microscopy (SEM) micrographs are shown in Figure 4). Cancer cells on the GAIN grew in flat sheets on the surface of oriented nanofiber matrixes but did not directly infiltrate the scaffold because cancer cells are much larger than the scaffold nanopores. However, the cell flatness significantly differed from that on the 2D controls: cells enveloped the fibers of scaffolds displayed a different cell–substrate interaction. These results prove that all cancer cells were able to proliferate well on the C3 and C4 GAIN scaffolds, and also that the GAIN scaffolds are sufficiently biocompatible for cancer cell lines, providing a proper surface for cell adhesion, growth, and spreading.
Figure 4.
SEM images of cancer cells on C3–H and C4–H scaffolds shown in two magnifications: scale bar = 5 μm for images in the first and the third rows, and 0.2 μm for the second and the fourth rows of images.
Next, the molecular patterning of cancer cells grown on the horizontal GAIN scaffolds (C3–H and C4–H) compared to the usual 2D cell culture was studied. Figure 5 demonstrates a heat map of gene expressions of all cancer cell lines used for the current study. All represented data at the heat maps shown in logarithmic color scale are at least with p < 0.01. For visual guidance, common genes groups in Figure 5 are arranged and colored by their primary functions, namely: cancer progression (CDH1, CDH2, SNAI1, VIM), angiogenesis (HGF, bFGF, VEGFA), proliferation (c-KIT, MKi67, PAI1), oncogenesis (ERBB2, TP53), and tissue degradation (MMP1, MMP9, TIMP1).
Figure 5.
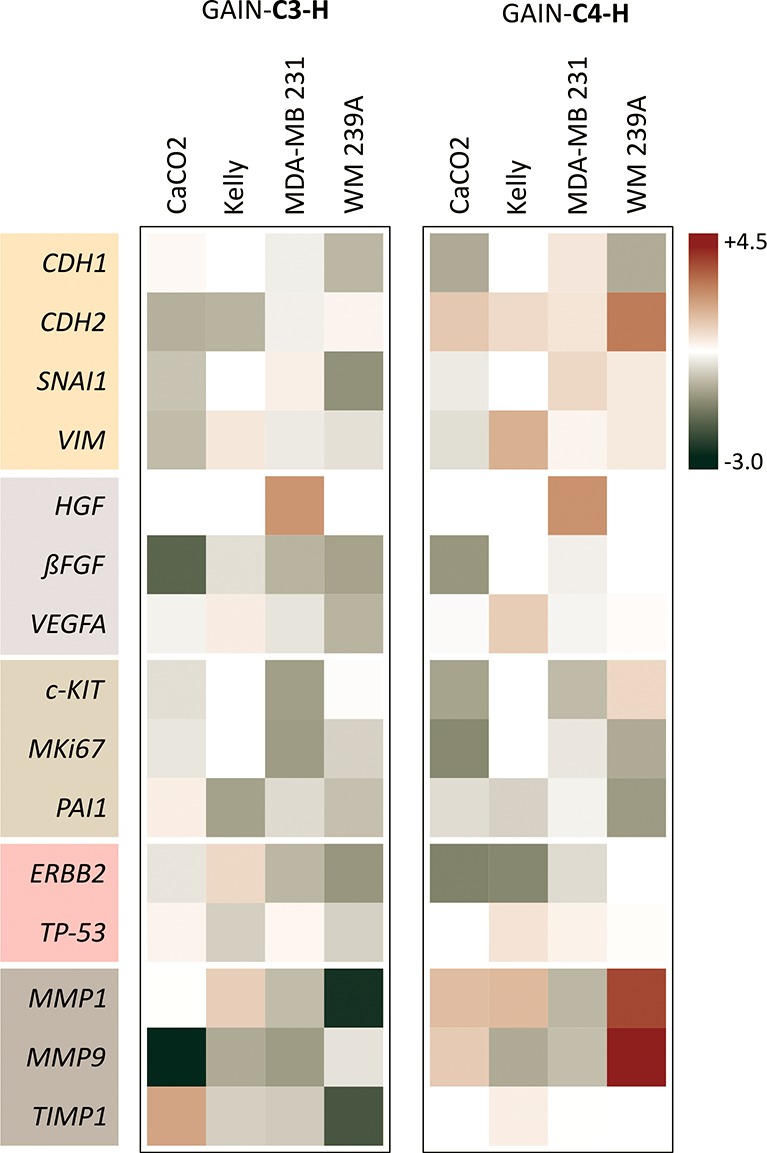
Heat map of grouped gene expression for all cancer cell types on C3–H and C4–H scaffolds vs control (natural logarithm scale: red = highest +4.3; green = lowest −3.0). Only statistically relevant data with at least p < 0.01 are shown.
