Abstract
Recent years have witnessed the emergence of bacterial semiorganelle encapsulins as promising platforms for bio-nanotechnology. To advance the development of encapsulins as nanoplatforms, a functional and structural basis of these assemblies is required. Encapsulin from Brevibacterium linens is known to be a protein-based vessel for an enzyme cargo in its cavity, which could be replaced with a foreign cargo, resulting in a modified encapsulin. Here, we characterize the native structure of B. linens encapsulins with both native and foreign cargo using cryo-electron microscopy (cryo-EM). Furthermore, by harnessing the confined enzyme (i.e., a peroxidase), we demonstrate the functionality of the encapsulin for an in vitro surface-immobilized catalysis in a cascade pathway with an additional enzyme, glucose oxidase. We also demonstrate the in vivo functionality of the encapsulin for cellular uptake using mammalian macrophages. Unraveling both the structure and functionality of the encapsulins allows transforming biological nanocompartments into functional systems.
Keywords: encapsulin, bacterial compartments, cryo-electron microscopy, nanoreactors, nanoplatforms
Protein-based nanocages have gained great interest in nanotechnology and have been developed into effective delivery agents (nanocarriers) and nanoreactors.1−3 In recent years, a class of nanocages from bacteria called the encapsulins has emerged as promising nanoplatforms, characterized by their robust nature.4−9 Nevertheless, compared to other protein-based nanocages, such as viral capsids,1,10 ferritins,11,12 and heat-shock proteins,13 the development of encapsulins for nanoplatforms is only in its infancy.6 While cages like viral capsids confine genetic material, previous studies highlighted that the encapsulins specifically confine functional and fully folded protein-based cargo, such as an enzyme or a ferritin-like protein, in that way being recognized as a bacterial semiorganelle. The encapsulin from Brevibacterium linens naturally houses a dye-decolorizing peroxidase (DyP) within its cavity, which is involved in oxidative stress. DyP is assembled as a trimer of dimers, i.e., a 240 kDa hexamer. The specific encapsulation mechanism of DyP is mediated by its C-terminal end, which interacts specifically with a defined region of the encapsulin inner surface. We recently demonstrated that fusion of the DyP C-terminal end to a heterologous protein, such as the teal fluorescent protein (TFP), allows its packaging in these nanocontainers.14
Despite the recent sharp increase in the number of studies on encapsulins,4,5,7−9,14−20 structural characterization of encapsulins at the molecular level is yet to be explored. Encapsulins from Myxococcus xanthus (PDB entry 4PT2)17 and Pyrococcus furiosus (PDB entry 2E0Z)21 assemble into T = 3 (180-subunit, ∼32 nm diameter) nanocompartments, whereas encapsulin from Thermotoga maritima (PDB entry 3DKT)6 assembles into a T = 1 (60-subunit, 24 nm) particle. Both icosahedral particles pack ferritin-like proteins that protect cells from oxidative stress.6,17 Notably, the encapsulin fold is similar to the capsid protein fold of Hong Kong 97 (HK97)-like virions22 (including dsDNA bacteriophages and herpesviruses), which constitute the most successful self-replicating system on Earth. Based on these structural similarities, encapsulins and the HK97-like capsid are hypothesized to share a common evolutionary origin; encapsulins could be of viral origin, or viruses might have originated from a similar cellular assembly.4,6 The structural characteristics at the molecular level of encapsulins that confine enzymes is still poorly understood. Deciphering the structure is crucial to advance the understanding of enzyme confinement in bacteria and the functioning of encapsulins as semiorganelles for biochemical reactions. To characterize the protein structure, as an alternative to the well-known protein crystallization, cryo-electron microscopy (cryo-EM) recently emerged as a powerful technique.
Here, we determine the structure of the encapsulin from B. linens with native DyP cargo (DyP-E) and foreign TFP cargo (TFP-E) using cryo-EM reconstruction. We also investigate the stability of the DyP-E system under various environmental changes to demonstrate the robust nature of the encapsulin particles. To put this robust nature into use and with the aim to substantiate experimentally that B. linens encapsulins are reconfigurable systems that function outside of its native environment (i.e., as bacterial semiorganelles), we separately performed an in vitro catalytic cascade involving immobilized encapsulins (using DyP-E) and an in vivo/cellular uptake study (using TFP-E). The two independent studies employing two different cargo systems, which are genetically reconstituted into B. linens encapsulins, demonstrate the use of encapsulins as versatile and reconfigurable nanoplatforms.
Results and Discussion
Characterization of Encapsulin Stability and Structure
To investigate the robust nature of B. linens encapsulin,20 we characterized the stability of native DyP-E against pH changes and higher ionic strength. The stability of encapsulin particles in seven different pH values was examined (i.e., acidic condition at pH 3–5, native condition at pH 7.5, basic condition at pH 9–11). Acetate buffer was used to prepare pH 3, pH 4, and pH 5 buffers, Tris-HCl buffer was used for pH 7.5 and pH 9 buffers, and phosphate buffer was used for pH 10 and pH 11 buffers. For each pH value, the encapsulin samples were incubated overnight and subsequently characterized using size-exclusion chromatography (SEC), dynamic light scattering (DLS), and transmission electron microscopy (TEM), respectively. Upon mixing and overnight incubation of encapsulin in pH 3 and pH 4 buffers, particle aggregation was suspected to occur as the solutions became cloudy. Centrifugation of the solutions resulted in white pellets, and therefore these samples were not further analyzed.
Apart from pH 3 and pH 4, the encapsulin particles seemed to be stable at all tested pH values, which is in agreement with a previous report.23 Characterization with SEC (Figure 1A,B), DLS (Figure 1C), and TEM (Figure 1D), respectively, showed that uniform and spherical structures were present in all samples. Notably, the elution profiles from SEC were different at basic pH (pH 9 and 10); instead of eluting at V = 11–12 mL (characteristic for native encapsulin particles), the encapsulin incubated at higher pH eluted at a lower volume (V = 9 mL), suggesting an increase in particle size, possibly due to aggregation. TEM analysis (Figure 1D) showed that typical spherical particles were still present at basic pH, although a tendency to aggregate was observed. Aggregation of particles was clearly seen when the incubation at elevated pH was prolonged to 2 days (Figure 1C), which could be partially reversed by adjusting the pH back to around neutral. Furthermore, we also noted that a second peak at V = 15 mL appeared in the case of encapsulin at pH 11 (Figure 1B), indicating the formation of smaller particles as it eluted at a higher volume than intact particles (V = 11–12 mL). This phenomenon was not observed previously at lower pH values. The species eluting at V = 15 mL was confirmed to be encapsulin-based by SDS-PAGE (Figure 1E, red arrow) since the protein band showed up at the same position as the band of native encapsulin at pH 7.5. Hence, it is concluded that native B. linens encapsulin partially disassembles at pH 11. Similar particle disintegration at elevated pH has been reported for T. maritima encapsulin.15
Figure 1.
