Abstract
For decades, hemagglutinin (HA) protein structure and its refolding mechanism have served as a paradigm for understanding protein-mediated membrane fusion. HA trimers are in a high-energy state and are functionally activated by low pH. Over the past decade, HA stability (or the pH at which irreversible conformational changes are triggered) has emerged as an important determinant in influenza virus host range, infectivity, transmissibility, and human pandemic potential. Here, we review HA protein structure, assays to measure its stability, measured HA stability values, residues and mutations that regulate its stability, the effect of HA stability on interspecies adaptation and transmissibility, and mechanistic insights into this process. Most importantly, HA stabilization appears to be necessary for adapting emerging influenza viruses to humans.
Keywords: pandemic, virus entry, interspecies adaptation, virus transmission, fusion glycoprotein, influenza A virus
Influenza A virus reservoirs and interspecies transmission pathways
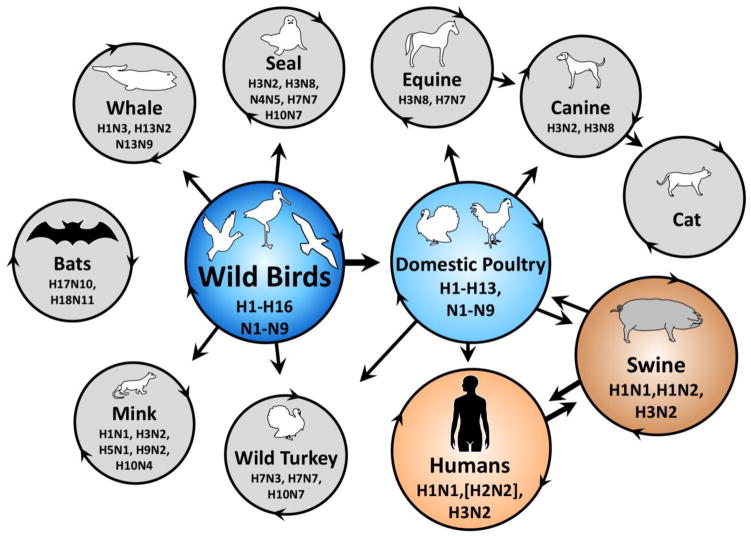
Influenza A viruses originate primarily from a reservoir of wild, aquatic birds (reviewed in [1]). Bats most likely constitute a separate reservoir [2]. Influenza A viruses are classified into antigenic subtypes according to their hemagglutinin (HA) and neuraminidase (NA) envelope glycoproteins. The HA protein binds sialic-acid-terminating surface receptors and actively causes membrane fusion in endosomes during virus entry. The NA protein destroys these receptors to reduce extracellular virion aggregation and superinfection. Many combinations of 16 HA (H1–H16) and 9 NA (N1–N9) subtypes have been found in wild birds, while H17N10 and H18N11 subtypes were recently discovered in bats. From wild birds, influenza A viruses transmit to wild terrestrial and aquatic species including domestic poultry, which permit further transmission to a variety of mammals including swine and humans (Figure 1). Current epidemic strains in swine include H1N1, H1N2, and H3N2 strains. Human epidemics over the past century have included H1N1, H2N2, and H3N2. Interspecies transmission of influenza A viruses occurs more readily from wild to domestic birds and between swine and humans than between avian and mammalian hosts. This is due to host-range restrictions (reviewed in [3]). The past three human pandemic viruses (1957 H2N2, 1968 H3N2, and 2009 H1N1) originated in swine; avian-origin human pandemics may also occur [4]. A knowledge of mechanisms by which influenza viruses acquire the ability to sustain transmission in a new host is key to understanding, attempting to prevent, and preparing for future pandemics.
Figure 1. Model of influenza A virus transmission.
Endemic circulation of representative subtypes is shown within circles. Major pathways of transmission between species are denoted by straight arrows. Diverse influenza A viruses of 16 known HA and 9 known NA subtypes circulate in a reservoir of wild aquatic birds (dark blue), occasionally transmitting to other wild and domestic species. Bats host two recently identified subtypes, H17N10 and H18N11. A major pathway for the transfer of genetic diversity occurs from wild birds to domestic poultry (light blue). Infections in domestic poultry sporadically spread to farm animals and humans. Influenza viruses frequently transmit between swine (dark orange) and humans (orange). Examples of endemic strains circulating within a species are described within circles. Examples of sporadic (non-endemic) transmission of influenza viruses between species have been omitted for simplicity including recent human outbreaks such as H5N1 and H7N9.
Viral factors enabling adaptation to humans and ferrets
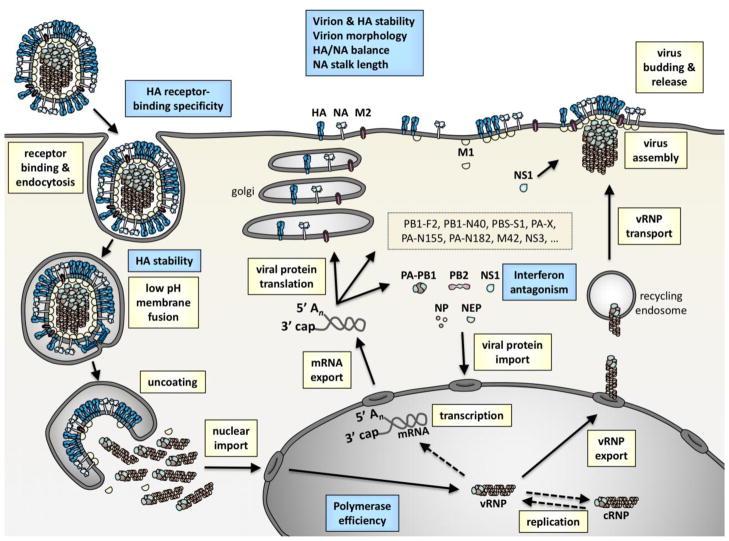
Traits associated with human pandemic potential have been identified by comparing human pandemic viruses to non-pandemic viruses (reviewed in [5–8]) and by studying airborne transmissibility in surrogate animal models such as ferrets and guinea pigs (reviewed in [3, 9–11]). These traits have been ascribed to several steps during the influenza virus replication cycle (Figure 2). Outside of the cell, influenza virions must be efficiently released and remain infectious while transiting between and within hosts. This is achieved in part by forming a filamentous morphology [12], by penetrating human mucus and disaggregating virions with a longer NA stalk length [13], and by having balanced HA receptor-binding avidity and NA receptor-destroying activity (reviewed in [14]). While a polybasic HA cleavage site that allows intracellular cleavage is not required for interspecies adaptation, it promotes influenza dissemination and increased virulence (reviewed in [15]). Human- and ferret-adapted HA proteins preferentially bind to α-2,6-linked sialic acid receptors, which are present in the mammalian upper respiratory tract, over binding to the α-2,3 form, which is abundant in the avian enteric tract (reviewed in [16–18]). Polymerase complex efficiency is enhanced in human cells by mutations that increase activity at the lower temperature of the mammalian respiratory tract and those that promote interactions with human host factors (reviewed in [19]). Species-specific NS1 protein binding to human host factors increases influenza virus replication by antagonizing interferon production [20]. During the last decade, HA stability has been shown to promote adaptation to humans and ferrets in part by helping virions resist inactivation in mildly acidic environments and by allowing them to cause membrane fusion in late versus early endosomes (reviewed in [21, 22]). The focus of this review is to summarize recent advances in our understanding of the biological impact of HA stability and to identify key questions that remain unanswered.