The data clearly illustrate that the C3–H and C4–H scaffolds in some cases exhibit very different changes in gene expression for the same cancer cell line. For example, CDH2 expression was downregulated for C3–H GAIN but upregulated for C4–H scaffold; levels of MMP1 and MMP9 mRNAs were strongly decreased in WM239A and CaCO2 cells on C3–H scaffold but increased in case of cells grown on C4–H. Evidently, the main reason for these variations relates to the topological properties of the scaffold material and could be biologically relevant for various aspects and stages of cancer progression. Because CDH2 and both matrix metalloproteinases (MMP1, MMP9) are associated with changes in ECM environment, their expression will be affected by the properties of the substrate surface.
The other valuable observation is that all cancer cell types reacted differently with the GAIN material. While the expression of HGF was significantly upregulated for MDA-MB231 breast cancer, other cancers did not change their expression in result of the contact with the GAIN. Also, TIMP1 expression was increased for CaCO2 on the C3–H but was decreased for all other cells. These deviations are cancer-type specific and point out the highly divergent and complex nature of cancer. Understanding the common features and special characteristics of a particular type of cancer in similar mechanical environment independent of external biological stimulus represents a valuable and unprecedented tool for cancer research. An interesting feature of the GAIN scaffolds is that they do not affect the expression levels of TP53: an important and the most studied tumor suppressor. Thus, lots of genes have significantly changed their expression profiles while growing cancer cell lines on the GAIN scaffolds, thereby providing a new biologically “clean” platform for understanding the mechano-transductor impact on tumor progression.
The more detailed studies were performed to understand the impact of two scaffolds type (C3–H or C4) on the expression profile of MDA-MB231 breast cancer cells, where various cultivation time (24 and 72 h) and the effect of anticancer drug (Mitomycin C) were analyzed. All data were normalized to cells grown on the control and are represented in a natural logarithm scale (Figure 6). The differences in gene expression at 24 and 72 h show that cells actively metabolize on the surface of the GAIN substrate, dynamically reacting on external environment.
Figure 6.

Gene expression heat map for MDA-MB231 vs type of GAIN scaffold and time (a: red = highest +2.3; green = lowest −2.0) and chemotherapy with Mitomycin C at 24 h exposure (b; red = highest +4.3; green = lowest −3.3). Only statistically relevant data with at least p < 0.01 are shown.
The most significant differences were observed in the case of cancer progression group of genes, where at 24 h the expression of CDH1, CDH2, and SNAI1 were induced while decreased after 72 h growth on the GAIN. The same pattern of gene expression at that time showed HGF strongly decreasing its level of expression.
Thus, various topology and accordingly microenvironment of substrates in dynamic time-varying manner could selectively trigger changes in cancer cell fate and thereby elicidate knowledge about the nature of cancer. To extend the scaffold application as a unique model for pharmacological agent testing, we performed MDA-MB231 treatment by chemotherapeutic drug Mitomycin C and analyzed the received data. In general, both scaffolds proved themselves similarly in response to anticancer treatment (Figure 6). It is nteresting to note that expression of angiogenic genes HGF, bFGF, and VEGF was induced at 24 h after Mitomycin C treatment.
