Abstract
The protein phosphatase Phlpp1 is an essential enzyme for proper chondrocyte function. Altered Phlpp1 levels are associated with cancer and degenerative diseases such as osteoarthritis. While much is known about the post-transcriptional mechanisms controlling Phlpp1 levels, transcriptional regulation of the Phlpp1 gene locus is underexplored. We previously showed that CpG methylation of the PHLPP1 promoter is lower in osteoarthritic cartilage than in normal cartilage, and indirectly correlates with gene expression. Here we further defined the effects of DNA methylation on PHLPP1 promoter activity in chondrocytes. We cloned a 1791 bp fragment of the PHLPP1 promoter (−1589:+202) and found that the first 500 bp were required for maximal promoter activity. General methylation of CpG sites within this fragment significantly blunts transcriptional activity, whereas site-specific methyltransferases HhaI or HpaII decreases transcriptional activation by approximately 50%. We located putative FoxO consensus sites within the PHLPP1 promoter region. Inhibition of DNA methylation by incorporation of 5-azacytidine increases Phlpp1 mRNA levels, but FoxO inhibition abolishes this induction. To determine which FoxO transcription factor mediates Phlpp1 expression, we performed overexpression and siRNA-mediated knock down experiments. Overexpression of FoxO3a, but not FoxO1, increases Phlpp1 levels. Likewise, siRNAs targeting FoxO3a, but not FoxO1, diminished Phlpp1 levels. Lastly, FoxO inhibition increases glycosaminoglycan staining of cultured chondrocytes and leads to concomitant increases in FGF18 and HAS2 expression. Together, these data demonstrate that CpG methylation and FoxO3a regulate PHLPP1 expression.
Keywords: osteoarthritis, cartilage Has2, CpG methylation, GAG
Cartilage formation and degeneration are under epigenetic control, by processes including DNA methylation, post-translational modification of histones and non-coding RNAs [Goldring and Marcu, 2012]. While all forms of epigenetic regulation maintain cartilage homeostasis, the impact of DNA methylation on osteoarthritis progression and cartilage development is increasingly recognized [Bomer et al., 2015; den Hollander et al., 2014; Ezura et al., 2009; Fernandez-Tajes et al., 2014; Jeffries et al., 2014; Kumar and Lassar, 2014; Moazedi-Fuerst et al., 2014; Rushton et al., 2014; Taylor et al., 2016]. DNA methylation of clustered CpG dinucleotides within gene promoters frequently leads to transcriptional repression due to transcription factor obstruction and/or by recruitment of silencing proteins [Deaton and Bird, 2011; Klose and Bird, 2006].
The PH-domain leucine rich protein phosphatase (Phlpp) 1 gene product is an atypical protein phosphatase that inactivates anabolic cell signaling pathways including Akt and PKC. As such, Phlpp1 is a critical regulator of cellular and tissue homeostasis. Alterations in Phlpp1 function and/or expression are associated with many pathological conditions, including cancer and degenerative diseases [Grzechnik and Newton, 2016]. Cellular levels of Phlpp1 are controlled by transcriptional and translational regulation and by ubiquitin-dependent proteolysis [Grzechnik and Newton, 2016]. Translational regulation of Phlpp1 levels is accomplished via mTORC1 and multiple microRNAs [Beezhold et al., 2011; Cai et al., 2013; Chang et al., 2014; Efeyan and Sabatini, 2010; Jiang et al., 2015a; Jiang et al., 2015b; Kim et al., 2016; Liu et al., 2011]. In contrast, protein degradation is βTCrP-dependent; however, mechanisms that control Phlpp1 transcription are underexplored [Gangula and Maddika, 2013; Gao et al., 2013; Li et al., 2009; Li et al., 2013; Sowa et al., 2009; Vera et al., 2015].
We previously showed that chondrocytes from OA patients exhibit markedly increased PHLPP1 expression as compared to chondrocytes within articular cartilage obtained from femoral neck fracture repair specimens [Bradley et al., 2015a]. Moreover, we demonstrated lower CpG methylation of the PHLPP1 promoter region in OA chondrocytes and that in vitro methylation of the PHLPP1 promoter decreases its transcriptional activity [Bradley et al., 2015a]. Phlpp1 deficiency increases cartilage content and Fgf18 expression by immature chondrocytes during cartilage development when FoxO1 is inactivated [Bradley et al., 2015b]. Moreover, FoxO1 blocks Fgf18 expression [Bradley et al., 2015b].
In this paper, we further examine the effects of DNA methylation on PHLPP1 promoter activity and identify several transcription factors that modulate PHLPP1 expression. We refine the sequence required for full transcriptional activation of the PHLPP1 promoter to approximately the first six hundred base pairs and show that DNA methylation attenuates activity of this fragment. Furthermore, we show that Phlpp1 promoter activity and mRNA levels are decreased with FoxO inhibition. This is accompanied by increased chondrocyte GAG production and elevated expression of FGF18 and HAS2. These data further support the crucial role of DNA methylation in controlling PHLPP1 transcription and suggest that FoxO transcription factors may facilitate PHLPP1 expression.