Figure 2. Influenza virus replication cycle and properties influencing adaptation to humans and ferrets.
Major steps during replication are denoted in yellow boxes. These include receptor binding, endocytosis, low-pH-induced membrane fusion, uncoating, nuclear import, transcription, mRNA export, viral protein translation, viral protein import into the nucleus, viral genome replication, viral ribonucleoprotein (vRNP) export, vRNP transport to the plasma membrane, virus assembly, virus budding, and virus release. Properties identified in interspecies adaptation are denoted by blue boxes. Extracellular adaptive properties include the stability of virions and the HA protein, virion morphology, balance of HA binding and NA receptor-destroying activities, and NA stalk length. Intracellular adaptive properties include receptor-binding specificity by the HA protein, HA stability, polymerase efficiency, and interferon antagonism.
HA protein structure and activation
The HA protein is translated as an uncleaved HA0 precursor protein, folded as a trimer, and glycosylated and acylated [23–25]. Uncleaved HA0 is unable to cause membrane fusion [26] and must first be protease-cleaved into a fusion-competent HA1/HA2 complex (Figure 3). Highly pathogenic avian influenza (HPAI) viruses have polybasic HA0 cleavage sites with an R-X-R/K-R furin recognition sequence that is recognized intracellularly in the trans-Golgi network [27, 28]. The HA0 proteins of human and low pathogenic avian influenza (LPAI) viruses are cleaved extracellularly by trypsin-like proteases or other soluble proteases [29]. The mature HA1/HA2 complex contains a membrane-proximal stalk domain containing the N- and C-terminal portions of HA1 and the entirety of HA2 and a membrane-distal receptor-binding domain containing receptor-binding and vestigial esterase subdomains (Figure 3B). After virus budding and HA0 cleavage, an influenza virus is capable of infecting a new host cell. To do so, the HA protein binds sialic-acid containing receptors and then the virion is internalized by endocytosis (clathrin-mediated or clathrin-independent) or macropinocytosis [30–32]. During endocytosis an influenza virion is exposed to sequentially lower pH in early endosomes (pH 6.0–6.5), late endosomes (pH 5.0–5.5), and lysosomes (pH 4.6–5.0) [33]. Cleaved HA1/HA2 is trapped in a high-energy (metastable) conformation and is triggered biologically by low pH to undergo irreversible conformational changes that cause membrane fusion [34–37]. If the virion is exposed to sufficiently low pH outside of a host or host cell, the HA protein is prematurely activated to refold irreversibly such that the virion becomes inactivated.
Figure 3. Prefusion structure of the HA protein and residues known to affect its stability.
(A) HA domain structure. HA1 domains include the fusion (F1, blue), vestigial esterase (VE, yellow), and receptor-binding domain (RBD, green). HA2 includes a stalk domain (orange), transmembrane region (TM, white), and cytoplasmic tail (CT, white). Solid circles identify residues to which stabilizing or destabilizing mutations have been identified in review articles [21, 22] and primary manuscripts [54, 60–62, 66, 70, 74–87]. (B) Domain insertion in the HA protein, adapted from [116]. (C) Prefusion structure of one HA monomer. (D) Prefusion structure of an HA trimer. Residues regulating stability (black balls) are located throughout the trimer in the receptor-binding pocket (R.B.P.), between HA1 heads (head-head), between the HA1 head the stalk (head-stalk), in the B loop and adjacent helix C (B loop area), in the core of the coiled coil (core), in the HA1 stalk, between helix A and the coiled coil (helix A), in and around the fusion peptide pocket (fus. pep. & pocket), and in the membrane-proximal region (M.P.R.). In panel D, two protomers are colored gray. Structures were generated using MacPYMOL using A/California/4/2009 (H1N1) protein data bank structure 3UBE [117].
Experimental techniques to measure HA stability
HA stability may be measured as the ability of a bulk sample of influenza virus to resist inactivation after exposure to buffers of varying pH. In an acid inactivation assay, aliquots of virus are incubated in pH-adjusted buffers typically ranging from 4.5 to 7.0, reneutralized, and assayed for infectivity by a standard assay such as TCID50 [38]. In this assay, HA stability is defined as the inflection point of the two-state (infective/noninfective) readout as pH is decreased, or pH50. While the biological trigger for HA activation is low pH, HA refolding can also be triggered by other destabilizing agents such as heat and urea [39, 40]. Thus, a heat inactivation assay may be performed using infectivity or hemagglutination assay as a readout [38]. As heat and thermal stability have been correlated in some [40] but not all [38] comparisons, acid-induced HA protein inactivation may be considered a primary assay and thermal inactivation, which typically occurs at temperatures greater than 50 °C, a surrogate because it is conducted under non-physiological conditions.
HA stability may also be measured as the ability of influenza virions, infected cells, or HA-transfected cells to cause membrane fusion. Virions, influenza-infected cells, HA-expressing cells, and/or target cells, erythrocytes, or liposomes can be labeled with fluorescent probes and then exposed to buffers of varying pH. Lipid and/or contents mixing are then measured by microscopy, fluorescence dequenching, or fluorescence resonance energy transfer (FRET) (reviewed in [41]). Membrane fusion may also be measured by cell-to-cell fusion assays whereby influenza-infected or HA-transfected cells are pulsed by pH-adjusted buffers. In such an assay, HA activation pH is defined as the highest incubation pH at which cell-to-cell fusion occurs. Typical readouts for cell-to-cell fusion include syncytia formation, dye transfer, and reporter gene expression. Membrane fusion of individual virions may also by measured by total internal reflection fluorescence microscopy [42].
The pH at which the HA protein undergoes conformational changes can be measured by a trypsin-susceptibility assay. Prefusion HA trimers are resistant to proteolysis after exposure to trypsin while postfusion HA is susceptible. Therefore, HA proteins can be incubated in media at varying pH, reneutralized, incubated with trypsin, and then resolved by SDS-PAGE to determine the exposure pH at which HA degradation occurs [43, 44]. Conformation-specific monoclonal antibodies and flow cytometry can also be used to measure the pH at which the HA protein is activated [45–50].
HA stability values
HA activation pH values typically fall within a range of pH 4.8–6.2 (reviewed in [21, 22]). A comparison of HA activation pH values for 159 surveillance samples measured by syncytia assay in our laboratory reveals some avian and swine subtypes have average HA activation pH values higher than those of humans (Figure 4A). Similar to independent findings [51–53], human pandemic H1N1 (pH1N1) isolates from 2009 had activation pH values of 5.5–5.6, while those recovered from 2010–2012 ranged from 5.2–5.4 [54] and were more stable structurally [55]. Human H1N1, H2N2, and H3N2 viruses from the 20th Century were previously found by other laboratories to also have relatively stable HA proteins, ranging from pH 5.0–5.4 [38, 53, 56]. As shown in Figure 4A, the average HA activation pH value of 5.7 for emerging H5N1, H7N7, H7N9, and H9N2 strains isolated from humans was significantly higher (P < 0.05) than an average of 5.4 for human pH1N1 viruses [57–59]. Other laboratories have also reported activation pH values for H5N1, H5N6, and H7N9 human isolates as 5.6–5.7 [60–62], 5.6 [63], and 5.6–5.8 [64, 65], respectively. Thus, emerging influenza viruses have been found to often have less stable HA proteins than those that are human adapted.