Characterization of encapsulin at different pH values. (A) Size-exclusion profiles of encapsulin at acidic, native, and basic pH monitored at λ = 280 nm, revealing a single peak of encapsulin (V = 11–12 mL or V = 9 mL) for each profile. (B) Size-exclusion profiles of encapsulin at native and highly basic pH, showing an extra peak (number 3) at V = 15 mL apart from the typical encapsulin peak (numbers 1 and 2). (C) DLS-based size distributions of encapsulin at acidic, native, and basic pH. (D) TEM images of encapsulin at acidic, native, and basic pH, revealing intact spherical particles under all conditions. (E) Denaturing gel electrophoresis revealing the encapsulin protein band (∼28 kDa) present for all samples. Lanes 1, 2, and 3 correspond to peaks 1, 2, and 3 in B, respectively; lane 4 corresponds to the protein marker.
To investigate the effect of increasing ionic strength, DyP-E at native conditions (pH 7.5) and elevated pH (pH 9) were incubated with either 1 M NaCl, 1 M MgCl2, or 1 M CaCl2 in the solution. At both pH 7.5 and pH 9, the encapsulin particles eluted at a similar elution volume compared to the samples with no salt added (Figure S1). This finding indicates that the increase in ionic strength (up to the tested range) does not significantly compromise the stability of encapsulin particles. This is in stark contrast, for instance, with the Cowpea Chlorotic Mottle plant virus (CCMV), which already disassembles when the ionic strength is increased to ∼1 M at an elevated pH.24 As for several (nano)technological applications nonaqueous conditions are required, the effect of the addition of organic solvents was examined. Here, we chose two of the most common organic solvents added to protein solutions (usually for chemical modifications),25,26 which are ethanol and DMSO. On the basis of characterization with TEM, we could still observe the presence of intact, spherical particles upon the addition of up to 40% DMSO to the encapsulin solution at pH 7.5 (Figure S2). However, the number of intact particles was noticeably low, and most of them were already disassembled/degraded into smaller fragments, even already in the presence of 20% DMSO. Similarly, addition of 40% ethanol also resulted in predominantly degraded fragments, and only a very small amount of intact particles could be observed (Figure S2). On the contrary, addition of up to 20% ethanol did not seem to affect the structures, as they were highly comparable to the native encapsulin structures (Figure S2).
We used 3D cryo-EM to analyze the native structures of encapsulin from B. linens loaded with its natural cargo, DyP (DyP-E), or with TFP (TFP-E). Nonloaded encapsulin (nl-E) was included as a control (Figure 2A–C). Whereas DyP- and TFP-loaded encapsulin particles were homogeneous, with an internal density corresponding to the cargo, nl-E showed two types of particles, either empty or containing some material in the internal cavity at a 1:5 ratio (Figure 2A, white and black arrows, respectively). The presence of material inside nl-E is attributed to random/statistical packaging of cellular components that might occur during in vivo assembly; such a phenomenon was also indicated by previously reported broad distribution of masses of nl-E particles analyzed by native mass spectrometry.14,20 A 3D reconstruction (3DR) was obtained for each of the three sets of particles (Figure 2D–F). Based on a 0.5 Fourier shell correlation (FSC) threshold, the resolution was 11.4 Å for nl-E and 13.5 and 15 Å for TFP-E and DyP-E, respectively. These values suggested that structural integrity was better preserved in nl-E than in cargo-loaded encapsulins, in accordance with previous biophysical analyses.20 Our biochemical analysis further confirmed that nl-E particles do not undergo partial disassembly at elevated pH (i.e., a tendency to aggregate was observed at basic pH, instead of forming smaller particles, Figure S3). The outer diameter, determined from spherically averaged radial density plots of the 3DR, was 228 Å for nl-E and TFP-E, but DyP-E measured 232 Å. Despite this ∼2% size increase, the inner diameter was identical for the three maps. Encapsulin maps were icosahedral shells with an average thickness of 28–30 Å, and the particle is based on a T = 1 lattice.
Figure 2.
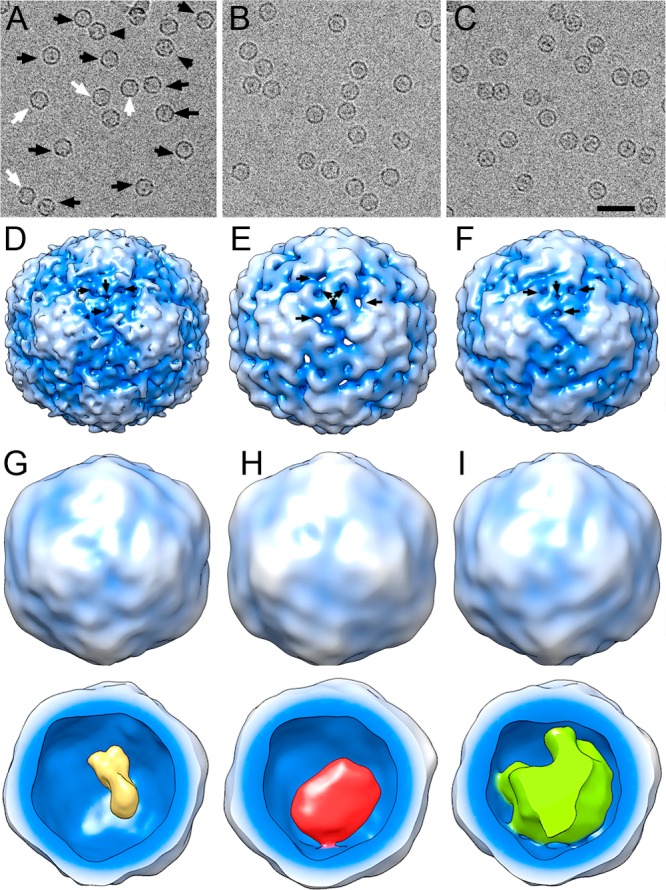
Cryo-EM images and 3DR of nonloaded and DyP- and TFP-loaded encapsulin from B. linens. (A–C) Cryo-electron micrographs of purified nonloaded encapsulin (nl-E) (A), DyP-loaded encapsulin (DyP-E) (B), and TFP-loaded encapsulin (TFP-E) (C). In A, white and black arrows indicate empty nl-E and nl-E with packed material, respectively. Bar, 500 Å. (D–F) 3DR with icosahedral symmetry of nl-E (D), DyP-E (E), and TFP-E (F) viewed along a 2-fold axis of symmetry contoured at 2σ above the mean density. Arrows indicate pores on the outer surface. (G–I) 3DR with no symmetry of nl-E (G), DyP-E (H), and TFP-E (I). Top row, the radially color-coded outer surfaces viewed along a 2-fold axis of symmetry contoured at 2σ above the mean density; bottom row, 3DR with the front half of the protein shell removed. Cargo is yellow (nl-E), red (DyP-E), or green (TFP-E).