2. Methods
2.1 Chondrocyte Cell Culture, Transfection and Treatments
T/C28-a2 and ATDC5 cells were maintained in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic [Goldring, 2004]. Cells were seeded at a density of 2.7 × 104 cells/cm2 for RNA isolation or 1.5x104 cells/cm2 for transcription assays and incubated overnight. Cells were then treated as described within the text and figure legends. The FoxO inhibitor AS1842856 (Calbiochem) was reconstituted in DMSO then diluted 1:1000 in culture medium to obtain final working concentration of 1 μM. DMSO diluted 1:1000 in culture medium was used as a vehicle control condition. The FoxO inhibitor AS1842856 is a cell-permeable oxodihydroquinoline that primarily inhibits Forkhead box O family member Foxo1 (IC50 = 33 nM), but also affects the transcriptional activity of functionally related Foxo3a and Foxo4 (70%, 20%, and 3% inhibition, respectively at [AS184256] = 100 nM) [Nagashima et al., 2010]. To inhibit DNA methylation, 1 μM 5-azacytidine or vehicle (PBS) was added on day 1 of culture. Cells were transfected with pcDNA3, pcDNA-FoxO1A3 and/or pcDNA3-FoxO3a (Addgene no. 13508 and 10709 respectively) with Lipofectamine (Invitrogen) using a 1:3 (Lipofectamine:DNA) ratio. For knock down experiments, ON-TARGET plus siRNA smart pools targeting FoxO1, FoxO3 or a control siRNA were purchased from Dharmacon. ATDC5 cells were transfected with each siRNA using Lipofectamine RNAiMax at a 1:1 ratio. T/C28-a2, ATDC5 and primary immature chondrocytes were also cultured in 10 μl micromasses of 2x105 cells. After 1 hour, each well containing micromasses was flooded with DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic containing either the FoxO inhibitor or vehicle. Micromass cultures were incubated for three days and then collected for analyses.
2.2 Generation of PHLPP1-CpG-Free Reporter Plasmids
The −1589:+202 fragment of the PHLPP1 promoter (chromosome 18: 60376310- 60378100) was previously used to construct a CpG-free luciferase reporter [Bradley et al., 2015a; Hashimoto et al., 2009]. The full length promoter was truncated through digestions of HindIII and the following restriction enzymes: −1447:+202 (Smll), −1135:+202 (BsrGI), −967:+202 (Pmll), −792:+202 (BstBI), −431:+202 (Pvull), −53:+202 (SacII). All fragments were subcloned into a CpG-free luciferase reporter.
2.3 Plasmid Methylation and Dual Luciferase Assays
pCpG-Free-PHLPP1-Luc promoter fragment plasmids were treated in vitro with the CpG methyltransferases HhaI, HpaII, HhaI and HpaII, or M. SssI (10 units/μg DNA). Mock treated plasmids were used as control. pCpG-Free-PHLPP1-Luc plasmids were transfected into T/C28-a2 cells with pRL-LUC using lipofectamine reagent (1:3 ratio, Invitrogen). Dual luciferase activity assays (Promega) were performed 48 to 72 hours post-transfection as indicated in figure legends. Each assay condition was tested as 3 to 4 technical replicates within each experiment. All experiments were repeated three times with representative data shown.
2.4 Identification of Putative Transcription Factor Binding Sites
The −431:+202 sequence of the PHLPP1 promoter was scanned using the JASPAR Core Vertebrata transcription factor binding profile database at a relative profile threshold score of 95%. Identified consensus sequences were verified as present within the PHLPP1 promoter region.
2.5 RNA isolation and real-time PCR
Total RNA was extracted from cells using TRIzol (Invitrogen) and chloroform, and 2 μg was reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen). The resulting cDNAs were used to assay gene expression via real-time qPCR using the following gene-specific primers: PHLPP1 (5′-AGCTGAAAGCCATCCCCAACA-3′, 5′-GCTCAGGTCCACACACTTGA-3′), GADD45 (5′-AGACCGAAAGGATGGATAAGGTGG-3′, 5′-AGAGCCACATCTCTGTCGTC-3′), FGF18 (5′-GACAAGTATGCCCAGCTCCTA-3′, 5′-CATCAGGGCCGTGTAGTTGT-3′), HAS2 (5′-GACCAAGAGCTGAACAAGATGC-3′, 5′-GGTGTGATGCCAAAAAGGCA-3′), and GAPDH (5′-GACCTGACCTGCCGTCCTAGAAA-3′, 5′-CCTGCTTCACCACCTTCTTGA-3′). Fold changes in gene expression for each sample were calculated using the 2−ΔΔCq method relative to control after normalization of gene-specific Cq values to GAPDH Cq values [Razidlo et al.]. Each experiment was performed in triplicate and repeated at least three times. Results from a representative experiment are shown.
2.6 Western blotting
Cell lysates were collected in a buffered SDS solution (0.1% glycerol, 0.01% SDS, 0.1 m Tris, pH 6.8) on ice. Total protein concentrations were obtained with the Bio-Rad DC assay (Bio-Rad). Proteins (15 μg) were then resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Western blotting was performed with antibodies (1:2000 dilution) for Phlpp1 (Millipore, catalog #07-1341), FoxO1, FoxO3a (Cell Signaling Technologies, catalog #2880 and 2497), and tubulin (Developmental Studies Hybridoma Bank, Iowa City, IA) and corresponding secondary antibodies conjugated to horseradish peroxidase (HRP) (Santa Cruz Biotechnology, Santa Cruz, CA). Antibody binding was detected with the Supersignal West Femto Chemiluminescent Substrate (Pierce). Each experiment was repeated at least two times, and data from a representative experiment are shown.
