Abstract
Purpose
Granulosa cell (GC) number in follicles is a simple characteristic of follicles. The present study examined the hypothesis that follicular fluid (FF) determines GC number and oocyte developmental ability and revealed the molecular background determining the number of follicular GCs.
Methods
FF was collected from antral follicles (3–5 mm in diameter), after which the number of GCs per follicle was determined for each donor gilt using real time PCR targeting single copy gene. GCs were analyzed by next-generation RNA sequencing and IPA pathway analysis.
Results
When oocyte cumulus cell-oocyte-complexes (COCs) were cultured in maturation medium containing 10% of each individual FF, the rate of development to the blastocyst stage was significantly correlated with the number of GCs in the donor gilt. In addition, when GCs were cultured in medium containing FF, the proliferative activity of the GCs was also significantly correlated to the number of GCs in the donor gilt. Moreover, when the FFs were categorized based on the number of GCs in the follicle, it was found that supplementation of culture media with GC-rich FF improved the developmental ability of oocytes compared to those supplemented with GC-poor FF. RNA sequencing and a pathway analysis of GCs from GC-rich and -poor follicles revealed the key regulatory pathway determining GC number in follicles.
Conclusion
GC number may be a useful marker for “good” follicles and oocytes, and the characteristics of the FFs determine granulosa cell number and oocyte developmental ability.
Keywords: Follicular fluid, Granulosa cells, Gene expression, Oocytes
Introduction
Follicular development is closely associated with the proliferation of granulosa cells (GCs). GCs provide crucial metabolites and energy to the developing oocytes through gap junctional communication, thereby helping the oocyte to develop [1]. It is well-known that the number of GCs varies between follicles, as well as between donors. However, studies using slaughter house-derived ovaries often used mixed cumulus cell-oocyte-complexes (COCs) derived from tens of donors, and accordingly the significance of the number of GCs in follicles, as well as the molecular background underlying the number of the GCs, have been largely ignored. During follicle growth, oocytes accumulate mRNA, proteins, and lipids and undergo changes in chromatin configuration with high levels of acetylated histones. The high ATP content, lipid content, and histone acetylation reflect a high developmental competence [2–8]. Recently, it has been reported that the number of GCs surrounding the oocytes is closely related to oocyte energy status, as measured by ATP content, lipid content, and the extent of histone acetylation. [9]. This correlation was confirmed by other studies: when oocyte-GC-complexes (OGCs) collected from early antral follicles (EAFs) from pigs and cows were cultured in vitro for 2 weeks, the number of GCs surrounding the oocytes was closely correlated with the developmental ability of the oocytes, ATP content, lipid content, and histone acetylation levels of lysine [10, 11]. The notion that an increase in GC number surrounding oocytes improves the developmental ability of oocytes has been demonstrated in in bovine EAFs [10]. These data suggest that GC number is an indication of successful follicle development, as well as of oocyte quality. One question that remains is what determines the GC number in follicles?
Follicular fluid (FF) is the sole environment surrounding the oocytes and GCs, and it has been reported to profoundly affect oocyte maturation and fertilization [12]. FF components are derived from GC secretion and circulation. Maternal physical conditions including aging and poor energy balance have been reported to negatively affect the component of FF, which adversely affects oocyte developmental ability [13, 14]. Furthermore, if follicular components become affected by low GC activities, they could further affect GCs and oocyte developmental ability, creating vicious cycle. However, FF contains a myriad of factors, and a clear elucidation of the molecular background associated with proper follicle development still remains unclear.
In the present study, we tested the hypothesis that the ability of FF to support oocyte maturation and subsequent embryo development, as well as GC proliferation, is closely linked to the number of GCs associated with the FF. We also sought to shed some light on the mechanism which determines the differential number of GCs in follicles using next-generation sequencing (NGS). The data presented here provide an understanding of the properties and qualities of FF.
Materials and methods
Chemicals and media
All chemicals were purchased from Nacalai Tesque (Kyoto, Japan) unless otherwise indicated. The medium used for in vitro maturation (IVM) was porcine oocyte medium (POM) [15] supplemented with 3-mg/mL polyvinyl alcohol, 0.5-mM L-cysteine, 10-ng/mL epidermal growth factor (Sigma-Aldrich), 10-IU/mL equine chorionic gonadotropin (Aska Pharma Co. Ltd., Tokyo, Japan), and 10-IU/mL human chorionic gonadotropin (Fuji Pharma Co. Ltd., Tokyo, Japan). The medium used for in vitro culture (IVC) was PZM-3 [16].