We earlier showed that GAIN-type scaffolds have a substantial mechanical anisotropy, as measured by the tangential elastic modulus, reaching 400 GPa/5 kPa = 80 × 106.30,46 This range spans for example over the elasticity of F-actin filaments (∼1–5 GPa16), involved in cells adhesion and taxis. This is combined with matched diameter of the nanofibers in GAIN (Figure 1) to actin filaments spacing (20–30 nm) and microtubules (∼25 nm),16,24,28 being significantly (500–1000 times) smaller than average cell sizes. Therefore, sterically one cell filament may bound to a single nanofiber in orthogonal direction, with simultaneous attachment of many filaments to the same single nanofiber along its longer dimension. From the point of view of focal adhesion, a million times difference in requirement of the cell membrane compliance to the substrate causes substantially larger displacements of nanofibers in the perpendicular direction (leading to lower stresses in the cell membrane). In the direction in parallel to the nanofibers the cell membrane would face substantially larger stress (less nanofiber displacement) and therefore the membrane will undergo much higher deformation. As a result, these two different magnitudes of mechanical transduction signals give a superposition of “too high” and “too low” stiffness stimuli, causing the cell to alter its gene expression in a nonconventional way without exposure to chemical or biochemical species.
Conclusions
In accordance with the working hypothesis, the results indicate that unique composition and structure of the GAIN scaffolds create the unique changes in microenvironment, which are not present in the standard 2D cultures, but likely to be similar for fibrous substrates. This assertively causes different expression of respective oncogenes and tumor suppressor genes. For instance, variation of HGF, c-KIT, CDH2, MMP1, and MMP9 levels are the most evident among the complete set of cells and conditions studied. For breast cancer cells,47 time significantly upregulates HGF expression, following by HGF radical downregulation by a few orders of magnitude. This feature was, however, fully overridden with Mitomycin C. Many studies cited do underline that cancer cells prefer substrates with lower stiffness, but as seen from these data, such assumption would be highly oversimplified, if at all extendable from 2D to 3D environments.48 A variety of signals and their differences are clearly measured and could not be allocated to one or another stiffness type. When cells are facing highly anisotropic substrate, one can imagine that a number of contradictory messages are being transduced into the nucleus via cell cytoskeleton and membrane tension. In this case, cells react in a unique way that is impossible for the growth of cells in plane.
The combined effects of time, substrate type, and drug addition are highly versatile and should be studied in much more detail. Despite the high reproducibility of the effects, the underpinning mechanisms of action are demanding more detailed studies and must be dependent on the precise scientific question raised. However, it is clear that GAIN scaffolds could be used as the new advanced tool for cancer research, drug development, or in cancer stem cell studies.30,46
All received data show that GAIN scaffolds do influence differences in cancer gene expression in time, material type, and in response to anticancer treatment. The expression of several genes was changed vs 2D control, and the whole pattern of mRNA levels was significantly affected. In whole, GAIN scaffolds, providing different topology, are suitable advanced models for cancer research studies, allowing specific gene expression up- and down-regulation by local mechano-transductive pathways. Whereas GAIN nanostructure itself does not mirror live tissues, its fibrillar nature has relevancy to many tissue types. The study has confirmed GAIN scaffolds to provide a suitable microenvironment of nanotopological features, transducing biomechanical cues in another way into an impact of specific gene expressions, which knowledge is of a paramount importance for development of novel cancer therapy. Here, we showed the opportunity to use GAIN scaffolds for analysis of effects of anticancer drugs on mechano-transductor microenvironment and corresponding changes of gene expression. Changed distinct morphology of cancer cells grown on GAIN due to synthetic microenvironment of substrate affects cytoskeleton structure and focal adhesion molecules expression, thereby changing cancer cell migration, proliferation, and behavior, makes GAIN an interesting alternative model for cancer research to enhance the physiological relevance of cancer in vitro model with multiple cell types. Although the detailed analyses and thorough correlation between scaffold mechano-physical properties and biological responses are necessary to better understand all aspects of scaffold–cell interaction, GAIN scaffolds might be considered as a valuable tool for new drug discovery.