2.7 Statistical analysis
Data obtained are the mean ± standard error of the mean (SEM). p values were determined with the Student’s t-test when only one experimental comparison was made. For assessment of significance with greater than two conditions, a one-way analysis of variance was performed. p < 0.05 was considered statistically significant.
3. Results
3.1 PHLPP1 Promoter Activity is Repressed by CpG Methylation
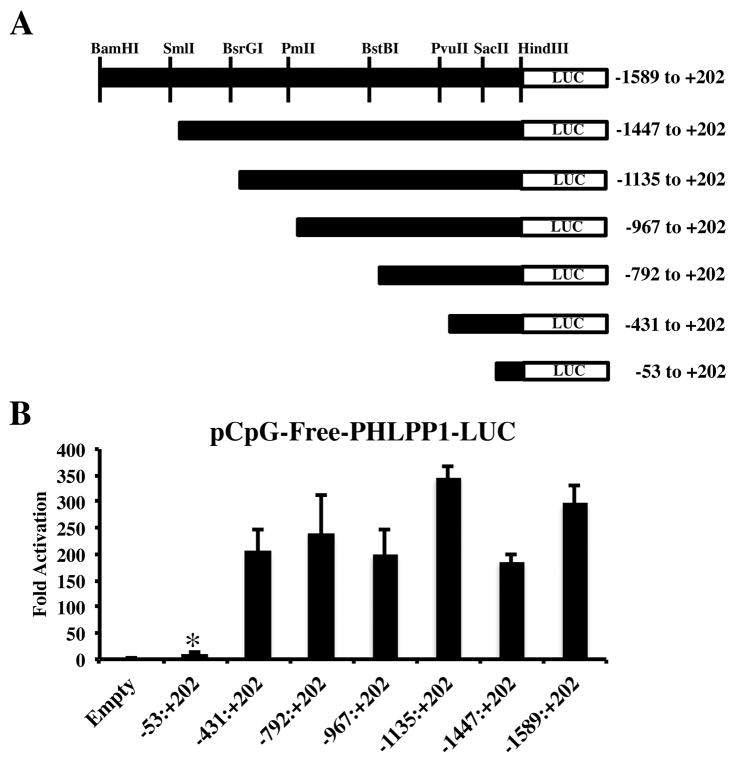
Our previous work demonstrated elevated PHLPP1 expression by OA articular chondrocytes correlated with demethylation of the PHLPP1 promoter [Bradley et al., 2015a]. Likewise, in vitro methylation of a PHLPP1 promoter fragment decreased transcriptional activity [Bradley et al., 2015a]. Here we further explored the effects of CpG methylation on PHLPP1 promoter activity to determine potential mechanisms of transcriptional control. We first constructed deletion mutants of the full-length (−1589:+202) PHLPP1 promoter construct which were subcloned into the pCpG-Free luciferase reporter (Figure 1A) [Bradley et al., 2015a; Hashimoto et al., 2009]. The −53:+202 deletion construct retained transcriptional activity, but was markedly blunted compared to the full-length PHLPP1 promoter (Figure 1B). In contrast, luciferase activity of the pCpG-free-PHLPP1-LUC −431:+202 fragment or any larger fragments was not statistically different from the full-length reporter (Figure 1B). These data indicate that most transcriptional regulation of the PHLPP1 promoter is imparted within the −431:+202 fragment.
Figure 1. Methylation blunts human PHLPP1 promoter activity.
A 1791 bp fragment (−1589:+202) was cloned into the pCpG-Free-LUC reporter. Deletions of the full-length promoter were generated. (A) Diagram of constructs. (B) Luciferase activity of each Phlpp1 promoter deletion construct.