Ovary collection
Ovaries were collected from individual gilts from a local slaughterhouse (Kanagawa Meat Center, Kanagawa, Japan), placed in phosphate-buffered saline (PBS) containing 10-IU/mL penicillin G potassium and 0.1-mg/mL streptomycin sulfate, and transported to the laboratory within 1 h. During transport, the temperature of the ovaries was maintained at 37 °C. The ovaries were discarded after intended use. This study was approved by the Ethical Committee for Animal Experimentation of Tokyo University of Agriculture.
Oocytes and follicular fluid collection
Ovaries containing middle-sized (3–5 mm in diameter) antral follicles (AFs) were selected, and the follicle contents were carefully aspirated from 20 AFs (3–5 mm in diameter) in ovaries from each gilt using a needle (21G) connected to a syringe. Follicles that had abnormal features (follicles with red or white coloring) were not aspirated. Therefore, there were no morphological and size differences among the follicles used in this study. Additionally, follicle aspiration was conducted by the same operator. COCs were collected under a stereomicroscope, and the remaining follicular contents were centrifuged (1000 ×g for 1 min) to separate the GCs from the FF. The FFs from individual gilts were further centrifuged (7000 ×g for 10 min) and stored at − 20 °C until use. GC pellets following centrifugation were used to determine the average GC number per follicle for each gilt as described below.
Measurement of average GC number per follicle using real time PCR
DNA was extracted from the GC pellets (as described above) using 1 mL of extraction buffer (Tris-HCl, 20 mM; Nonidet-40, 0.9%, Tween 20, 0.9%; and proteinase K, 0.4 mg/mL) by heating at 55 °C for 30 min followed by 98 °C for 5 min. PCR was performed using a Rotor-Gene 6500 real-time rotary analyzer (Qiagen GmbH, Hilden, Germany) with primers targeting the single-copy gene (GCG glucagon, NC_010457) and Ssofast-TM EvaGreen Supermix (Bio-Rad, Hercules CA, USA). Primers used for PCR were 5′-agcagaatcaacaccatcggt-3′ and 5′-tggctccacccatagaatgc-3′ (154 bp), which were designed using Primer3Plus (http://sourceforge.net/projects/primer3/) and the NCBI database. PCR was performed with an initial denaturation at 95 °C for 1 min, followed by 40 cycles at 98 °C for 5 s and 60 °C for 10 s. A standard curve was generated for each run using tenfold serial dilutions representing the copy number of the external standard. The external standard was the PCR product of the corresponding gene cloned into a vector using a Zero Blunt TOPO PCR cloning kit (In vitrogen, Carlsbad, CA, USA), and the PCR product was sequenced before use. Amplification efficiencies of all trials were > 1.98. Using the copy number of the single copy gene and the number of follicles aspirated (20), we calculated the total GCs per follicle (relative total GCs) for each gilt using the following formula: GC number per follicle = (copy number of single copy gene / 2) / 20.
Preparation of GC-rich and -poor FFs
Follicular contents were aspirated from 20 randomly selected AFs from the ovaries of 25 individual gilts, and the GC numbers from each gilt were determined as described above. Based on the GC number, gilts with the five highest and five lowest average GC counts per follicle were selected, together with the corresponding FF samples. These five GC-rich and five GC-poor FF samples were mixed in equal amounts to create two batches, namely a of GC rich-FF (GC-rich FF) and a GC-poor FF (GC-poor FF), respectively. Five batches were prepared from different ovary series (total 125 gilts: 25 gilts × 5). In addition, the average GC number for GC-rich and GC-poor FF was 816,254 ± 27,458 and 357,302 ± 43,707, respectively.