Materials and Methods
Scaffolds Preparation
Self-assembled metal oxide fibers network with an average single-fiber diameter of ∼40 or ∼10 nm (transmission electron microscopy, high-resolution scanning electron microscopy) and length of 2–10 cm with 85–95% oriented porosity (Brunauer–Emmett–Teller; mercury porosimetry) was produced using bottom-up approach of controlled liquid phase oxidation.33,34 The graphene shells were grown on these substrates in a single-step process of catalyst-free chemical vapor deposition (CVD) at 1000 °C and atmospheric pressure, using a mixture of nitrogen (N2) and methane (CH4) gases, Figure 1. Graphene on the nanofibers was augmented to the underlying oxide fiber surface during the manufacturing process and was not functionalized in any way.34 Two types of the GAIN scaffolds were made. The C3 type is based on ∼40 nm diameter alumina nanofibers with augmented graphene coating to 10–15 nm (Figure 1). The type C4 consists of base nanofibers with 7–10 nm diameter covered with a thicker graphene coating toward total diameter of 30–50 nm diameter but with a surface topology different than that for C3 type (Figure 1).
Cell Cultures
Four different human cancer cell lines were used: breast cancer (MDA-MB231), colorectal cancer (CaCO2), melanoma (WM239A), and neuroblastoma (Kelly), all purchased from ATCC. Cells were grown in DMEM high glucose with 10% FBS, 1 mg/mL penicillin, and 0.1 mg/mL streptomycin at 37 °C in 5% CO2. GAIN scaffolds of both types C3 and C4 were pretreated for 3 days before the cell seeding by complete medium with changing the medium for fresh every 24 h to saturate them by active components adsorbed from liquid phase. This medium change was done before seeding the cells, and no media changes were made during cell culturing. Cells were analyzed on expression of the cancer-related markers during the time (24 and 72 h of cells growth on scaffolds) and the response of grown on the GAIN cells to cancer-specific drug treatment.
For visualization of cells on the GAIN scaffold, specific to filamentous actin (F-actin) phalloidin tagged by FITC (Sigma) was applied. Similar cells grown on a flat glass at the same density and cell culture conditions were considered as the controls. For analysis, the cells were fixed by 4% PFA, washed by PBS following by permeation with 0.3% TRITON X-100 in PBS for 5 min. Phalloidin-FITC (1:100) staining lasted for 18 h at 4 °C for GAIN scaffolds and 2 h at RT for controls. To stain the cell’s nucleus, cells were incubated for 10 min with Hoechst 33342 (Invitrogen, 1 μg/mL). After a final wash, the phalloidin-stained cells were analyzed by Nikon Eclipse 80i microscope.
For study of cancer cell behavior on the surface of C3 and C4 scaffolds, cancer cell lines (MDA-MB231, CaCO2, WM239A, and Kelly) were used, and their cancerous gene expression was analyzed at 24 and 72 h after experiment initiation. Cells were seeded at 150 000 cells/well in 12-well plate and grew in DMEM high glucose medium with 10% FBS and 1% antibiotic at 37 °C and 5% CO2 atmosphere. The medium was changed every 24 h during 3 preceding days before seeding the cells. To assess RNA quality, RNA gel electrophoresis was performed, and 1 μg of RNA was taken for cDNA synthesis. The primers for gene expression analysis were designed by NCBI/Primer-BLAST program, with primers located in different exons, and the length of the final product did not extend 100 bp. Primers were blasted to verify their specificity. For each gene expression analysis, three independent studies were performed.