At the moderate resolutions achieved, some features are distinguished; the capsid is formed by 12 slightly outward-protruding pentamers, each consisting of five curved, elongated subunits. Particles have ∼5–8 Å diameter holes that extend through the capsid. These pores were located around the 3-fold axes (Figure 2D–F, arrows) and might serve as channels through which solutes can be interchanged with the cytoplasm. To infer cargo organization, 3DRs of DyP-E, TFP-E, and nl-E particles were calculated without imposing icosahedral symmetry to avoid smearing cargo features (Figure 2G–I). Although the three maps showed smooth topography (average resolution ∼24 Å), the cargo density exhibited distinct features. Whereas DyP-related density was connected to the inner surface of DyP-E at one 3-fold axis, the TFP-related density occupied a large part of the inner cavity that, depending on the map contour, was connected at six or 12 locations. The internal density of nl-E capsids was much smaller, ∼30 and 15% less compared to those of DyP-E and TFP-E, respectively.
Encapsulin maps for B. linens were similar to the map for the T. maritima T = 1 encapsulin particle, which has a smaller diameter (230–240 Å). To evaluate this structural resemblance and considering a similar encapsulin fold, we performed docking analysis of the T. maritima encapsulin crystallographic model (PDB entry 3DKT) into the cryo-EM density maps of nonloaded and loaded T = 1 encapsulins. A total of 60 monomers of T. maritima encapsulin accounted well for the protein shell of B. linens nl-E (Figure 3A). This model includes the C-terminal extension of the packaged protein, a ferritin-like protein (Flp) from T. maritima (Figure 3A, yellow). In addition, most pores in the T. maritima encapsulin shell colocalized with the pores in B. linens nl-E (Figure 3B, dashed circles). In the functional context of native DyP-E, we were able to pinpoint the location of both the encapsulated DyP (i.e., connected to one of the 3-fold axes) and the pores (i.e., mostly at the 3-fold axes); we could deduce that the pores at the corresponding 3-fold axis can function as direct gates for substrates from the cytoplasm to access the confined enzyme. Together with previous results from the analyses on particle stability, these findings indicate that the encapsulin acts as a robust shell encasing the confined protein cargo, while at the same time allowing molecular flux through the shell resulting from the porous structure.
Figure 3.
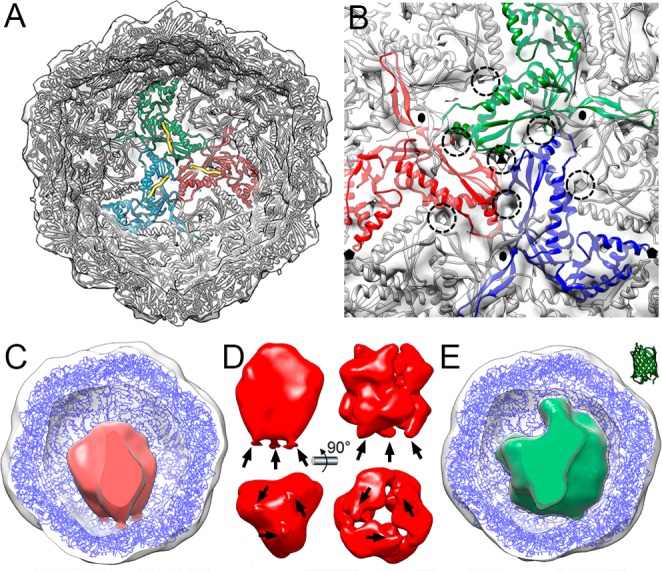
Pseudoatomic models of nl-E, DyP-E, and TFP-E. (A) nl-E inner surface viewed along a 3-fold axis, with docked T. maritima encapsulin atomic coordinates. Encapsulin monomers at the 3-fold axis are depicted in red, green, and blue, and their corresponding Flp C-terminal ends in yellow. (B) nl-E outer surface viewed along a 3-fold axis. There are numerous pores at and around the 3-fold axis (dashed circles). The cargo density has been removed computationally. Black symbols indicate icosahedral symmetry axes. (C) DyP-E map with C3 symmetry from inside, with docked T. maritima encapsulin (blue) and DyP density (red). (D) DyP cryo-EM density extracted from the map shown in C; arrows indicate connections to the inner encapsulin surface (left). DyP density calculated from negative staining electron microscopy6 (right). (E) TFP-E map from inside, with docked T. maritima encapsulin (blue) and TFP density (green). The TFP atomic structure (2HQK) is shown at the same scale (top right).
The role of the DyP C-terminal extension is, as it is for Flp, to target the cargo protein to the binding sites on the encapsulin interior. Considering that DyP is organized as a trimer of dimers, we calculated a 3DR of DyP-E with C3 symmetry (Figure 3C). The map resolution was slightly improved compared to the map calculated with no symmetry (25 Å versus 28 Å). Furthermore, DyP was solved as a density with three connections that match with the location of the Flp C-terminal extensions (Figure 3D, left). The B. linens DyP, expressed in the absence of encapsulin, was used to calculate a map from negatively stained particles (Figure 3D, right, EMD entry 1530).6 This recombinantly expressed DyP is similar to our native encapsulated DyP model. The cryo-EM hexameric DyP map highlighted the structural differences between the two trimers; the trimer connected to the encapsulin capsid interior probably has ordered C-terminal extensions, whereas the other three C-terminal extensions are probably disordered. The cryo-EM-packed hexameric DyP was observed as an elongated density (∼114 Å long × ∼ 92 Å wide; ∼3.3 × 105 Å3 volume), compatible with the dimensions and volume of a recombinant hexameric DyP (∼104 × 104 Å; 2.85 × 105 Å3). The cryo-EM TFP-related density was ∼5.1 × 105 Å3 (Figure 3E); a TFP monomer (PDB entry 2HQK; a TFP derived from Clavularia sp.)27 is 4.7 × 104 Å3, which suggested that 10–12 TFP copies/particle are encapsulated, compatible with previous mass spectrometry analysis.14 The internal volume of the B. linens encapsulin particle (∼2.6 × 106 Å3) does not limit loading of the natural cargo, a single hexameric DyP (240 kDa). DyP loading capacity is instead constrained by the shape and oligomeric state of the cargo enzyme. Although the DyP hexamer has six C-terminal extensions that are able to interact with their cargo-binding sites, its quaternary structure indicates that only three of these extensions are bound. By similar reasoning, our TFP-E map indicates that the TFP monomers are evenly distributed in the encapsulin interior, and the TFP-E particle can contain 10–12 copies (∼400 kDa) of monomeric TFP.