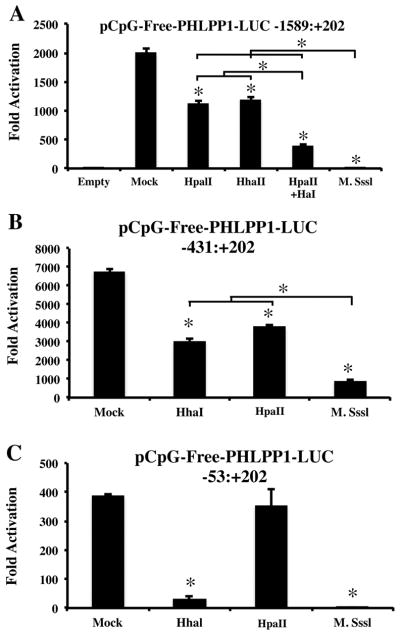
To further explore the impact of CpG methylation on PHLPP1 promoter transcriptional activity, we treated the full-length, −431:+202 or −53:+202 promoter fragments in vitro with the general CpG methyltransferase M. SssI, or the site-specific methyltransferases HhaI and/or HpaII. Mock treated plasmids were used as negative controls for each experiment. General methylation of the full-length PHLPP1 promoter markedly attenuated luciferase activity as previously observed [Bradley et al., 2015a]. Methylation with either site-specific methyltransferase reduced transcriptional activation by approximately 50%, whereas methylation using both HhaI and HpaII further decreased luciferase activity (Figure 2A). However, treatment with the general CpG methyltransferase M. SssI decreased transcriptional activity of the full-length promoter significantly as compared to either site-specific methyltransferase alone or in combination (Figure 2A). Since the −431:+202 PHLPP1 promoter fragment retained maximal transcriptional activity, we assessed the effects of CpG methylation using this luciferase reporter. Treatment with site specific methyltransferases HhaI or HpaII decreased transcriptional activation of the −431:+202 fragment by approximately 50%; no significant difference was observed when comparing treatment with HhaI to HpaII (Figure 2B). Treatment with M. SssI further decreased luciferase activity (Figure 2B). Lastly we determined the effects of each CpG methyltransferase on luciferase activity of the smallest PHLPP1 promoter fragment (−53:+202), as this fragment retained minimal transcriptional activity. HhaI and M. SssI significantly decreased transcriptional activity of this reporter; however, HpaII did not suppress luciferase activity (Figure 2C). These data indicate that specific CpG dinucleotides within the −431:+202 PHLPP1 promoter act to repress PHLPP1 expression.
Figure 2. Specific CpG dinucleotides within the −431:+202 PHLPP1 promoter dictate repression of PHLPP1 expression.
(A) The full length PHLPP1 promoter fragment was treated with the methyltransferases HpaII, HhaI HpaII + HhaI or M. SssI and luciferase activity of each fragment was evaluated. *p < 0.05 (B) The −431:+202 PHLPP1 promoter fragment was treated with the methyltransferases HpaII, HhaI or M. SssI and luciferase activity of each fragment was evaluated. *p < 0.05 (C) The −53:+202 PHLPP1 promoter fragment was treated with the methyltransferases HpaII, HhaI or M. SssI and luciferase activity of each fragment was evaluated. *p < 0.05
3.2 FoxO TFs regulate Phlpp1 expression in a methyl-sensitive manner
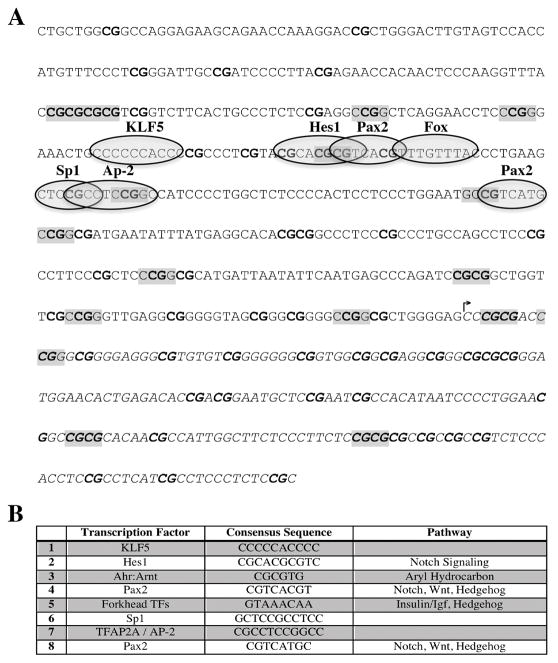
We used the JASPAR Core Vertebrata transcription factor binding profile database to identify putative transcription factor binding sites within the −431:+202 region of the PHLPP1 promoter that overlap and therefore can be blocked by CpG methylation. The PHLPP1 promoter sequence contains 16 HhaI and HpaII methylation sites, 70 CpG dinucleotides, and multiple transcription factor binding sites (Figure 3A). A single HhaI site is present within the −53:+202 sequence and accounts for the reduction in transcriptional activity observed in Figure 2C. A Sp1 consensus sequence that is known the regulate PHLPP1 transcription in a DNA methyl-dependent fashion [Dong et al., 2014] was identified. In addition, bindings sites for Klf5, Hes1, Pax2, Ap-2 and Fox transcription factors were identified. These transcription factors are induced by the Notch, Wnt, Hedgehog and Insulin/Igf pathways as indicated in Figure 3B.
Figure 3. The human PHLPP1 proximal promoter.
(A) CpG sites within the PHLPP1 −431: +202 promoter fragment are noted in bold. HhaI (GCGC) and HpaII (CCGG) methyltransferase sensitive sites are highlighted. The arrow denotes the transcription start site. (B) Table listing potential transcription factors with consensus sequences within the PHLPP1 −431:+202 promoter fragment and pathways that induce activity of each respective transcription factor.