Cells Analysis
For mRNA analysis, RNA was extracted directly from scaffolds by TRIzol (Ambion) reagent. Each experiment was done in three independent replicas. RNA extraction was performed according to the manufacturer’s recommendations. cDNAs were synthesized from DNase-treated (Ambion) RNA by RevertAid Reverse Transcriptase (Thermo Fisher Scientific) with addition of RiboLock (Thermo Fisher Scientific) according to the manufacturer’s recommendations. cDNA quality was verified by RT-PCR by using GAPDH primers. RT-qPCR was performed in triplicates using EvaGreen qPCR mix plus no Rox (Solis Biodyne, Estonia) and the LightCycler480 RT-PCR System (Roche Applied Science). Amplifications were carried out using the program: 95 °C for 15 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 20 s, and 72 °C for 30 s. The fold of change was calculated relatively to the control (cells grown without scaffolds) after normalization to GAPDH expression, using 2–ΔΔCt method, where ΔCt = (gene of interest Ct) – (GAPDH Ct), and ΔΔCt = (ΔCt treated) – (ΔCt of control). Only statistically relevant data with p < 0.01 at least were chosen for presentation. For analyses, four distinct groups of genes were selected. The first group is primarily responsible for invasiveness and EMT of cancer (CDH1: GAATCCAAAGCCTCAGGTCAG and CCCACCTCTAAGGCCATCT; CDH2: ATCGTGTCTCAGGCTCCAAG and GGATTGCCTTCCATGTCTGT; VIM: AGACAGGTGCAGTCCCTCAC and TCTTCCATTTCACGCATCTG; SNAI1: GCCTTCAACTGCAAATACTGC and CTTCTTGACATCTGAGTGGGTC). The second group is related to cancer angiogenesis (growth of new blood vessels, which is an important component in the progression of cancer), namely VEGFA: CCTGGTGGACATCTTCCAGGAGTACC and GAAGCTCATCTCTCCTATGTGCTGGC; HGF: GCCTGAAAGATATCCCGACA and CCCTGTAGCCTTCTCCTTGA; bFGF: ATCAAAGGAGTGTGTGCTAACC and ACTGCCCAGTTCGTTTCAGTG). The third group of genes has a role in the cells proliferation: PAI1: CGATGGCCATTACTACGACA and GTTGGTGAGGGCAGAGAGAG; MKi67: CAAGACTCGGTCCCTGAAAA and TTGCTGTTCTGCCTCAGTCTT; c-KIT: GGCGACGAGATTAGGCTGTT and CATTCGTTTCATCCAGGATCTCA); and in oncogenesis: TP-53 (TGGAGGAGCCGCAGTCAGATCC and TTGCTTGGGACGGCAAGGGG) and ERBB1 (GCAGGACCAAGCAAATTGAG and CTTCCAGACCAGGGTGTTGT). The fourth group is associated with the participation in tissue degradation: MMP1 (TGCTCATGCTTTTCAACCAG and ATGCCATCAATGTCATCCTG); MMP9 (CATTTCGACGATGACGAGTTGT and CGGGTGTAGAGTCTCTCGC); TIMP1 (GGGTTCCAAGCCTTAGGGG and TTCCAGCAATGAGAAACTCCTC).
Analysis of the Anticancer Drug Effect
To try the application of C3 and C4 scaffolds as a platform for drug discovery studies, control and GAIN-growth breast cancer MDA-MB231 cells were treated by chemotherapeutic agent Mitomycin C, and received data were analyzed for changes in gene expression. For analysis, Mitomycin C (Sigma) in final concentration of 25 μg/mL was applied for MDA-MB231 cells. The scheme of the study was first to grow MDA-MB231 on the C3–H and C4–H GAIN scaffolds for 24 h, change the medium for low FBS (5% instead of 10%) and exclude the antibiotic; add the drug, and collect the cell after 24 h by TRIzol for RNA extraction.
Acknowledgments
This research was supported by the Estonian Research Council under the personal research grant PUT1063 (I.H.) and PROTOTRON foundation (Estonia) grant under VIROGAIN and ULTRINIA (www.ultrinia.com) projects. M.G. is grateful to Dr. Y. Bilotsky (Seqvera Ltd., Finland, www.seqvera.com) for suggestions about the nonlinear anisotropic mechano-transduction mechanisms and pathways.