DyP-E and TFP-E have lower breaking force than nl-E, according to atomic force microscopy (AFM) studies, which implies that DyP- and TFP-loaded encapsulins are less stable.20 The estimated resolution values for the calculated maps, which are inversely related to the number of particles in each map, also suggested that structural integrity was better preserved in nl-E than in cargo-loaded encapsulins. As suggested,20 binding of the three cargo-anchoring sequences to the shell interior could distort locally icosahedral symmetry for DyP-E. This local distortion in a 3-fold symmetry axis might be needed for optimal function of the hexameric DyP enzyme complex. For TFP-E, binding of multiple TFP (up to 10–12 copies) to the interior surface of the encapsulin pocket results in a similar overall destabilization of the shell.
In Vitro Study of Encapsulin as Nanoreactors on the Surface
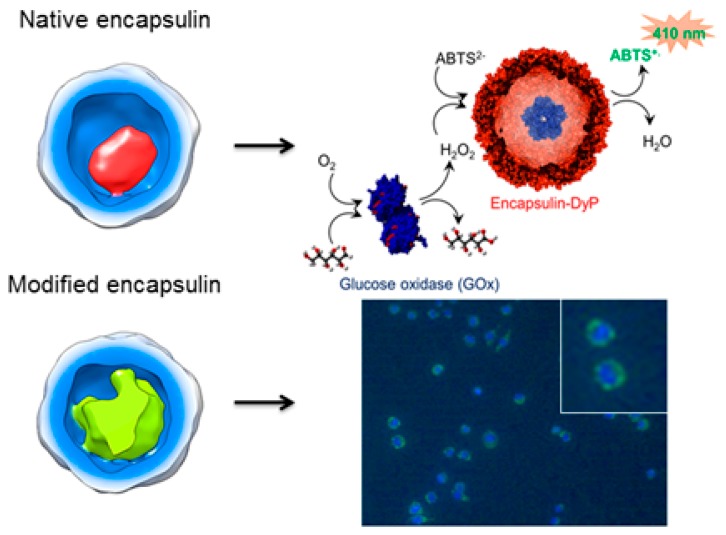
To demonstrate the functionality of encapsulins for in vitro applications, we investigated the performance of native DyP-E as a bionanoreactor. The confined DyP inside the encapsulin cavity allows the particles to catalytically oxidize dye molecules, for example 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) dye, while reducing hydrogen peroxide to water.19,28 We immobilized the encapsulin particles on a glass surface in order to ease the particle recovery as well as to demonstrate the catalysis on a surface, which is commonly preferred for in vitro application such as protein-based sensors.29,30 In comparison to tethering enzymes directly onto the surface, the immobilized encapsulin would offer a robust and chemically modifiable shell for, in principle, a variety of confined enzymes. The glass surface was first modified with pentafluorophenyl silicate (Figure 4A), which introduces surface reactivity and allows chemical reaction with lysine residues present on the encapsulin surface.31 We have previously reported that the lysine residues of B. linens encapsulin are indeed available for chemical modification.18
Figure 4.
Surface-immobilized encapsulin as a bionanoreactor. (A) Immobilization strategy of encapsulin particles on a glass surface using PFPS molecules as linkers. (B) SEM image of surface-immobilized encapsulin showing the presence of nanometer-sized particles (∼24 nm) on the surface. (C) Schematic representation of a tandem system consisting of GOx and DyP-E. The initial substrate glucose is oxidized by GOx while producing H2O2, which in turn is converted by DyP into water while producing green-colored ABTS radical. (D) Catalytic assay of the tandem system monitored at λ = 410 nm based on the absorption of the ABTS radical cation.
To confirm that the particles were covalently immobilized onto the surface using this approach, we performed a contact angle measurement on the encapsulin-modified glass surface, which resulted in the contact angle of 55–60° as opposed to 75–85° for a PFPS-modified glass surface (before adding the encapsulin). The decrease in the contact angle after encapsulin addition indicated a successful immobilization, as the presence of encapsulin changed the surface properties into a more polar state (in comparison to the surface with PFPS only). Furthermore, as shown in Figure 4B, we were also able to visualize the encapsulin particles on the surface using scanning electron microscopy (SEM). On the basis of multiple images recorded with SEM, we estimated the percentage of coverage area to be 35 ± 5% with up to 1.16 × 1011 encapsulin particles immobilized on the surface (area: 1 × 1.5 cm2).
After surface attachment, we investigated the catalytic activity by carrying out a cascade reaction on the surface. The encapsulin enzymatic activity was tested based on the oxidization of ABTS dye while reducing H2O2 produced by an additional enzyme called glucose oxidase (GOx) that was added into the solution (Figure 4C). We chose the well-studied tandem GOx-peroxidase system, as it has been proven useful as a biosensor for glucose detection,28,32,33 thus providing our system with a possible functionality. The oxidation of ABTS into its green-colored radical form was monitored at λ = 410 nm for up to 6 min (Figure 4D) at different glucose concentrations. In our experiments, GOx (1 μM) was added as the final compound into the reaction mixture, as it kick-starts the entire cascade reaction. Noticeably, the DyP-catalyzed formation of ABTS-based radical did not start immediately after GOx addition (t = 0, Figure 4D), which is attributed to GOx-catalyzed H2O2 production and diffusion to the surface-immobilized, encapsulated DyP. As expected, this “lag phase” was shortened by increasing the concentration of glucose as more H2O2 is produced with increasing glucose concentration. In a similar way the “lag phase” decreased when the surface-immobilized encapsulin was added into a premixed glucose–GOx solution with H2O2 already present (Figure S4), which did not change the reaction rate.