Figure 4. HA activation pH values for influenza A viruses.
(A) Highest pH values of overlaid media that trigger virus-infected cells to form syncytia, or fuse, grouped by host species. Isolates from wild birds include H1N1 and H5Nx (H5N2 and H5N8) [54, 68]. Isolates from poultry include H5Nx, H5N1, and H9N2 [57, 58, 68, 72, 73, 98]. Swine isolates include H1N1, H1N2, and H3N2 [54, 66]. Human isolates include zoonotic infections (H5N1, H7N7, H7N9, and H9N2) and 2009 pandemic H1N1 (pH1N1) viruses [54]. Mean values for each group are shown by horizontal bars. All data was collected using the same methods in the same laboratory. Statistical significance between the human pH1N1 group and the other groups was determined by one-way ANOVA analysis followed by a Tukey post-hoc test: *P < 0.05, **P < 0.01, ***P < 0.001. (B) Relationship between HA activation pH and transmissibility between mallards. A/chicken/Vietnam/C58/2004 (C58) (H5N1) wild-type (WT, closed circle) and HA1-H18Q (closed square) were transmissible between mallards, while CH58 HA2-K58I (open circle) was loss-of-function for transmissibility [73]. (C) Relationship between HA activation pH and transmissibility between ferrets by the airborne route. Viruses incapable of ferret airborne transmission included A/Tennessee/560-1/2009 (H1N1, closed circle) containing an HA1-Y17H mutation [54], A/Indonesia/5/2005 (H5N1, open square) wild-type [60, 95], and an H5N1 reassortant virus containing the wild-type HA protein from A/Vietnam/1203/2004 (VN1203, gray triangle) [96]. Airborne transmissible gain-of-function viruses had stabilized HA proteins including pH1N1 HA2-R106K, H5N1 HA1-H110Y, and H5N1 HA1-T318I. H3 numbering is used.
Average HA activation pH for H1N1, H1N2, and H3N2 swine isolates were 5.6–5.7 [54, 66] (Figure 4A), higher than the average for human pH1N1. However, the ranges of HA activation pH for the swine viruses overlapped with the range for human pH1N1. Inactivation pH values of Eurasian avian-like swine viruses have been reported by others to range from 5.5–5.9, while the pH50 values for RBC hemolytic activity for the same viruses ranged from 5.1–5.4 [67]. Discrepancies between the data for the two assays are unclear, although many in the field have discontinued use of the RBC-hemolysis assay to measure HA protein activation. Overall, swine influenza viruses appear to support a relatively broad range of HA activation pH.
With respect to avian species, trends in HA stability have been more diverse. North American H5N2 and H5N8 isolates recovered in 2015 were substantially higher with HA activation pH values of 6.0 [68] (Figure 4A). In contrast, H9N2 chicken isolates from 1994–2003 had an average activation pH of 5.4 and a range of 5.3–5.5 [58], which was similar to that for human pH1N1. Other H9N2 chicken isolates have been reported to have HA inactivation pH values between 5.0 and 5.8 [69, 70]. A broad survey of chicken isolates from multiple HA subtypes found a wide range of 4.9–5.9 for RBC hemolytic activity [71]. Broad ranges in HA stability have also been observed for influenza viruses recovered from wild birds. We previously found H5Nx and H1N1 wild bird isolates had HA activation pH values that ranged from 5.9–6.0 and 5.5–6.1, respectively [54, 68] (Figure 4A). Others have observed Eurasian duck and coot H1N1 viruses to range from 5.0–5.4 [67], while mallard, duck, coot, whooper swan, and peregrine falcon isolates of varying subtypes have broadly ranged from pH 4.9–5.8 [53, 71]. Overall, reported HA stability values for avian isolates have varied greatly. This suggests either a lack of preference for HA stability in avian species altogether or preferences may depend on individual influenza virus genetic constellations and particular avian hosts, as has been reported for related H5N1 viruses in mallards and chickens [72, 73].
Mutations to many amino-acid residues alter HA stability
Mutations to over 50 HA residues across H1, H2, H3, H5, and H7 subtypes have been identified that alter HA stability. HA stability mutants have been summarized in two 2014 review articles [21, 22] and detailed in other primary manuscripts not included in the previous reviews [54, 60–62, 66, 70, 74–88]. HA stability-altering residues are located throughout the primary sequence in both HA1 and HA2 subunits, and tend to be positioned in regions of the molecule that undergo large-scale changes in structure during HA protein refolding [25, 89] (Figure 3). Activation pH-altering mutations do not appear to alter the prefusion HA protein backbone in x-ray crystal structures [72, 90, 91]. Therefore, high-resolution structures and computer modeling may not reliably predict HA stability. Genetic prediction of HA activation pH is complicated further due to observations that the NA and M proteins can also modulate HA stability in some cases [73, 92–94]. Thus, phenotypic assays are most likely needed to determine HA stability in surveillance studies.
Impact of HA stability on interspecies adaptation and transmissibility
General interest in HA stability within the influenza field exploded in 2012 after demonstration that H5N1 influenza viruses could acquire gain-of-function airborne transmissibility in ferrets, in part, by HA stabilization [95, 96]. Previously, we had found that wild-type viruses with HA activation pH values of 5.6–6.0 were shown to have greater fitness, virulence, and transmissibility in chickens and mallards than related mutant viruses with stabilized HA proteins that are activated at pH 5.2 [72, 73]. This observation extends to other H5N1 viruses [97]. These same HA-stabilized loss-of-function mutants in avian species were shown to be gain-of-function for replication in the upper respiratory tracts of mice and ferrets [57, 98] and airborne transmissibility in ferrets [95]. Related studies on reassortant viruses harboring H5 HA proteins support the notion that an acid-stable HA protein is necessary to support airborne transmissibility in ferrets but insufficient in the absence of specificity for α(2,6)-linked sialic acid receptors [96, 99]. For these limited studies, an HA activation pH greater than 5.5 supported replication and transmission in avian hosts while a value of 5.5 or less was favored for upper respiratory tract replication and airborne transmissibility in ferrets (Figure 4B,C). An opposite effect was found with respect to virus replication in the lungs and pathogenicity. HA-stabilized H5N1 viruses that supported higher virus replication in the upper respiratory tracts of ferrets were simultaneously found to be attenuated for replication in the lungs and were less lethal [57, 95, 96]. Thus, greater transmissibility afforded by HA stabilization does not necessarily dictate greater, or even equivalent, pathogenicity.