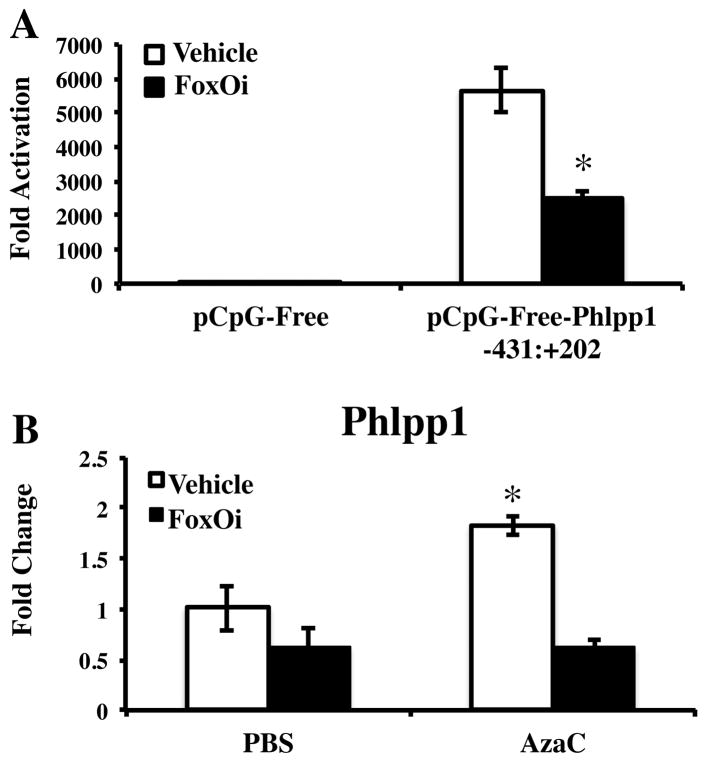
Because we previously showed increased FoxO levels with Phlpp1 deficiency, we investigated if FoxO transcription factors impacted PHLPP1 expression. The FoxO inhibitor AS1842856 significantly decreased luciferase activity of the −431:+202 PHLPP1 reporter (Figure 4A). No change in PHLPP1 promoter activity occurred when cells were treated with Wnt3a, Forskolin or DAPT under these conditions (data not shown). Inhibition of DNA methylation enhances Phlpp1 expression in ATDC5 cells (Figure 4B). To determine if this methylation-sensitive increase in Phlpp1 transcription was dependent on FoxO activity, we co-treated cells with the FoxO inhibitor. Induction of Phlpp1 levels produced by inhibition of DNA methylation was abolished by FoxO inhibition (Figure 4B). These data suggest that FoxO TFs regulate Phlpp1 expression in a methyl-sensitive manner.
Figure 4. FoxO inhibition attenuates Phlpp1 transcription in a methylation-sensitive manner.
(A) The −431:+202 PHLPP1 promoter fragment was transfected into T/C28-a2 cells and cells were treated with the FoxO inhibitor AS1842856 (1 μM) 24 hours following transfection. Dual luciferase assays were performed 72 hours post-transfection. *p < 0.05 as compared to DMSO control (B) ATDC5 cells were treated with 1 μM 5-azacytidine for 24 hours, after which the FoxO inhibitor AS1842856 (1 μM) was added for 24 hours. Expression of Phlpp1 was then determined by qPCR. *p < 0.05
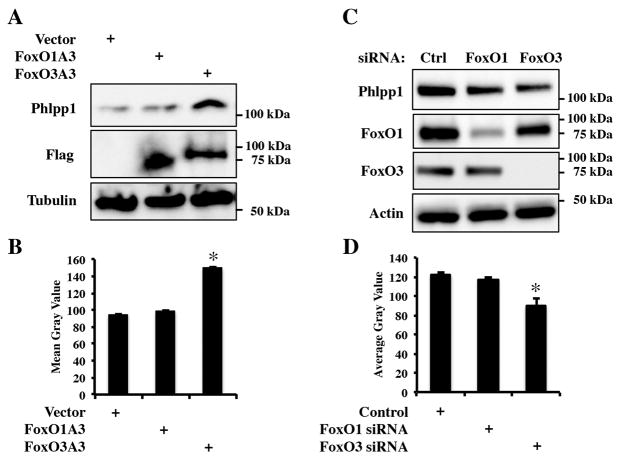
3.3 FoxO3a promotes Phlpp1 expression
The FoxO inhibitor AS184256 inhibits several FoxO isoforms, but targets FoxO1 and FoxO3 with greatest potency. To determine which isoform modulated Phlpp1 expression, FoxO1 or FoxO3a constructs were transfected into ATDC5 cells (Figure 5A,B). FoxO1 did not alter Phlpp1 levels, but expression of FoxO3 enhanced Phlpp1 levels. Next FoxO1 and FoxO3a levels were suppressed with siRNA pools (Figure 5C,D). FoxO1 siRNAs did not alter Phlpp1 levels, but siRNAs targeting FoxO3a reduced Phlpp1 expression. These results demonstrate that FoxO3a promotes Phlpp1 expression.
Figure 5. FoxO3a promotes Phlpp1 expression.
(A) ADTC5 cells were transfected with expression constructs as indicated and harvested after 48 hours. Western blotting was performed. (B) Mean gray values for each ban in panel A were measured. (C) ADTC5 cells were transfected with siRNAs as indicated and harvested after 48 hours. Western blotting was performed. (D) Mean gray values for each ban in panel C were measured.