On the basis of the maximum slope (max dA410/dt), we calculated the reaction rates (i.e., the rate of the ABTS radical cation being formed) to be 2.67 μM/min for 0.5 mM glucose, 1.57 μM/min for 3.2 mM glucose, and 1.83 μM/min for 5 mM glucose. As the activity at lower concentrations is minimal (see Supporting Information section S4), we assume that 0.5 mM glucose was sufficient to achieve the maximum reaction rate on the surface (i.e., Vmax = 2.67 μM/min), particularly since increasing glucose concentrations did not lead to a higher reaction rate and likely resulted in substrate inhibition instead.34 Considering that the number of the encapsulins immobilized is equal to the number of confined DyP since each particle encapsulates one DyP (i.e., 1.16 × 1011 particles), we could calculate the apparent turnover number of (Vmax/DyP concentration) to be kcat = 115.4 s–1 (within the reaction volume of 500 μL, the total protein concentration = 3.86 × 10–10 M). The obtained turnover number is comparable in magnitude to reported turnover numbers for (nonencapsulated) DyP in the literature (i.e., kcat = 224 s–1),35 indicating that the surface immobilization did not lead to a significant activity loss. We have demonstrated here the catalytic activity of surface-immobilized encapsulin in vitro and the use of B. linens encapsulins as bionanoreactors.
Cellular Uptake of Encapsulin
To evaluate the use of B. linens encapsulins for in vivo applications, we investigated the uptake of the encapsulin with a fluorescent protein cargo (TFP-E) by mammalian J774 macrophages. A more detailed biochemical analyses of TFP-E particles has been reported in previous work by our group.14 Notably, the modified particles remain intact and stable over five years of storage at 4 °C (Figure S5), further confirming their robust nature even with a non-native cargo.
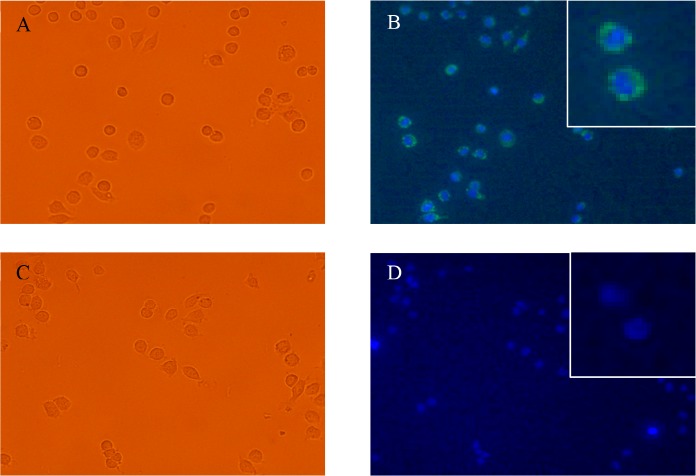
After the addition of TFP-E to the macrophage cells and subsequent incubation for 4 h, images were taken with a fluorescent microscope to visualize the TFP cargo (excitation at λ = 460–490 nm, emission at λ = 525 nm) and the nuclei of the cells with Hoechst-staining (excitation at λ = 360–370 nm, emission at λ = 460 nm) (Figure 5). The negative control, a sample of cells that were not treated with any encapsulin particles and that remain present after the removal of culture medium, does not show any fluorescence signal (Figure 5C,D). For 0.033 μM TFP-E, the treated macrophages show a high fluorescent signal as shown in Figure 5A,B. The green-colored signal that is found around the nucleus indicates that the TFP cargo was taken up by the macrophages. On the basis of the fluorescence signal from the cells (Figure 5B, inset), TFP-E appears to enter the macrophages effectively, but does not enter the nucleus.
Figure 5.
Fluorescence microscopy images of J774 macrophages treated with TFP-E. (A) Bright-field image of macrophages treated with 0.033 μM TFP-E. (B) Fluorescence microscopy images of macrophages treated with 0.033 μM TFP-E showing fluorescence from TFP cargo protein taken up by the cells. The TFP (green-colored) does not enter the nucleus (inset). (C) Bright-field image of the negative control where macrophages were treated with PBS only. (D) Fluorescence microscopy images showing no green fluorescence in cells treated with PBS only.
This study shows that the encapsulins can be used to confine a foreign protein cargo and preserve their structural integrity for a prolonged time in vitro, and the cargo can be taken up by mammalian cells. Further studies are needed to identify whether the spherical particles stay intact inside the cells as well as the uptake mechanism.
Conclusion
We have provided structural insight into the encapsulins from B. linens using cryo-EM, confirming that the native icosahedral, cage-like assemblies confine a single peroxidase DyP within its cavity. Together with particle stability analyses, our findings indicate the encapsulin acts as a robust shell protecting the confined DyP, while at the same time allowing molecular flux through the shell due to the porous structure. Using the same analysis, we also characterized the structure of modified encapsulin particles with non-native cargo (i.e., a fluorescent protein). Compared to the empty encapsulin (nl-E) the protein cages that contain a cargo are slightly larger, which is in line with our previous analysis.20 The data further confirm the directive role the C-terminal fragments on the cargo proteins play in their inclusion,6,14 allowing the design and preparation of encapsulin containers with a variety of guest systems.
To investigate the functionality of encapsulins as a promising agent for nanoplatforms, we demonstrated a tandem in vitro catalysis, proving that the encapsulin is able to play a role as bionanoreactors on the surface as well as the in vivo cellular uptake of modified encapsulins by mammalian macrophages. Our results provide both a functional and structural basis of B. linens encapsulins for the development of encapsulins as functional assemblies for nanotechnology, for instance as therapeutic agents and biosensors.
Methods
Materials
B. linens encapsulin was recombinantly expressed in E. coli and purified based on reported procedures.14,18 Encapsulin samples were stored in “encapsulin storage buffer” containing 20 mM Tris-HCl, 150 mM NH4Cl, and 1 mM β-mercaptoethanol at pH 7.5 at 4 °C. For pH variation studies, 100 mM acetate buffer (CH3COONa–CH3COOH) was used to prepare pH 3, pH 4, and pH 5 buffers, 20 mM Tris-HCl buffer was used for pH 7.5 and pH 9 buffers, and 10 mM phosphate buffer (Na2HPO4–NaOH) was used for pH 10 and pH 11 buffers. All chemicals were purchased from Sigma-Aldrich unless stated otherwise. All incubations and reactions were conducted at room temperature unless stated otherwise.