Recently, HA stabilization was linked to the human pandemic potential of 2009 pH1N1 influenza virus [53, 54]. This work showed pre-2009 H1N1 viruses from the Classical swine lineage had HA activation pH values ranging from 5.5–6.0, early 2009 human pH1N1 isolates had values of approximately 5.5, and human-adapted 2010–2012 pH1N1 viruses had HA proteins stabilized to 5.2–5.4. Experiments in ferrets recapitulated these findings during which a mutant with a destabilized HA protein (pH 6.0) was loss-of-function for airborne transmissibility but was able to regain ferret airborne transmissibility after acquiring two mutations that stabilized the HA protein to pH 5.3 (Figure 4C). These same pH1N1 viruses containing HA stability mutations were also used to investigate the importance of this property for virus replication and transmissibility in swine [66]. Viruses with destabilized (pH 6.0), intermediate (5.5), or a stabilized (5.3) HA proteins replicated and transmitted by contact between swine. Thus, swine may support the propagation of influenza viruses varying widely in HA activation pH, perhaps serving as an intermediate host for the adaptation of both receptor-binding specificity and HA stability.
Further surveillance studies on the species-specificity of HA stability and experimental studies on animal infections using viruses with stability mutations are needed for a broader understanding of the importance of this property in interspecies adaptation. Several other studies increase enthusiasm that HA may broadly affect influenza virus properties. These include revelations that adaptation of H1N1 and H3N2 viruses to murine lungs has been linked to an increase in HA activation pH [100–103] and that an LPAI H7N3 turkey isolate had a lower HA activation pH than a related duck virus [104].
Potential mechanisms
Increased environmental persistence of influenza viruses, including recent swine and human H1N1 strains, is promoted by near-neutral pH, colder temperatures, and low salinity [105]. For H5N1 viruses containing HA point mutations altering its stability, a lower HA activation pH was associated with greater environmental persistence at 28 °C [73]. Thus, a more stable HA protein could extend virus half-life during transit between hosts.
Stable HA proteins may also resist extracellular inactivation within the mammalian respiratory tract (Figure 5). Extracellular pH in the nasal cavity has been measured to be mildly acidic with an average of 6.3 in healthy adults and 5.9 in children [106–108]. Average extracellular pH in the nasopharynx, soft palate, oropharynx, trachea, and lungs has been measured to be near-neutral (Figure 5), but can become transiently acidic in those suffering gastroesophageal reflux disease (GERD), especially in the oropharynx [109]. We hypothesize that avian-like influenza viruses with HA proteins activated at pH values approaching 6.0 are more readily inactivated in the nasal cavity than human-adapted viruses containing stabilized HA proteins. Further studies are needed to determine respiratory pH values in the upper respiratory tracts of other mammals such as ferrets and swine and to determine if extracellular pH in the nasal cavity exerts a pressure that promotes selection of acid-stabilizing HA mutations.
Figure 5. Respiratory tract pH values.
Compilation of reported respiratory pH values. Values for healthy humans include those from the nasal cavity [106, 108], nasopharynx [118], soft palate [119], oropharynx [120], trachea [121, 122], and lungs [123–125]. Limited data is available for non-human species. Reported values for animals include those for the ferret trachea [126], swine trachea [127–129], and mouse nasal cavity [123] and trachea [130, 131]. Values for co-morbidities include those for cystic fibrosis patients [107, 132, 133], healthy children [107, 119, 134, 135], and respiratory infected [135, 136]. Reported values include average values (bold type) and the range of data in parentheses. For illustrative purposes, reported values are shaded using a spectral color scheme (bottom left).
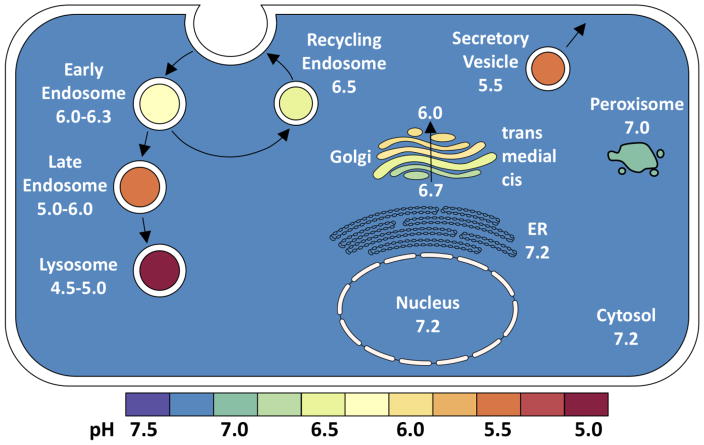
Additionally, HA stability most likely influences the site of membrane fusion during endocytosis, thereby altering intracellular host cell responses that in turn regulate infectivity. HA proteins have activation pH values within a range of acidity to which influenza virions are exposed in early and late endosomes (Figure 6). For A/PR/8/34 (H1N1), which has a relatively stable HA activated at pH 5.0–5.1 [53], increased glucose exposure that decreases endosomal pH has been shown to amplify replication [110]. For other influenza viruses, lower HA activation pH values have been associated with blocked entry in macrophages [111] and increased sensitivity to the IFN-induced antiviral state via functional interaction with IFITM2 and IFITM3 proteins in late endosomes [87]. Thus, HA stability has been shown to alter tropism and entry-related host responses, although further studies are needed to define precisely the affected cellular pathways and downstream consequences.
Figure 6. Intracellular pH values.
pH values for various subcellular compartments in a prototypic mammalian cell [137, 138]. For illustrative purposes, pH values are shaded using the same spectral color scheme (bottom) as in Figure 5.
A role for HA stabilization in vaccine development
Several studies suggest vaccines can be enhanced by optimizing HA stability. A/PR/8/34 (H1N1) has a relatively stable HA protein with an activation pH of 5.0–5.1 [53]. Replication of PR8 in Vero cells, which are used to produce vaccine seed stocks, was enhanced by a mutation in PR8 that increased the HA activation pH by at least 0.2 units [112]. With respect to live vaccine immunogenicity, an NS1-deleted H3N2 vaccine candidate with an HA activation pH of 5.8 was enhanced compared to related viruses with values of 6.2–6.3 [113]. For a similar NS1-deleted H5N1 virus in mice, a mutation that decreased the HA activation pH from 6.0 to 5.5 increased its infectivity and immunogenicity [114], most likely by increasing upper respiratory replication [98]. For the live-attenuated pH1N1 vaccine, an HA-stabilizing mutation that lowered its activation pH from 5.4 to 5.0 increased vaccine stability and infectivity [51]. Overall, there appear to be pH-optima for vaccine production, stability, and immunogenicity; thus, vaccines may be enhanced by strategic introduction of mutations that modulate HA stability.