Author Contributions
J.K., R.I., M.G., T.N., and I.H. wrote the main manuscript text. R.I. and I.H. prepared the GAIN scaffolds; J.K. and R.I. performed cell analytics and images acquisition, and M.G. composed the final manuscript, made heatmap data analysis, and evaluated the main mechanical anisotropic features of the scaffolds. R.I. and M.G. finalized manuscript figures. J.K. also made a detailed description of the cell analytics methods, which were proofread by T.N. in combination with results and conclusions. All authors have reviewed the manuscript.
The authors declare no competing financial interest.
References
- Ghajar C. M.; Bissell M. J. Tumor engineering: the other face of tissue engineering. Tissue Eng., Part A 2010, A16, 2153–2156. 10.1089/ten.tea.2010.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBarge M. A.; Parvin B.; Lorens J. B. Molecular deconstruction, detection, and computational prediction of microenvironment-modulated cellular responses to cancer therapeutics. Adv. Drug Delivery Rev. 2014, 69–70, 123–131. 10.1016/j.addr.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith J. Trial watch: phase II failures: 2008–2010. Nat. Rev. Drug Discovery 2011, 10, 328–329. 10.1038/nrd3439. [DOI] [PubMed] [Google Scholar]
- Meacham C. E.; Morrison S. J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S. I.; Greten F. R.; Karin M. Immunity, inflammation and cancer. Cell 2010, 140, 883–899. 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny P. A.; Bissell M. J. Tumor reversion: correction of malignant behavior by micro-environmental cues. Int. J. Cancer 2003, 107, 688–695. 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill B. J.; West J. L. Modeling the tumor extracellular matrix: tissue engineering tools repurposed towards new frontiers in cancer biology. J. Biomech. 2014, 47, 1969–1978. 10.1016/j.jbiomech.2013.09.029. [DOI] [PubMed] [Google Scholar]
- Claridge M. W.; Bate G. R.; Hoskins P. R.; Adam D. J.; Bradbury A. W.; Wilmink A. B. Measurement of arterial stiffness in subjects with vascular disease: are vessel wall changes more sensitive than increase in intima-media thickness?. Atherosclerosis 2009, 205, 477–480. 10.1016/j.atherosclerosis.2008.12.030. [DOI] [PubMed] [Google Scholar]
- Boyd N. F.; Lockwood G. A.; Martin L. J.; Knight J. A.; Byng J. W.; Yaffe M. J.; Tritchler D. L. Mammographic densities and breast cancer risk. Breast Dis. 1998, 10, 113–126. 10.3233/BD-1998-103-412. [DOI] [PubMed] [Google Scholar]
- Cil T.; Fishell T. E.; Hanna W.; Sun P.; Rawlinson E.; Narod S. A.; McCready D. R. Mammographic density and the risk of breast cancer recurrence after breast-conserving surgery. Cancer 2009, 115, 5780–5787. 10.1002/cncr.24638. [DOI] [PubMed] [Google Scholar]
- Paszek M. J.; Zahir N.; Johnson K. R.; Lakins J. N.; Rozenberg G. I.; Gefen A.; Reinhart-King C. A.; Margulies S. S.; Dembo M.; Boettiger D.; Hammer D. A.; Weaver V. M. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Erler J. T.; Weaver V. M. Three-dimensional context regulation of metastasis. Clin. Exp. Metastasis 2009, 26, 35–49. 10.1007/s10585-008-9209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Cuddihy M. J.; Kotov N. A. Three-dimensional cell culture matrices: state of the art. Tissue Eng., Part B 2008, 14, 61–89. 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- Tibbitt M. W.; Anseth K. S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. C.; Kiemele L.; Maller O.; O’Brien J.; Shankar A.; Fornetti J.; Schedin P. An in-solution ultrasonication-assisted digestion method for improved extracellular matrix proteome coverage. Mol. Cell. Proteomics 2009, 8, 1648–1657. 