Cryo-electron Microscopy and Image Processing
Nonloaded and DyP- and TFP-loaded B. linens encapsulin (5 μL) were applied to one side of Quantifoil R 2/2 holey grids, blotted, and plunged into liquid ethane in a Leica EM CPC cryofixation unit. Samples were analyzed in a Tecnai G2 electron microscope equipped with a field emission gun operating at 200 kV, and images were recorded under low-dose conditions with a FEI Eagle CCD at a detector magnification of 69 444× (2.16 Å/pixel sampling rate). Image processing operations were performed using Xmipp36 and Relion37 packages integrated in the Scipion platform.38 Graphic representations were produced by UCSF Chimera.39 The Xmipp automatic picking routine was used to select 6785, 19 591, and 29 680 individual particle images of nonloaded and DyP- and TFP-loaded encapsulin, respectively. A 1.2–4.1 μm defocus range was determined for each image with CTFfind4.40 Particle images were extracted, normalized, and downsampled to a factor of 2, with a final sampling ratio of 4.32 Å/pixel. Using the Relion routine, a two-dimensional (2D) classification was performed to discard low-quality particles, and 5357, 15 941, and 19 779 isometric particles were selected for nonloaded and DyP- and TFP-loaded encapsulin, respectively. A 3D classification was run using the structure of Thermotoga maritima encapsulin (PDB entry 3DKT), low-pass filtered to 40 Å, as an initial model. When icosahedral symmetry was imposed, a single class with 4246, 12 833, and 15 976 particles was obtained for nonloaded and DyP- and TFP-loaded encapsulin, respectively. When assuming no symmetry for nonloaded and DyP- and TFP-loaded encapsulin, three classes were obtained, but no significant differences were observed between them at the resolutions achieved, and particles were refined together. DyP-loaded encapsulin particles were analyzed considering C3 symmetry, and a single class with 9930 particles was selected. These data sets were used to obtain the final 3DR using the Relion autorefinement routine. Resolutions of 3D with icosahedral symmetry were estimated from two independent half data sets using the 0.5 (or 0.3) criterion of the Fourier shell correlation, and the values for nonloaded and DyP- and TFP-loaded encapsulin were 11.4 (10.7), 15.1 (13.5), and 13.5 (12) Å, respectively. Similarly, resolution values for asymmetric 3DR were 27.3 (23.8), 28.3 (25), and 24.2 (22.7) Å. For the DyP-loaded encapsulin with C3 symmetry, resolution was 24.7 (22.8) Å. The Chimera fitting tool was used to dock the atomic crystallographic model T. maritima encapsulin into our cryo-EM encapsulin maps. The encapsulin 3DR are deposited in the Electron Microscopy Data Bank (http://www.ebi.ac.uk/pdbe/emdb) with accession no. EMD-3608 (nl-E), EMD-3612 (DyP-E), and EMD-3615 (TFP-E) for maps with icosahedral symmetry; EMD-3609 (nl-E), EMD-3614 (DyP-E) and EMD-3616 (TFP-E) for maps without imposing icosahedral symmetry; and EMD-3613 (DyP-E with C3 symmetry).
Characterization of Encapsulin Stability
Size-exclusion profiles of encapsulins upon pH and ionic strength variation were obtained by injecting and running 500 μL of each sample (∼15 μM) into a Superose 6 preparative column 10/100 GL (GE Healthcare FPLC Äkta purifier 900 with a 24 mL bed volume). The size-exclusion analysis was repeated two times. The hydrodynamic size distribution of the particles was determined using a Nanotrac Wave (Microtrac) particle analyzer. For particle imaging with TEM, 5 μL of a sample was applied onto Formvar-carbon-coated grids, and the liquid was drained after 30 s. Afterward, 5 μL of a staining solution consisting of uranyl acetate (1% w/v) was added onto the grids, and the liquid was drained after 1 min. For protein characterization with denaturing gel electrophoresis (SDS-PAGE), experiments using 12% polyacrylamide gel were conducted based on procedures in the literature,41 and Bio-Safe Coomassie (Bio-Rad) was used for visualization of protein bands.
Immobilization of Encapsulin onto a Glass/Si Surface
Substrates were rinsed with water, activated by immersion/cycling piranha solution (H2SO4–H2O2, 3:1), rinsed with water and ethanol, and then dried with a steam of nitrogen. Perfluorophenyl-11-(triethoxysilyl)undecanoate (PFPS) was deposited by substrate immersion into a PFPS solution (dichloromethane (DCM), 10 mM) for 24 h at room temperature under an argon atmosphere. The glass wafer was rinsed with DCM to remove unreacted reagent and dried in a stream of N2. Encapsulin particles were deposited from buffered solution (0.2 M phosphate buffer, pH 7.2). The particles were drop-coated on the flat glass substrate overnight in a closed vessel to avoid solvent evaporation. Afterward, the glass was rinsed with buffer solution to remove nonimmobilized particles.
Catalysis of Surface-Immobilized Encapsulin
A 200 μL amount of glucose (three different final concentrations: 0.5, 3.2, and 5 mM) and 200 μL of ABTS (2.5 mM) in Tris-HCl buffer pH 7.5 were placed inside a cuvette together with the glass surface with the immobilized encapsulin particles. Afterward, 100 μL of GOx (1 μM) in PBS buffer pH 7.4 was added into the cuvette to start the reaction, and the absorption value at λ = 410 nm was immediately recorded for at least 6 min using a PerkinElmer Lamba 850 UV–visible spectrometer. As control experiments, a PFPS-modified glass surface without encapsulin particles was used for similar kinetic studies.
The reaction rate based on radical ABTS production is calculated as follows:
where A is the absorbance of radical ABTS at λ = 410 nm, ε is the extinction coefficient of radical ABTS at λ = 410 nm (36 000 M–1 cm–1), and b is the cuvette path length (1 cm). dA/dt corresponds to the maximum slope of the kinetic plots in Figures 4D and S4 derived using OriginPro 9.0 software. The apparent turnover number kcat was calculated based on Vmax/DyP concentration. Although the particles were immobilized on the glass surface, the catalytic assay was performed in solution (i.e., the modified surface was fully immersed inside a cuvette). The DyP concentration was calculated based on the number of particles on the surface and the volume of the reaction. The catalytic assay was repeated at least two times.
Cell Experiments
B. linens encapsulins containing mTFP (monomeric teal fluorescent protein) were recombinantly produced in E. coli and purified using the same protocol established for native B. linens encapsulin. The concentration of TFP-E was 0.30 μM. Murine macrophage cells (J774) were cultivated in DMEM. Following the cultivation, 100 μL of ∼2000 cells was plated per well on a 96-well plate, and either 0 or 10 μL of TFP-E was added into the well (0 or 0.03 μM TFP-E, respectively). The treated cells were incubated at 37 °C and 5% CO2 for 4 h to allow the cells to take up the particles. After 3.5 h of incubation, Hoechst nucleus stain was added to a final concentration of 0.5 μg/mL. Following the incubation, the medium was removed and the cells were rinsed 2× with phosphate buffer saline (10 mM PBS, pH 7.4) to further remove nonabsorbed species. A 100 μL amount of HEPES was added to the cells prior to visualization. For the visualization based on fluorescence, the cells were imaged using a fluorescence microscope (Olympus TH4-200 with an X-Cite series 120pc Q laser from Lumen Dynamics, excitation at λ = 460–490 nm, emission at λ = 525 nm).