Concluding Remarks
In summary, there is a growing list of HA activation and inactivation values for circulating influenza A viruses, and these values can vary substantially between different HA subtypes and infected hosts. There is also an expanding literature cataloguing a growing number of mutations that alter HA stability. While incomplete, we have noted a trend whereby HA proteins from avian and swine hosts vary widely in HA stability and human- and ferret-adaptation is associated with HA stabilization. Our understanding of the mechanism by which HA stability helps determine species-specific replication, virulence, and transmissibility is incomplete but may include aspects that are both extracellular (susceptibility to inactivation outside the cell) and intracellular (differential endosomal entry sites and differential triggering of host cell responses). To enhance surveillance of emerging influenza viruses, we recommend adding phenotypic screening of HA stability in addition to gene sequencing and current assays that measure serological cross-reactivity and receptor-binding specificity. Engineering mutations that optimize HA stability appears promising as a method to improve vaccine production and immunogenicity. As stability-altering resistance mutations may decrease virus fitness, targeting the major antigen of influenza virus with broadly neutralizing antibodies and antiviral agents, such as a low-pH intranasal spray [115], has become increasingly more attractive. However, many questions remain to be addressed to gain a more complete understanding of the biological importance of HA stability and how this understanding can be exploited to help predict and control future influenza virus pandemics.
Highlights.
The structure and acid-induced activation mechanism of the influenza virus hemagglutin (HA) protein has served as a paradigm for protein-mediated membrane fusion over the past four decades.
Recently, several laboratories have shown that in order to adapt to humans and ferrets, influenza viruses need to acquire mutations that stabilize the HA protein.
HA stability has been linked to pandemic potential and should be considered in surveillance, risk assessment tools, and pre-pandemic planning.
In general, the preference for a stable or unstable HA protein varies by species. Thus, HA stability has been recently discovered as a novel trait associated with the ability of influenza viruses to cross species barriers.
A knowledge of optimal HA activation pH values and mutations affecting this property may help investigators generate enhanced vaccines and therapeutics.
Outstanding Questions.
Can influenza A viruses with relatively unstable HA proteins maintain airborne transmissibility in humans and ferrets via compensatory mutations that alter other molecular, cellular, and virological properties in HA or other viral genes? If so, what are they and how does this work?
Does an apparent requirement of a stabilized HA for ferret airborne transmissibility and human pandemic potential extend beyond the H1 and H5 subtypes?
How does host response depend on HA stability and on host species and cell type? What specific host-cell genes are up- and down-regulated based on the endosomal location of influenza virus-mediated membrane fusion?
Does HA stability affect the retention of infectivity as influenza virions transit in aerosolized particles between hosts? Does a role for HA stability in aerosolized transmission vary based on environmental conditions?
What advantage does an unstable HA protein afford H5N1 influenza virus replication and transmission in an avian host? How do influenza viruses avoid inactivation in the highly acidic avian enteric tract?
Broadly neutralizing antibodies and universal vaccines often target the HA stalk region, which helps regulate HA stability and contains the fusogenic nanomachine. How do resistance and escape mutants to these therapeutics and vaccines affect HA stability? Would a strict requirement for an optimal range of HA activation pH limit resistance and escape mutants to these promising antiviral agents?
Which HA residues are so tightly coupled to maintaining the metastable prefusion state that they are less likely to be mutated and, thus, constitute a more attractive universal vaccine epitope beyond candidate residues identified by amino-acid conservation?
Acknowledgments
We used the sequence-determines-credit (SDC) approach to authorship where the sequence of authors reflects declining importance of their contributions. This work was funded, in part, by the National Institute of Allergy and Infectious Diseases under Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400006C, St. Jude Children’s Research Hospital, and the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yoon SW, et al. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol. 2014;385:359–75. doi: 10.1007/82_2014_396. [DOI] [PubMed] [Google Scholar]
- 2.Tong S, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann G, Kawaoka Y. Transmission of influenza A viruses. Virology. 2015;479–480:234–46. doi: 10.1016/j.virol.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Runstadler J, et al. Connecting the study of wild influenza with the potential for pandemic disease. Infect Genet Evol. 2013;17:162–87. doi: 10.1016/j.meegid.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard M, et al. Avian influenza A viruses: from zoonosis to pandemic. Future Virol. 2014;9(5):513–524. doi: 10.2217/fvl.14.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrauwen EJ, Fouchier RA. Host adaptation and transmission of influenza A viruses in mammals. Emerg Microbes Infect. 2014;3(2):e9. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipsitch M, et al. Viral factors in influenza pandemic risk assessment. Elife. 2016;5:e18491. doi: 10.7554/eLife.18491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munoz O, et al. Genetic Adaptation of Influenza A Viruses in Domestic Animals and Their Potential Role in Interspecies Transmission: A Literature Review. Ecohealth. 2016;13(1):171–98. doi: 10.1007/s10393-014-1004-1. [DOI] [PubMed] [Google Scholar]
- 9.Lowen AC, et al. Transmission in the guinea pig model. Curr Top Microbiol Immunol. 2014;385:157–83. doi: 10.1007/82_2014_390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belser JA, Tumpey TM. H5N1 pathogenesis studies in mammalian models. Virus Res. 2013;178(1):168–85. doi: 10.1016/j.virusres.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouvier NM. Animal models for influenza virus transmission studies: a historical perspective. Curr Opin Virol. 2015;13:101–8. doi: 10.1016/j.coviro.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seladi-Schulman J, et al. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. J Virol. 2013;87(24):13343–53. doi: 10.1128/JVI.02004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blumenkrantz D, et al. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J Virol. 2013;87(19):10539–51. doi: 10.1128/JVI.00967-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaymard A, et al. Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin Microbiol Infect. 2016;22(12):975–983. doi: 10.1016/j.cmi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Bertram S, et al. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol. 2010;20(5):298–310. doi: 10.1002/rmv.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33(8):823–41. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong XL, et al. Receptor Binding Properties of the Influenza Virus Hemagglutinin as a Determinant of Host Range. In: Compans RW, Oldstone MBA, editors. Influenza Pathogenesis and Control - Vol I. Springer Int Publishing Ag; 2014. pp. 63–91. [DOI] [PubMed] [Google Scholar]
- 18.Gambaryan AS, Matrosovich MN. What adaptive changes in hemagglutinin and neuraminidase are necessary for emergence of pandemic influenza virus from its avian precursor? Biochemistry (Mosc) 2015;80(7):872–80. doi: 10.1134/S000629791507007X. [DOI] [PubMed] [Google Scholar]