10.1074/mcp.M900039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007, 3, 413–478. 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z.-D.; Ji X.-Y.; Qazi H.; Tarbell J. M. Interstitial flow promotes vascular fibroblast, myofibroblast, and smooth muscle cell motility in 3-D collagen I via upregulation of MMP-1. Am. J. Physiol. - Heart and Circul. Physiol. 2009, 297, H1225–H1234. 10.1152/ajpheart.00369.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields J. D.; Emmett J.D. M.S.; Dunn D. B.; Joory K. D.; Sage L. M.; Rigby H.; Mortimer P. S.; Orlando A.; Levick J. R.; Bates D. O. Chemokine-mediated migration of melanoma cells towards lymphatics – a mechanism contributing to metastasis. Oncogene 2007, 26, 2997–3005. 10.1038/sj.onc.1210114. [DOI] [PubMed] [Google Scholar]
- Helmlinger G.; Netti P. Solid stress inhibits the growth of multicellular tumor spheroids. Nat. Biotechnol. 1997, 15, 778–783. 10.1038/nbt0897-778. [DOI] [PubMed] [Google Scholar]
- Butcher D. T.; Alliston T.; Weaver V. W. A tense situation: forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122. 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocen R.; Gasik M.; Gantar A.; Novak S. Viscoelastic behaviour of hydrogel-based composites for tissue engineering under mechanical load. Biomed. Mater. 2017, 12, 025004. 10.1088/1748-605X/aa5b00. [DOI] [PubMed] [Google Scholar]
- Ulrich T. A.; de Juan Pardo E. M.; Kumar S. T. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009, 69, 4167–4174. 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander N. R.; Branch K. M.; Parekh A.; Clark E. S.; Iwueke I. C.; Guelcher S. A.; Weaver A. M. Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 2008, 18, 1295–1299. 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D. E.; Janmey P.; Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Tilghman R. W.; Cowan C. R.; Mih J. D.; Koryakina Y.; Gioeli D.; Slack-Davis J. K.; Blackman B. R.; Tschumperlin D. J.; Parsons J. T. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS One 2010, 5, e12905. 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel T.; Schinkel G.; Lendlein A. Design and preparation of polymeric scaffolds for tissue engineering. Expert Rev. Med. Devices 2006, 3, 835–851. 10.1586/17434440.3.6.835. [DOI] [PubMed] [Google Scholar]
- Fong E. L. S.; Lamhamedi-Cherradi S.-E.; Burdett E.; Ramamoorthy V.; Lazar A. J.; Kasper F. K.; Farach-Carson M. C.; Vishwamitra D.; Demicco E. G.; Menegaz B. A.; Amin H. M.; Mikos A. G.; Ludwig J. A. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 6500–6505. 10.1073/pnas.1221403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyckmans J.; Boudou T.; Yu X.; Chen C. S. A hitchhiker’s guide to mechanobiology. Dev. Cell 2011, 21, 35–47. 10.1016/j.devcel.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasik M. Understanding biomaterial-tissue interface quality: combined in vitro evaluation. Sci. Technol. Adv. Mater. 2017, 18, 550–562. 10.1080/14686996.2017.1348872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantseva J.; Ivanov R.; Gasik M.; Neuman T.; Hussainova I. Graphene-augmented nanofiber scaffolds demonstrate new features in cells behaviour. Sci. Rep. 2016, 6, 30150. 10.1038/srep30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. Y. H.; Lim C. T. Biomechanics approaches to studying human diseases. Trends Biotechnol. 2007, 25, 111–118. 10.1016/j.tibtech.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Yao X.; Peng R.; Ding J. Cell-material interactions revealed via material techniques of surface patterning. Adv. Mater. 2013, 25, 5257–5286. 10.1002/adma.201301762. [DOI] [PubMed] [Google Scholar]