Data and Schematic Representation
All data plotting and mathematical calculations were performed with OriginPro 9.0 software. Protein structures are rendered using PyMOL 1.3 software, and chemical structures are drawn using ChemBioDraw Ultra 12.0 software.
Acknowledgments
We acknowledge financial support from the ERC Consolidator Grant (Protcage) and the Indonesia Endowment Fund for Education (LPDP). This work was supported in part by grants from the Spanish Ministry of Economy and Competitivity (BFU2014-55475 to J.R.C.) and the Comunidad Autónoma de Madrid (S2013/MIT-2807 to J.R.C.). We are grateful to Dr. E. G. Keim, M. A. Smithers, and R. van der Hee (MESA+ Institute for Nanotechnology, University of Twente) for assistance with TEM, SEM imaging, and cell culture. We are grateful to Prof. N. Ban and Dr. M. Sutter for the kind donation of plasmids and to Dr. J. Snijder for fruitful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b07669.
Native encapsulin (DyP-E) response to variation of ionic strength and addition of organic solvents, analysis of empty encapsulins (nl-E), cascade catalysis of GOx and surface-immobilized DyP-E, and stability of modified encapsulin (TFP-E) over time (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Uchida M.; Klem M. T.; Allen M.; Suci P.; Flenniken M.; Gillitzer E.; Varpness Z.; Liepold L. O.; Young M.; Douglas T. Biological Containers: Protein Cages as Multifunctional Nanoplatforms. Adv. Mater. 2007, 19, 1025–1042. 10.1002/adma.200601168. [DOI] [Google Scholar]
- Peek L. J.; Middaugh C. R.; Berkland C. Nanotechnology in Vaccine Delivery. Adv. Drug Delivery Rev. 2008, 60, 915–928. 10.1016/j.addr.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J.; Lee N. K.; Kim I. S. Bioengineered Protein-Based Nanocage for Drug Delivery. Adv. Drug Delivery Rev. 2016, 106, 157–171. 10.1016/j.addr.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Giessen T. W. Encapsulins: Microbial Nanocompartments with Applications in Biomedicine, Nanobiotechnology and Materials Science. Curr. Opin. Chem. Biol. 2016, 34, 1–10. 10.1016/j.cbpa.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Choi B.; Moon H.; Hong S. J.; Shin C.; Do Y.; Ryu S.; Kang S. Effective Delivery of Antigen-Encapsulin Nanoparticle Fusions to Dendritic Cells Leads to Antigen-Specific Cytotoxic T Cell Activation and Tumor Rejection. ACS Nano 2016, 10, 7339–7350. 10.1021/acsnano.5b08084. [DOI] [PubMed] [Google Scholar]
- Sutter M.; Boehringer D.; Gutmann S.; Guenther S.; Prangishvili D.; Loessner M. J.; Stetter K. O.; Weber-Ban E.; Ban N. Structural Basis of Enzyme Encapsulation into a Bacterial Nanocompartment. Nat. Struct. Mol. Biol. 2008, 15, 939–947. 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- Moon H.; Lee J.; Min J.; Kang S. Developing Genetically Engineered Encapsulin Protein Cage Nanoparticles as a Targeted Delivery Nanoplatform. Biomacromolecules 2014, 15, 3794–3801. 10.1021/bm501066m. [DOI] [PubMed] [Google Scholar]
- Tamura A.; Fukutani Y.; Takami T.; Fujii M.; Nakaguchi Y.; Murakami Y.; Noguchi K.; Yohda M.; Odaka M. Packaging Guest Proteins into the Encapsulin Nanocompartment from Rhodococcus erythropolis N771. Biotechnol. Bioeng. 2015, 112, 13–20. 10.1002/bit.25322. [DOI] [PubMed] [Google Scholar]
- Corchero J. L.; Cedano J. Self-Assembling, Protein-Based Intracellular Bacterial Organelles: Emerging Vehicles for Encapsulating, Targeting and Delivering Therapeutical Cargoes. Microb. Cell Fact. 2011, 10, 92. 10.1186/1475-2859-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir N.; Streatfield S. J.; Yusibov V. Virus-Like Particles as a Highly Efficient Vaccine Platform: Diversity of Targets and Production Systems and Advances in Clinical Development. Vaccine 2012, 31, 58–83. 10.1016/j.vaccine.2012.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Z.; Tang W.; Todd T.; Xie J. Ferritins as Nanoplatforms for Imaging and Drug Delivery. Expert Opin. Drug Delivery 2014, 11, 1913–1922. 10.1517/17425247.2014.941354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K.; Hoiczyk E.; Okuda M. Ferroxidase-Mediated Iron Oxide Biomineralization: Novel Pathways to Multifunctional Nanoparticles. Trends Biochem. Sci. 2016, 41, 190–203. 10.1016/j.tibs.2015.11.011. [DOI] [PubMed] [Google Scholar]
- Choi S. H.; Kwon I. C.; Hwang K. Y.; Kim I. S.; Ahn H. J. Small Heat Shock Protein as a Multifunctional Scaffold: Integrated Tumor Targeting and Caspase Imaging within a Single Cage. Biomacromolecules 2011, 12, 3099–3106. 10.1021/bm200743g. [DOI] [PubMed] [Google Scholar]
- Rurup W. F.; Snijder J.; Koay M. S. T.; Heck A. J. R.; Cornelissen J. J. L. M. Self-Sorting of Foreign Proteins in a Bacterial Nanocompartment. J. Am. Chem. Soc. 2014, 136, 3828–3832. 10.1021/ja410891c. [DOI] [PubMed] [Google Scholar]
- Cassidy-Amstutz C.; Oltrogge L.; Going C. C.; Lee A.; Teng P.; Quintanilla D.; East-Seletsky A.; Williams E. R.; Savage D. F. Identification of a Minimal Peptide Tag for In Vivo and In Vitro Loading of Encapsulin. Biochemistry 2016, 55, 3461–3468. 10.1021/acs.biochem.6b00294. [DOI] [PubMed] [Google Scholar]