- 19.Manz B, et al. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol. 2013;87(13):7200–9. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajsbaum R, et al. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8(11):e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mair CM, et al. Receptor binding and pH stability - how influenza A virus hemagglutinin affects host-specific virus infection. Biochim Biophys Acta. 2014;1838(4):1153–68. doi: 10.1016/j.bbamem.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Russell CJ. Acid-Induced Membrane Fusion by the Hemagglutinin Protein and Its Role in Influenza Virus Biology. In: Compans RW, Oldstone MBA, editors. Influenza Pathogenesis and Control - Vol I. 2014. pp. 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, et al. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95(3):409–17. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 24.Shaw ML, Palese P. Orthomyxoviridae. In: Knipe DM, Howley PM, editors. Fields virology. 6. Lippincott Williams and Wilkins; 2013. pp. 1151–1185. [Google Scholar]
- 25.Wilson IA, et al. Structure of the Hemagglutinin Membrane Glycoprotein of Influenza-Virus at 3-a Resolution. Nature. 1981;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 26.Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258(1):1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 27.Garten W, et al. Proteolytic activation of the influenza virus hemagglutinin: The structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981;115(2):361–74. doi: 10.1016/0042-6822(81)90117-3. [DOI] [PubMed] [Google Scholar]
- 28.Webster RG, Rott R. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell. 1987;50(5):665–6. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 29.Kido H, et al. Isolation and Characterization of a Novel Trypsin-Like Protease Found in Rat Bronchiolar Epithelial Clara Cells - a Possible Activator of the Viral Fusion Glycoprotein. Journal of Biological Chemistry. 1992;267(19):13573–13579. [PubMed] [Google Scholar]
- 30.de Vries E, et al. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7(3):e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakadamyali M, et al. Endocytosis of influenza viruses. Microbes Infect. 2004;6(10):929–36. doi: 10.1016/j.micinf.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. Journal of Virology. 2002;76(20):10455–10464. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellman I, et al. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 34.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73(4):823–32. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 35.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 36.Weber T, et al. Evidence for H(+)-induced insertion of influenza hemagglutinin HA2 N-terminal segment into viral membrane. J Biol Chem. 1994;269(28):18353–8. [PubMed] [Google Scholar]
- 37.Wharton SA, et al. Electron microscopy of antibody complexes of influenza virus haemagglutinin in the fusion pH conformation. Embo j. 1995;14(2):240–6. doi: 10.1002/j.1460-2075.1995.tb06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholtissek C. Stability of Infectious Influenza-a Viruses at Low Ph and at Elevated-Temperature. Vaccine. 1985;3(3):215–218. doi: 10.1016/0264-410x(85)90109-4. [DOI] [PubMed] [Google Scholar]
- 39.Carr CM, et al. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruigrok RWH, et al. Conformational-Changes in the Hemagglutinin of Influenza-Virus Which Accompany Heat-Induced Fusion of Virus with Liposomes. Virology. 1986;155(2):484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- 41.Hoekstra D, Klappe K. Fluorescence assays to monitor fusion of enveloped viruses. Methods Enzymol. 1993;220:261–76. doi: 10.1016/0076-6879(93)20088-k. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton BS, et al. Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses. 2012;4(7):1144–68. doi: 10.3390/v4071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinhauer DA, et al. Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc Natl Acad Sci U S A. 1996;93(23):12873–8. doi: 10.1073/pnas.93.23.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skehel JJ, et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci U S A. 1982;79(4):968–72. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed ML, et al. Amino acid residues in the fusion peptide pocket regulate the pH of activation of the H5N1 influenza virus hemagglutinin protein. J Virol. 2009;83(8):3568–80. doi: 10.1128/JVI.02238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinhauer DA, et al. Amantadine selection of a mutant influenza virus containing an acid-stable hemagglutinin glycoprotein: evidence for virus-specific regulation of the pH of glycoprotein transport vesicles. Proc Natl Acad Sci U S A. 1991;88(24):11525–9. doi: 10.1073/pnas.88.24.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster RG, et al. Molecular changes in A/Chicken/Pennsylvania/83 (H5N2) influenza virus associated with acquisition of virulence. Virology. 1986;149(2):165–73. doi: 10.1016/0042-6822(86)90118-2. [DOI] [PubMed] [Google Scholar]
- 48.Yewdell JW, et al. Monoclonal anti-hemagglutinin antibodies detect irreversible antigenic alterations that coincide with the acid activation of influenza virus A/PR/834-mediated hemolysis. J Virol. 1983;48(1):239–48. doi: 10.1128/jvi.48.1.239-248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White JM, Wilson IA. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J Cell Biol. 1987;105(6 Pt 2):2887–96. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nestorowicz AN, et al. Conformational changes in influenza virus haemagglutinin and its monomer detected by monoclonal antibodies. Vaccine. 1985;3(3 Suppl):175–81. doi: 10.1016/0264-410x(85)90099-4. [DOI] [PubMed] [Google Scholar]
- 51.Cotter CR, et al. A single amino acid in the stalk region of the H1N1pdm influenza virus HA protein affects viral fusion, stability and infectivity. PLoS Pathog. 2014;10(1):e1003831. doi: 10.1371/journal.ppat.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurer-Stroh S, et al. A new common mutation in the hemagglutinin of the 2009 (H1N1) influenza A virus. PLoS Curr. 2010;2:RRN1162. doi: 10.1371/currents.RRN1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galloway SE, et al. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog. 2013;9(2):e1003151. doi: 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russier M, et al. Molecular requirements for a pandemic influenza virus: An acid-stable hemagglutinin protein. Proc Natl Acad Sci U S A. 2016;113(6):1636–41. doi: 10.1073/pnas.1524384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, et al. Structural stability of influenza A(H1N1)pdm09 virus hemagglutinins. J Virol. 2014;88(9):4828–38. doi: 10.1128/JVI.02278-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costello DA, et al. Variations in pH sensitivity, acid stability, and fusogenicity of three influenza virus H3 subtypes. J Virol. 2015;89(1):350–60. doi: 10.1128/JVI.01927-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaraket H, et al. Increased acid stability of the hemagglutinin protein enhances H5N1 influenza virus growth in the upper respiratory tract but is insufficient for transmission in ferrets. J Virol. 2013;87(17):9911–22. doi: 10.1128/JVI.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Group SW. Assessing the fitness of distinct clades of influenza A (H9N2) viruses. Emerg Microbes Infect. 2013;2(11):e75. doi: 10.1038/emi.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaraket H, et al. Mammalian adaptation of influenza A(H7N9) virus is limited by a narrow genetic bottleneck. Nat Commun. 2015;6:6553. doi: 10.1038/ncomms7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linster M, et al. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell. 2014;157(2):329–339. doi: 10.1016/j.cell.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe Y, et al. Characterization of H5N1 influenza virus variants with hemagglutinin mutations isolated from patients. MBio. 2015;6(2):15. doi: 10.1128/mBio.00081-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanson A, et al. Identification of Stabilizing Mutations in an H5 Hemagglutinin Influenza Virus Protein. J Virol. 