- Aghayan M.; Hussainova I.; Gasik M.; Kutuzov M.; Friman M. Coupled thermal analysis of novel alumina nanofibers with ultra-high aspect ratio. Thermochim. Acta 2013, 574, 140–144. 10.1016/j.tca.2013.10.010. [DOI] [Google Scholar]
- Ivanov R.; Mikli V.; Kübarsepp J.; Hussainova I. Direct CVD growth of foliated graphene closed shells on alumina nanofibers. Key Eng. Mater. 2016, 674, 77–80. 10.4028/www.scientific.net/KEM.674.77. [DOI] [Google Scholar]
- Bressan E.; Ferroni L.; Gardin C.; Sbricoli L.; Gobbato L.; Ludovichetti F. S.; Tocco I.; Carraro A.; Piattelli A.; Zavan B. Graphene based scaffolds effects on stem cells commitment. J. Transl. Med. 2014, 12, 296. 10.1186/s12967-014-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Nayak T. R.; Hong H.; Cai W. Graphene: a versatile nanoplatform for biomedical applications. Nanoscale 2012, 4, 3833–3842. 10.1039/c2nr31040f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka S.; Sant V.; Sant S. Graphene-based nanomaterials for drug delivery and tissue engineering. J. Controlled Release 2014, 173, 75–88. 10.1016/j.jconrel.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Ding X.; Liu H.; Fan Y. Graphene-based materials in regenerative medicine. Adv. Healthcare Mater. 2015, 4, 1451–1468. 10.1002/adhm.201500203. [DOI] [PubMed] [Google Scholar]
- Dubey N.; Bentini R.; Islam I.; Cao T.; Castro Neto A. H.; Rosa V. Graphene: a versatile carbon-based material for bone tissue engineering. Stem Cells Int. 2015, 2015, 804213. 10.1155/2015/804213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C.; Lim C. H. Y. X.; Shi H.; Tang L. A. L.; Wang Y.; Lim C. T.; Loh K. P. Origin of enhanced stem cell growth and differentiation on graphene and graphene oxide. ACS Nano 2011, 5, 7334–7341. 10.1021/nn202190c. [DOI] [PubMed] [Google Scholar]
- Pinto A. M.; Goncalves I. C.; Magalhães F. D. Graphene-based materials biocompatibility: a review. Colloids Surf., B 2013, 111, 188–202. 10.1016/j.colsurfb.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Hu W.; Peng C.; Lv M.; Li X.; Zhang Y.; Chen N.; Fan C.; Huang Q. Protein corona-mediated mitigation of cytotoxicity of graphene oxide. ACS Nano 2011, 5, 3693–3700. 10.1021/nn200021j. [DOI] [PubMed] [Google Scholar]
- Mao H. Y.; Laurent S.; Chen W.; Akhavan O.; Imani M.; Ashkarran A. A.; Mahmoudi M. Graphene: promises, facts, opportunities, and challenges in nanomedicine. Chem. Rev. 2013, 113, 3407–3424. 10.1021/cr300335p. [DOI] [PubMed] [Google Scholar]
- Sanchez V. C.; Jachak A.; Hurt R. H.; Kane A. B. Biological interactions of graphene-family nanomaterials: an interdisciplinary review. Chem. Res. Toxicol. 2012, 25, 15–34. 10.1021/tx200339h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatin S.; Hussainova I.; Ivanov R.; Colavita P. Quantifying graphitic edge exposure in graphene-based materials and its role in oxygen reduction reactions. ACS Catal. 2016, 6, 5215–5221. 10.1021/acscatal.6b00945. [DOI] [Google Scholar]
- Kazantseva J.; Hussainova I.; Ivanov R.; Neuman T.; Gasik M. Hybrid graphene–ceramic nanofibre network for spontaneous neural differentiation of stem cells. Interface Focus 2018, 8, 20170037. 10.1098/rsfs.2017.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S.; Duan X.; Lo P. K.; Liu S.; Liu X.; Chen H.; Wang Q. Expansion of breast cancer stem cells with fibrous scaffolds. Integr. Biol. (Camb) 2013, 5, 768–777. 10.1039/c3ib20255k. [DOI] [PubMed] [Google Scholar]
- Voutouri C.; Polydorou C.; Papageorgis P.; Gkretsi V.; Stylianopoulos T. Hyaluronan-derived swelling of solid tumors, the contribution of collagen and cancer cells, and implications for cancer therapy. Neoplasia 2016, 18, 732. 10.1016/j.neo.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]