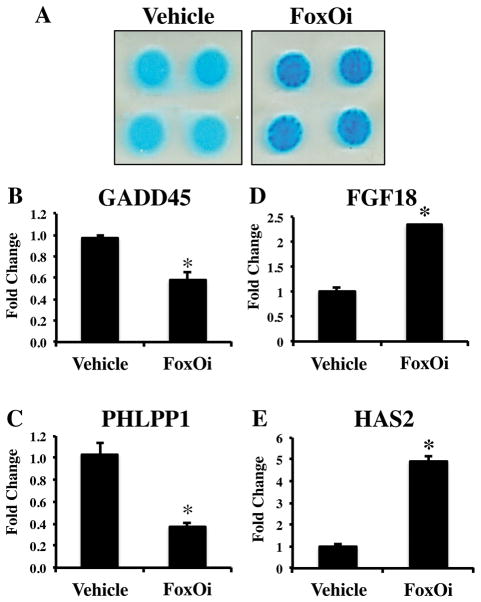
3.4 FoxO Inhibition Represses PHLPP1 Expression and Increases Cartilage Production
Since Phlpp1 deficiency or inhibition leads to increased cartilage matrix content [Bradley et al., 2015b], we also determined if FoxO inhibition increased GAG levels. T/C28-a2 cells were cultured in micromass and treated with the FoxO inhibitor. Micromasses treated with AS184256 stained more intensely with Alcian blue (Figure 6A). Similar results were obtained from micromasses of ATDC5 cells and primary immature articular chondrocytes (data not shown). Next we determined how FoxO inhibition affected expression of chondrogenic markers and Phlpp1. As expected, AS184256 decreased expression of the FoxO target gene Gadd45 (Figure 6B). Phlpp1 levels were also reduced with concomitant increases in Fgf18 and Has2 expression (Figure 6C–E). FoxO inhibition also elevated Fgf18 and Has2 transcripts in murine ATDC5 and primary immature articular chondrocytes (data not shown).
Figure 6. FoxO inhibition attenuates Phlpp1 transcription and increases cartilage production.
(A) T/C28-a2 cells were cultured in micromass and treated with the FoxO inhibitor (1 μM). After 3 days, cells were fixed and stained with Alcian blue. T/C28-a2 cells were also cultured in monolayer and treated with the FoxO inhibitor (1 μM) for 24 hours. Expression of (B) GADD45, (C) PHLPP1, (D) FGF18 and (E) HAS2 was determined by qPCR. *p < 0.05 as compared to DMSO control.
4. Discussion
Transcriptional activity of the PHLPP1 locus is an important mechanism controlling anabolic signaling pathways [Grzechnik and Newton, 2016]. Previous work showed that CpG methylation suppresses PHLPP1 promoter activity [Bradley et al., 2015a; Dong et al., 2014]. Here we demonstrate that the first six hundred base pairs of the PHLPP1 proximal promoter drive maximal transcription of the PHLPP1 locus in chondrocytes and that this sequence is highly sensitive to CpG DNA methylation. This proximal portion of the PHLPP1 promoter has a GC content of approximately 68 percent. The distal portion of the PHLPP1 promoter also contains CpG sites and is likewise sensitive to DNA methylation. The distal sites may have a weaker influence on PHLPP1 transcription, as longer promoter fragments did not enhance reporter activity.
The PHLPP1 promoter sequence contains putative transcription factor binding sites for Sp-1, Klf5, Hes1, Pax2, Ap-2 and Fox transcription factors. While Sp-1 is known to promote expression of PHLPP1 in a methyl-sensitive fashion, we also showed that FoxO3a, potentially in concert with FoxO1, influences PHLPP1 transcription [Dong et al., 2014]. Although binding of Pax2 to the PHLPP1 promoter could be affected by CpG methylation, we did not observe a change in PHLPP1 expression when activators of Pax2 transcriptional activity were altered.
Since we previously demonstrated that Phlpp1 regulates FoxO1 activity to control expression of Fgf18, we explored a potential feedback mechanism between FoxO and PHLPP1 [Bradley et al., 2015b]. Our data show that FoxO inhibition suppresses transcriptional activity of the Phlpp1 promoter and mRNA levels. This is accompanied by increased GAG staining and a concomitant increase in Fgf18 and Has2 mRNA levels. At concentrations used here, the FoxO inhibitor AS1842856 blocked transcriptional activity of FoxO1, 3 and 4 with decreasing efficiency respectively. We demonstrated that overexpression of FoxO3a enhanced expression of endogenous Phlpp1 levels, and that this effect was potentiated by the addition of FoxO1. Likewise, knockdown of FoxO3a reduced Phlpp1 levels; thus, FoxO1/3a isoforms may regulate transcription of the PHLPP1 locus. Moreover, mice lacking FoxO1 or FoxO1/3/4 in Col2Cre expressing cells exhibit increased chondrocyte proliferation and proteoglycan production, mirroring the effects of Phlpp1 deficiency [Bradley et al., 2015b; Matsuzaki et al., 2018]. FoxO deficient mice also exhibited OA-like changes with age, demonstrating that FoxO transcription factors are important for cartilage maintenance [Matsuzaki et al., 2018]. Reduced expression of FoxO transcription factors was shown to increase sensitivity of chondrocytes to oxidative stress [Akasaki et al., 2014b]. Enhanced chondrocyte cell death in response to oxidative stress is also a consequence of reduced FoxO expression and is thought to be due to reduced antioxidant- and autophagy-related pathways [Akasaki et al., 2014a]. Thus, FoxO inhibition may be beneficial in an OA disease state due to clearing of catabolic cells [Zhang et al., 2016]. FoxO TFs may also suppress activity of cartilage anabolic genes, as FoxO inhibition enhanced Fgf18 and Has2 expression and increased GAG staining.