- Contreras H.; Joens M. S.; McMath L. M.; Le V. P.; Tullius M. V.; Kimmey J. M.; Bionghi N.; Horwitz M. A.; Fitzpatrick J. A.; Goulding C. W. Characterization of a Mycobacterium tuberculosis Nanocompartment and Its Potential Cargo Proteins. J. Biol. Chem. 2014, 289, 18279–18289. 10.1074/jbc.M114.570119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh C. A.; Fontana J.; Nemecek D.; Cheng N. Q.; Aksyuk A. A.; Heymann J. B.; Winkler D. C.; Lam A. S.; Wall J. S.; Steven A. C.; et al. A Virus Capsid-Like Nanocompartment that Stores Iron and Protects Bacteria from Oxidative Stress. EMBO J. 2014, 33, 1896–1911. 10.15252/embj.201488566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putri R. M.; Fredy J. W.; Cornelissen J. J. L. M.; Koay M. S. T.; Katsonis N. Labelling Bacterial Nanocages with Photo-Switchable Fluorophores. ChemPhysChem 2016, 17, 1815–1818. 10.1002/cphc.201600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanpour R.; Bugg T. D. H. Assembly In Vitro of Rhodococcus jostii RHA1 Encapsulin and Peroxidase DypB to Form a Nanocompartment. FEBS J. 2013, 280, 2097–2104. 10.1111/febs.12234. [DOI] [PubMed] [Google Scholar]
- Snijder J.; Kononova O.; Barbu I. M.; Uetrecht C.; Rurup W. F.; Burnley R. J.; Koay M. S. T.; Cornelissen J. J. L. M.; Roos W. H.; Barsegov V.; et al. Assembly and Mechanical Properties of the Cargo-Free and Cargo-Loaded Bacterial Nanocompartment Encapsulin. Biomacromolecules 2016, 17, 2522–2529. 10.1021/acs.biomac.6b00469. [DOI] [PubMed] [Google Scholar]
- Akita F.; Chong K. T.; Tanaka H.; Yamashita E.; Miyazaki N.; Nakaishi Y.; Suzuki M.; Namba K.; Ono Y.; Tsukihara T.; et al. The Crystal Structure of a Virus-Like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses. J. Mol. Biol. 2007, 368, 1469–1483. 10.1016/j.jmb.2007.02.075. [DOI] [PubMed] [Google Scholar]
- Abrescia N. G.; Bamford D. H.; Grimes J. M.; Stuart D. I. Structure Unifies the Viral Universe. Annu. Rev. Biochem. 2012, 81, 795–822. 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- Valdes-Stauber N.; Scherer S. Isolation and Characterization of Linocin M18, a Bacteriocin Produced by Brevibacterium linens. Appl. Environ. Microbiol. 1994, 60, 3809–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavelle L.; Michel J. P.; Gingery M. The Disassembly, Reassembly and Stability of CCMV Protein Capsids. J. Virol. Methods 2007, 146, 311–316. 10.1016/j.jviromet.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Teramoto H.; Nakajima K.; Takabayashi C. Chemical Modification of Silk Sericin in Lithium Chloride/Dimethyl Sulfoxide Solvent with 4-Cyanophenyl Isocyanate. Biomacromolecules 2004, 5, 1392–1398. 10.1021/bm034537r. [DOI] [PubMed] [Google Scholar]
- Basle E.; Joubert N.; Pucheault M. Protein Chemical Modification on Endogenous Amino Acids. Chem. Biol. 2010, 17, 213–227. 10.1016/j.chembiol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Ai H. W.; Henderson J. N.; Remington S. J.; Campbell R. E. Directed Evolution of a Monomeric, Bright and Photostable Version of Clavularia cyan Fluorescent Protein: Structural Characterization and Applications in Fluorescence Imaging. Biochem. J. 2006, 400, 531–540. 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X.; Chen Y.-P.; Liu T.-F.; Zhou H.-C. Coupling Two Enzymes into a Tandem Nanoreactor Utilizing a Hierarchically Structured MOF. Chem. Sci. 2016, 7, 6969–6973. 10.1039/C6SC01438K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassolas A.; Blum L. J.; Leca-Bouvier B. D. Immobilization Strategies to Develop Enzymatic Biosensors. Biotechnol. Adv. 2012, 30, 489–511. 10.1016/j.biotechadv.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Homaei A. A.; Sariri R.; Vianello F.; Stevanato R. Enzyme Immobilization: An Update. J. Chem. Biol. 2013, 6, 185–205. 10.1007/s12154-013-0102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.; Yang L.; Traulsen C. H. -H.; Cornelissen J. J. L. M. Immobilization of Catalytic Virus-like Particles in a Flow Reactor. Chem. Commun. 2017, 53, 7632–7634. 10.1039/C7CC03024J. [DOI] [PubMed] [Google Scholar]
- Heller A.; Feldman B. Electrochemical Glucose Sensors and Their Applications in Diabetes Management. Chem. Rev. 2008, 108, 2482–2505. 10.1021/cr068069y. [DOI] [PubMed] [Google Scholar]
- Shlyahovsky B.; Li D.; Katz E.; Willner I. Proteins Modified with DNAzymes or Aptamers Act as Biosensors or Biosensor Labels. Biosens. Bioelectron. 2007, 22, 2570–2576. 10.1016/j.bios.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Nicol M. J.; Duke F. R. Substrate Inhibition with Glucose Oxidase. J. Biol. Chem. 1966, 241, 4292–4293. [PubMed] [Google Scholar]
- Salvachúa D.; Prieto A.; Martínez A. T.; Martínez M. J. Characterization of a Novel Dye-decolorizing Peroxidase (DyP)-Type Enzyme from Irpex lacteus and Its Application in Enzymatic Hydrolysis of Wheat Straw. Appl. Environ. Microbiol. 2013, 79, 4316–4324. 10.1128/AEM.00699-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini R.; Masegosa I. M.; San Martin M. C.; Marco S.; Fernandez J. J.; de la Fraga L. G.; Vaquerizo C.; Carazo J. M. Xmipp: An Image Processing Package for Electron Microscopy. J. Struct. Biol. 1996, 116, 237–240. 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- Scheres S. H. RELION: Implementation of a Bayesian Approach to Cryo-EM Structure Determination. J. Struct. Biol. 2012, 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rosa-Trevin J. M.; Quintana A.; Del Cano L.; Zaldivar A.; Foche I.; Gutierrez J.; Gomez-Blanco J.; Burguet-Castell J.; Cuenca-Alba J.; Abrishami V.; et al. Scipion: A Software Framework Toward Integration, Reproducibility and Validation in 3D Electron Microscopy. J. Struct. Biol. 2016, 195, 93–99. 10.1016/j.jsb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera--A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rohou A.; Grigorieff N. CTFFIND4: Fast and Accurate Defocus Estimation from Electron Micrographs. J. Struct. Biol. 2015, 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro A. L.; Vinuela E.; Maizel J. V. Molecular Weight Estimation of Polypeptide Chains by Electrophoresis in SDS-Polyacrylamide Gels. Biochem. Biophys. Res. Commun. 1967, 28, 815–820. 10.1016/0006-291X(67)90391-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.