2015;90(6):2981–92. doi: 10.1128/JVI.02790-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herfst S, et al. Human Clade 2.3.4.4 A/H5N6 Influenza Virus Lacks Mammalian Adaptation Markers and Does Not Transmit via the Airborne Route between Ferrets. mSphere. 2018;3(1):e00405–17. doi: 10.1128/mSphere.00405-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richard M, et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 2013;501(7468):560–3. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gabbard JD, et al. Novel H7N9 influenza virus shows low infectious dose, high growth rate, and efficient contact transmission in the guinea pig model. J Virol. 2014;88(3):1502–12. doi: 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russier M, et al. H1N1 influenza viruses varying widely in hemagglutinin stability transmit efficiently from swine to swine and to ferrets. PLoS Pathog. 2017;13(3):e1006276. doi: 10.1371/journal.ppat.1006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baumann J, et al. H1N1 Swine Influenza Viruses Differ from Avian Precursors by a Higher pH Optimum of Membrane Fusion. J Virol. 2015;90(3):1569–77. doi: 10.1128/JVI.02332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaplan BS, et al. Novel Highly Pathogenic Avian A(H5N2) and A(H5N8) Influenza Viruses of Clade 2.3.4.4 from North America Have Limited Capacity for Replication and Transmission in Mammals. mSphere. 2016;1(2):e00003–16. doi: 10.1128/mSphere.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peacock TP, et al. Variability in H9N2 haemagglutinin receptor-binding preference and the pH of fusion. Emerg Microbes Infect. 2017;6(3):e11. doi: 10.1038/emi.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong L, et al. Molecular mechanism of the airborne transmissibility of H9N2 avian influenza A viruses in chickens. J Virol. 2014;88(17):9568–78. doi: 10.1128/JVI.00943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okamatsu M, et al. Is the optimal pH for membrane fusion in host cells by avian influenza viruses related to host range and pathogenicity? Arch Virol. 2016;161(8):2235–42. doi: 10.1007/s00705-016-2902-z. [DOI] [PubMed] [Google Scholar]
- 72.DuBois RM, et al. Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity. PLoS Pathog. 2011;7(12):e1002398. doi: 10.1371/journal.ppat.1002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reed ML, et al. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J Virol. 2010;84(3):1527–35. doi: 10.1128/JVI.02069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gething MJ, et al. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102(1):11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Godley L, et al. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68(4):635–45. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 76.Kemble GW, et al. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J Virol. 1992;66(8):4940–50. doi: 10.1128/jvi.66.8.4940-4950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smeenk CA, et al. Mutations in the hemagglutinin and matrix genes of a virulent influenza virus variant, A/FM/1/47-MA, control different stages in pathogenesis. Virus Res. 1996;44(2):79–95. doi: 10.1016/0168-1702(96)01329-9. [DOI] [PubMed] [Google Scholar]
- 78.Gruenke JA, et al. New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J Virol. 2002;76(9):4456–66. doi: 10.1128/JVI.76.9.4456-4466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rachakonda PS, et al. The relevance of salt bridges for the stability of the influenza virus hemagglutinin. FASEB J. 2007;21(4):995–1002. doi: 10.1096/fj.06-7052hyp. [DOI] [PubMed] [Google Scholar]
- 80.Mair CM, et al. A histidine residue of the influenza virus hemagglutinin controls the pH dependence of the conformational change mediating membrane fusion. J Virol. 2014;88(22):13189–200. doi: 10.1128/JVI.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Byrd-Leotis L, et al. Influenza hemagglutinin (HA) stem region mutations that stabilize or destabilize the structure of multiple HA subtypes. J Virol. 2015;89(8):4504–16. doi: 10.1128/JVI.00057-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaverin NV, et al. Pleiotropic effects of amino acid substitutions in H5 hemagglutinin of influenza A escape mutants. Virus Res. 2015;210:81–9. doi: 10.1016/j.virusres.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 83.Rudneva IA, et al. Effects of hemagglutinin amino acid substitutions in H9 influenza A virus escape mutants. Arch Virol. 2016;161(12):3515–3520. doi: 10.1007/s00705-016-3038-x. [DOI] [PubMed] [Google Scholar]
- 84.Di Lella S, et al. Modulation of the pH Stability of Influenza Virus Hemagglutinin: A Host Cell Adaptation Strategy. Biophys J. 2016;110(11):2293–2301. doi: 10.1016/j.bpj.2016.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schrauwen EJ, et al. Amino Acid Substitutions That Affect Receptor Binding and Stability of the Hemagglutinin of Influenza A/H7N9 Virus. J Virol. 2016;90(7):3794–9. doi: 10.1128/JVI.03052-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wormann X, et al. Genetic characterization of an adapted pandemic 2009 H1N1 influenza virus that reveals improved replication rates in human lung epithelial cells. Virology. 2016;492:118–29. doi: 10.1016/j.virol.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 87.Gerlach T, et al. pH Optimum of Hemagglutinin-Mediated Membrane Fusion Determines Sensitivity of Influenza A Viruses to the Interferon-Induced Antiviral State and IFITMs. J Virol. 2017;91(11):16. doi: 10.1128/JVI.00246-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang W, et al. Intermonomer Interactions in Hemagglutinin Subunits HA1 and HA2 Affecting Hemagglutinin Stability and Influenza Virus Infectivity. J Virol. 2015;89(20):10602–11. doi: 10.1128/JVI.00939-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bullough PA, et al. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371(6492):37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 90.de Vries RP, et al. Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses. J Virol. 2014;88(1):768–73. doi: 10.1128/JVI.02690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weis WI, et al. The Structure of a Membrane-Fusion Mutant of the Influenza-Virus Hemagglutinin. Embo Journal. 1990;9(1):17–24. doi: 10.1002/j.1460-2075.1990.tb08075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang RT, et al. The function of the neuraminidase in membrane fusion induced by myxoviruses. Virology. 1980;107(2):313–9. doi: 10.1016/0042-6822(80)90299-8. [DOI] [PubMed] [Google Scholar]
- 93.Su B, et al. Enhancement of the influenza A hemagglutinin (HA)-mediated cell-cell fusion and virus entry by the viral neuraminidase (NA) PLoS One. 2009;4(12):e8495. doi: 10.1371/journal.pone.0008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Donnell CD, et al. The matrix gene segment destabilizes the acid and thermal stability of the hemagglutinin of pandemic live attenuated influenza virus vaccines. J Virol. 2014;88(21):12374–84. doi: 10.1128/JVI.01107-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herfst S, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–41. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Imai M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–8. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Richard M, et al. Mutations Driving Airborne Transmission of A/H5N1 Virus in Mammals Cause Substantial Attenuation in Chickens only when combined. Sci Rep. 2017;7(1):7187. doi: 10.1038/s41598-017-07000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zaraket H, et al. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus replication and pathogenesis in mice. J Virol. 2013;87(9):4826–34. doi: 10.1128/JVI.03110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shelton H, et al. Mutations in haemagglutinin that affect receptor binding and pH stability increase replication of a PR8 influenza virus with H5 HA in the upper respiratory tract of ferrets and may contribute to transmissibility. J Gen Virol. 2013;94(Pt 6):1220–9. doi: 10.1099/vir.0.050526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hartley CA, et al. Changes in the hemagglutinin molecule of influenza type A (H3N2) virus associated with increased virulence for mice. Arch Virol. 1997;142(1):75–88. doi: 10.1007/s007050050060. [DOI] [PubMed] [Google Scholar]