In summary, Phlpp1 transcription is attenuated by CpG methylation. We further find that FoxO transcription factors regulate activity of the PHLPP1 promoter. FoxO inhibition also increases cartilage content and expression of Fgf18 and Has2, both of which may contribute to enhanced cartilage production. Future work will be aimed at defining how DNA methylation affects binding of these transcription factors to the PHLPP1 promoter.
Acknowledgments
This work was made possible by research and training grants from the NIH (AR065397, AR068103, AR056950), and Regenerative Medicine Minnesota. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of these funding agencies. The authors thank Dr. David Razidlo and Mr. Xiaodong Li for technical assistance.
Contract grant sponsors: National Institutes of Health and Regenerative Medicine Minnesota; Contract grant numbers: AR065397, AR056950, AR068103, MRM 2015 6272-01-02
References
- Akasaki Y, Alvarez-Garcia O, Saito M, Carames B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014a;66:3349–58. doi: 10.1002/art.38868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaki Y, Hasegawa A, Saito M, Asahara H, Iwamoto Y, Lotz MK. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage. 2014b;22:162–70. doi: 10.1016/j.joca.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beezhold K, Liu J, Kan H, Meighan T, Castranova V, Shi X, Chen F. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol Sci. 2011;123:411–20. doi: 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomer N, den Hollander W, Ramos YF, Bos SD, van der Breggen R, Lakenberg N, Pepers BA, van Eeden AE, Darvishan A, Tobi EW, Duijnisveld BJ, van den Akker EB, Heijmans BT, van Roon-Mom WM, Verbeek FJ, van Osch GJ, Nelissen RG, Slagboom PE, Meulenbelt I. Underlying molecular mechanisms of DIO2 susceptibility in symptomatic osteoarthritis. Ann Rheum Dis. 2015;74:1571–9. doi: 10.1136/annrheumdis-2013-204739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley EW, Carpio LR, McGee-Lawrence ME, Castillejo Becerra C, Amanatullah DF, Ta LE, Otero M, Goldring MB, Kakar S, Westendorf JJ. Phlpp1 facilitates post-traumatic osteoarthritis and is induced by inflammation and promoter demethylation in human osteoarthritis. Osteoarthritis Cartilage. 2015a doi: 10.1016/j.joca.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley EW, Carpio LR, Newton AC, Westendorf JJ. Deletion of the PH-domain and leucine rich repeat protein phosphatase 1 (Phlpp1) increases fibroblast growth factor (Fgf) 18 expression and promotes chondrocyte proliferation. J Biol Chem. 2015b doi: 10.1074/jbc.M114.612937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Fang L, Huang Y, Li R, Yuan J, Yang Y, Zhu X, Chen B, Wu J, Li M. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73:5402–15. doi: 10.1158/0008-5472.CAN-13-0297. [DOI] [PubMed] [Google Scholar]
- Chang RM, Yang H, Fang F, Xu JF, Yang LY. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology. 2014;60:1251–63. doi: 10.1002/hep.27221. [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander W, Ramos YF, Bos SD, Bomer N, van der Breggen R, Lakenberg N, de Dijcker WJ, Duijnisveld BJ, Slagboom PE, Nelissen RG, Meulenbelt I. Knee and hip articular cartilage have distinct epigenomic landscapes: implications for future cartilage regeneration approaches. Ann Rheum Dis. 2014;73:2208–12. doi: 10.1136/annrheumdis-2014-205980. [DOI] [PubMed] [Google Scholar]
- Dong L, Jin L, Tseng HY, Wang CY, Wilmott JS, Yosufi B, Yan XG, Jiang CC, Scolyer RA, Zhang XD, Guo ST. Oncogenic suppression of PHLPP1 in human melanoma. Oncogene. 2014;33:4756–66. doi: 10.1038/onc.2013.420. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22:169–76. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezura Y, Sekiya I, Koga H, Muneta T, Noda M. Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis Rheum. 2009;60:1416–26. doi: 10.1002/art.24472. [DOI] [PubMed] [Google Scholar]
- Fernandez-Tajes J, Soto-Hermida A, Vazquez-Mosquera ME, Cortes-Pereira E, Mosquera A, Fernandez-Moreno M, Oreiro N, Fernandez-Lopez C, Fernandez JL, Rego-Perez I, Blanco FJ. Genome-wide DNA methylation analysis of articular chondrocytes reveals a cluster of osteoarthritic patients. Ann Rheum Dis. 2014;73:668–77. doi: 10.1136/annrheumdis-2012-202783. [DOI] [PubMed] [Google Scholar]
- Gangula NR, Maddika S. WD repeat protein WDR48 in complex with deubiquitinase USP12 suppresses Akt-dependent cell survival signaling by stabilizing PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1) J Biol Chem. 2013;288:34545–54. doi: 10.1074/jbc.M113.503383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Kun T, Sheng Y, Qian M, Kong F, Liu X, Yu Z, Zhang H, Zhang Q, Gu J, Zhang X. SGT1 regulates Akt signaling by promoting beta-TrCP-dependent PHLPP1 degradation in gastric cancer cells. Mol Biol Rep. 2013;40:2947–53. doi: 10.1007/s11033-012-2363-8. [DOI] [PubMed] [Google Scholar]