- 101.Keleta L, et al. Experimental evolution of human influenza virus H3 hemagglutinin in the mouse lung identifies adaptive regions in HA1 and HA2. J Virol. 2008;82(23):11599–608. doi: 10.1128/JVI.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Narasaraju T, et al. Adaptation of human influenza H3N2 virus in a mouse pneumonitis model: insights into viral virulence, tissue tropism and host pathogenesis. Microbes Infect. 2009;11(1):2–11. doi: 10.1016/j.micinf.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 103.Koerner I, et al. Altered receptor specificity and fusion activity of the haemagglutinin contribute to high virulence of a mouse-adapted influenza A virus. J Gen Virol. 2012;93(Pt 5):970–9. doi: 10.1099/vir.0.035782-0. [DOI] [PubMed] [Google Scholar]
- 104.Giannecchini S, et al. Comparison of in vitro replication features of H7N3 influenza viruses from wild ducks and turkeys: potential implications for interspecies transmission. J Gen Virol. 2006;87(Pt 1):171–5. doi: 10.1099/vir.0.81187-0. [DOI] [PubMed] [Google Scholar]
- 105.Poulson RL, et al. Environmental Stability of Swine and Human Pandemic Influenza Viruses in Water under Variable Conditions of Temperature, Salinity, and pH. Appl Environ Microbiol. 2016;82(13):3721–6. doi: 10.1128/AEM.00133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.England RJ, et al. Nasal pH measurement: a reliable and repeatable parameter. Clin Otolaryngol Allied Sci. 1999;24(1):67–8. doi: 10.1046/j.1365-2273.1999.00223.x. [DOI] [PubMed] [Google Scholar]
- 107.McShane D, et al. Airway surface pH in subjects with cystic fibrosis. Eur Respir J. 2003;21(1):37–42. doi: 10.1183/09031936.03.00027603. [DOI] [PubMed] [Google Scholar]
- 108.Man WH, et al. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15(5):259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wiener GJ, et al. Oropharyngeal pH monitoring for the detection of liquid and aerosolized supraesophageal gastric reflux. J Voice. 2009;23(4):498–504. doi: 10.1016/j.jvoice.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 110.Kohio HP, Adamson AL. Glycolytic control of vacuolar-type ATPase activity: a mechanism to regulate influenza viral infection. Virology. 2013;444(1–2):301–9. doi: 10.1016/j.virol.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 111.Marvin SA, et al. Influenza Virus Overcomes Cellular Blocks To Productively Replicate, Impacting Macrophage Function. J Virol. 2017;91(2):e01417–16. doi: 10.1128/JVI.01417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Murakami S, et al. Enhanced growth of influenza vaccine seed viruses in vero cells mediated by broadening the optimal pH range for virus membrane fusion. J Virol. 2012;86(3):1405–10. doi: 10.1128/JVI.06009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nakowitsch S, et al. Mutations affecting the stability of the haemagglutinin molecule impair the immunogenicity of live attenuated H3N2 intranasal influenza vaccine candidates lacking NS1. Vaccine. 2011;29(19):3517–24. doi: 10.1016/j.vaccine.2011.02.100. [DOI] [PubMed] [Google Scholar]
- 114.Krenn BM, et al. Single HA2 mutation increases the infectivity and immunogenicity of a live attenuated H5N1 intranasal influenza vaccine candidate lacking NS1. PLoS One. 2011;6(4):e18577. doi: 10.1371/journal.pone.0018577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rennie P, et al. Low pH gel intranasal sprays inactivate influenza viruses in vitro and protect ferrets against influenza infection. Respir Res. 2007;8:38. doi: 10.1186/1465-9921-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rosenthal PB, et al. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396(6706):92–6. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu R, et al. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol. 2012;86(2):982–90. doi: 10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brunworth JD, et al. Detecting nasopharyngeal reflux: a novel pH probe technique. Ann Otol Rhinol Laryngol. 2012;121(7):427–30. doi: 10.1177/000348941212100701. [DOI] [PubMed] [Google Scholar]
- 119.Aframian DJ, et al. The distribution of oral mucosal pH values in healthy saliva secretors. Oral Dis. 2006;12(4):420–3. doi: 10.1111/j.1601-0825.2005.01217.x. [DOI] [PubMed] [Google Scholar]
- 120.Ayazi S, et al. A new technique for measurement of pharyngeal pH: normal values and discriminating pH threshold. J Gastrointest Surg. 2009;13(8):1422–9. doi: 10.1007/s11605-009-0915-6. [DOI] [PubMed] [Google Scholar]
- 121.Fischer H, et al. Acid secretion and proton conductance in human airway epithelium. Am J Physiol Cell Physiol. 2002;282(4):C736–43. doi: 10.1152/ajpcell.00369.2001. [DOI] [PubMed] [Google Scholar]
- 122.Gatto LA. pH of mucus in rabbit trachea: cholinergic stimulation and block. Lung. 1985;163(2):109–15. doi: 10.1007/BF02713812. [DOI] [PubMed] [Google Scholar]
- 123.Jayaraman S, et al. Airway surface liquid pH in well-differentiated airway epithelial cell cultures and mouse trachea. Am J Physiol Cell Physiol. 2001;281(5):C1504–11. doi: 10.1152/ajpcell.2001.281.5.C1504. [DOI] [PubMed] [Google Scholar]
- 124.Steinmann E. La secretion bronchique et le pH. Bronches. 1956;6:126–9. [Google Scholar]
- 125.West JB. Regional differences in the lung. Chest. 1978;74(4):426–37. doi: 10.1378/chest.74.4.426. [DOI] [PubMed] [Google Scholar]
- 126.Kyle H, et al. Control of pH of airway surface liquid of the ferret trachea in vitro. J Appl Physiol (1985) 1990;68(1):135–40. doi: 10.1152/jappl.1990.68.1.135. [DOI] [PubMed] [Google Scholar]
- 127.Berkebile AR, McCray PB., Jr Effects of airway surface liquid pH on host defense in cystic fibrosis. Int J Biochem Cell Biol. 2014;52:124–9. doi: 10.1016/j.biocel.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Inglis SK, et al. Regulation of intracellular pH in Calu-3 human airway cells. J Physiol. 2002;538(Pt 2):527–39. doi: 10.1113/jphysiol.2001.012806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pezzulo AA, et al. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature. 2012;487(7405):109–13. doi: 10.1038/nature11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Song Y, et al. Sodium and chloride concentrations, pH, and depth of airway surface liquid in distal airways. J Gen Physiol. 2003;122(5):511–9. doi: 10.1085/jgp.200308866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Verkman AS, et al. Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease. Am J Physiol Cell Physiol. 2003;284(1):C2–15. doi: 10.1152/ajpcell.00417.2002. [DOI] [PubMed] [Google Scholar]
- 132.Abou Alaiwa MH, et al. Neonates with cystic fibrosis have a reduced nasal liquid pH; a small pilot study. J Cyst Fibros. 2014;13(4):373–7. doi: 10.1016/j.jcf.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schultz A, et al. Airway surface liquid pH is not acidic in children with cystic fibrosis. Nat Commun. 2017;8(1):1409. doi: 10.1038/s41467-017-00532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hue V, et al. Simultaneous tracheal and oesophageal pH monitoring during mechanical ventilation. Arch Dis Child. 1996;75(1):46–50. doi: 10.1136/adc.75.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Junqueira JC, Penna FJ. Nasopharyngeal pH and gastroesophageal reflux in children with chronic respiratory disease. J Pediatr (Rio J) 2007;83(3):225–32. doi: 10.2223/JPED.1634. [DOI] [PubMed] [Google Scholar]
- 136.Hilding A. The common cold. Arch Otolaryngol. 1930;12(2):133–50. [Google Scholar]
- 137.Casey JR, et al. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11(1):50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 138.Paroutis P, et al. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology (Bethesda) 2004;19:207–15. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]