- Goldring MB. Culture of immortalized chondrocytes and their use as models of chondrocyte function. Methods Mol Med. 2004;100:37–52. doi: 10.1385/1-59259-810-2:037. [DOI] [PubMed] [Google Scholar]
- Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012;18:109–18. doi: 10.1016/j.molmed.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik AT, Newton AC. PHLPPing through history: a decade in the life of PHLPP phosphatases. Biochem Soc Trans. 2016;44:1675–1682. doi: 10.1042/BST20160170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Oreffo RO, Gibson MB, Goldring MB, Roach HI. DNA demethylation at specific CpG sites in the IL1B promoter in response to inflammatory cytokines in human articular chondrocytes. Arthritis Rheum. 2009;60:3303–13. doi: 10.1002/art.24882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, James JA, Sawalha AH. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheumatol. 2014;66:2804–15. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhang Y, Guo Y, Yu C, Chen M, Li Z, Tian S, Sun C. MicroRNA-3127 promotes cell proliferation and tumorigenicity in hepatocellular carcinoma by disrupting of PI3K/AKT negative regulation. Oncotarget. 2015a;6:6359–72. doi: 10.18632/oncotarget.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wang C, Lei F, Zhang L, Zhang X, Liu A, Wu G, Zhu J, Song L. miR-93 promotes cell proliferation in gliomas through activation of PI3K/Akt signaling pathway. Oncotarget. 2015b;6:8286–99. doi: 10.18632/oncotarget.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Qiang L, Hayden MS, Sparling DP, Purcell NH, Pajvani UB. mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat Commun. 2016;7:10255. doi: 10.1038/ncomms10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Kumar D, Lassar AB. Fibroblast growth factor maintains chondrogenic potential of limb bud mesenchymal cells by modulating DNMT3A recruitment. Cell Rep. 2014;8:1419–31. doi: 10.1016/j.celrep.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu J, Gao T. beta-TrCP-mediated ubiquitination and degradation of PHLPP1 are negatively regulated by Akt. Mol Cell Biol. 2009;29:6192–205. doi: 10.1128/MCB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Stevens PD, Yang H, Gulhati P, Wang W, Evers BM, Gao T. The deubiquitination enzyme USP46 functions as a tumor suppressor by controlling PHLPP-dependent attenuation of Akt signaling in colon cancer. Oncogene. 2013;32:471–8. doi: 10.1038/onc.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stevens PD, Gao T. mTOR-dependent regulation of PHLPP expression controls the rapamycin sensitivity in cancer cells. J Biol Chem. 2011;286:6510–20. doi: 10.1074/jbc.M110.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T, Alvarez-Garcia O, Mokuda S, Nagira K, Olmer M, Gamini R, Miyata K, Akasaki Y, Su AI, Asahara H, Lotz MK. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med. 2018:10. doi: 10.1126/scitranslmed.aan0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazedi-Fuerst FC, Hofner M, Gruber G, Weinhaeusel A, Stradner MH, Angerer H, Peischler D, Lohberger B, Glehr M, Leithner A, Sonntagbauer M, Graninger WB. Epigenetic differences in human cartilage between mild and severe OA. J Orthop Res. 2014;32:1636–45. doi: 10.1002/jor.22722. [DOI] [PubMed] [Google Scholar]
- Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol. 2010;78:961–70. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- Razidlo DF, Whitney TJ, Casper ME, McGee-Lawrence ME, Stensgard BA, Li X, Secreto FJ, Knutson SK, Hiebert SW, Westendorf JJ. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS One. 5:e11492. doi: 10.1371/journal.pone.0011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton MD, Reynard LN, Barter MJ, Refaie R, Rankin KS, Young DA, Loughlin J. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66:2450–60. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SE, Li YH, Smeriglio P, Rath M, Wong WH, Bhutani N. Stable 5-Hydroxymethylcytosine (5hmC) Acquisition Marks Gene Activation During Chondrogenic Differentiation. J Bone Miner Res. 2016;31:524–34. doi: 10.1002/jbmr.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera J, Lartigue L, Vigneron S, Gadea G, Gire V, Del Rio M, Soubeyran I, Chibon F, Lorca T, Castro A. Greatwall promotes cell transformation by hyperactivating AKT in human malignancies. Elife. 2015:4. doi: 10.7554/eLife.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Mani SB, He Y, Hall AM, Xu L, Li Y, Zurakowski D, Jay GD, Warman ML. Induced superficial chondrocyte death reduces catabolic cartilage damage in murine posttraumatic osteoarthritis. J Clin Invest. 2016;126:2893–902. doi: 10.1172/JCI83676. [DOI] [PMC free article] [PubMed] [Google Scholar]