IVM, activation, and in vitro culture
During the 44-h maturation period, COCs were cultured in POM. IVM medium was supplemented with either 10% FF. After IVM, 25 oocytes were randomly selected, and the rate of maturation was examined (metaphase II). To determine the ability of oocytes to develop to the blastocyst stage, after culturing in IVM, oocytes were activated with single electrical pulse of 60 V for 0.1 ms using a NEPA21 (Nepa Gene Co., Ltd., Chiba, Japan) followed by culture in IVC medium containing 10-μg/mL cytochalasin B and cycloheximide for 6 h. After activation, the embryos were cultured for 7 days in IVC medium, and the rate of blastulation and the total blastocyst cell numbers were determined. Oocytes and embryos were fixed in 4% paraformaldehyde, mounted onto glass slides using an anti-fade reagent containing DAPI (ProLong Gold anti-fade reagent with DAPI; Invitrogen, Carlsbad, OR, USA), and observed using a fluorescence digital microscope (BZ-8000; Keyence, Tokyo, Japan). IVM was performed at 38.5 °C in an atmosphere containing 5% CO2 and 95% air, whereas in vitro embryo culture was performed at 38.5 °C in an atmosphere containing 5% O2, 5% CO2, and 90% N2.
Measurement of the ability of FFs from individual donors to support oocyte maturation and subsequent development
FFs from individual donor gilts were added to the IVM medium at a final concentration 10%. COCs were collected from at least 50 gilts and pooled. Forty randomly selected COCs were matured, followed by activation and IVC as described above. This experiment was repeated four times (40 COCs × 4), and the average developmental rate to the blastocyst stage (day 7) was determined for each FF.
GC proliferation assay
The GC suspension was transferred to 199 medium (Gibco, Waltham, MA, USA) containing 5% FCS, and the cells were incubated in a 96-well plate (100 μL/well; Falcon, New Jersey, USA) for 24 h at 38.5 °C with maximum humidity in an atmosphere of 5% CO2 in air. Following this, the culture medium was replaced with 199 medium containing 10% FF, and cell proliferative activity was assessed using a cell proliferation ELISA BrdU assay kit (Roche, Mannheim, Germany), according to the manufacturer’s protocol. The average of duplicate measurements was determined for each FF.
Measurement of cumulus cells surrounding the oocyte
After maturation, the COCs were vortexed to separate the oocytes and cumulus cells, and the cumulus cell suspension was centrifuged and resuspended in PBS. Total cell number was determined from the volume of the cell suspension and the cell concentration determined using a hemocytometer, and this number was then divided by the oocyte number.
Transcriptome analysis
Top five GC-rich and GC-poor donors were selected from 25 gilts, and corresponding GCs were subjected to NGS. Total RNA was extracted from each of the ten GC pellets using an RNAqueous kit (Life Technologies, Carlsbad, CA, USA). RNA quality was confirmed using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), and cDNA libraries were prepared using a TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Library quality and quantity were determined using an Agilent 2100 Bioanalyzer and KAPA Library Quantification Kit (KAPA Biosystems, Wilmington, MA, USA), respectively. Clusters were generated on a cBot (Illumina), and one lane of the multiplexed samples was sequenced as 100-bp reads (single read) on a HiSeq 2500 (Illumina). Image analysis, base calling, and quality filtering were performed using bcl2fastq2 (Illumina) following the manufacturer’s instructions. Sequence data were filtered to discard the adapter sequence, ambiguous nucleotides, and low-quality sequences. The remaining sequence data were aligned to the Swine genome sequence (susScr3) to count sequence reads. Using mapped sequence data, the expression values for each gene and statistical analysis of the differentially expressed genes were determined. Filtering, mapping, and subsequent analysis were performed using a CLC Genomics Workbench (Qiagen, Redwood City, CA, USA). Statistical significance was determined by the empirical analysis of the differential gene expression tool [17]. Differentially expressed genes (fold-change ≧ 2.0 and FDR adjusted P < 0.05 and q < 0.2; [18]) were used for further analyses. To predict upstream transcriptional regulators, genes that were significantly differentially were interpreted using the upstream regulator function of ingenuity pathway analysis (IPA, Qiagen). This determined how many known targets of each transcriptional regulator were present in the differentially expressed gene list, and calculated overlapping P values were used to measure statistically significant overlap. Fisher’s exact test was used in the analysis of gene set enrichment in each functional category, and significance was generally attributed to a P value less than 0.01. The gene expression analysis data have been registered (DRA006323).
Statistical analysis
All data were analyzed using an ANOVA followed by a post hoc Tukey’s test. Percentages were arcsine transformed before analysis. Correlation between the two groups was calculated, and values less than 0.05 were considered significantly different.
Results
The average number of GCs in follicles differs among gilts but are consistent within a gilt
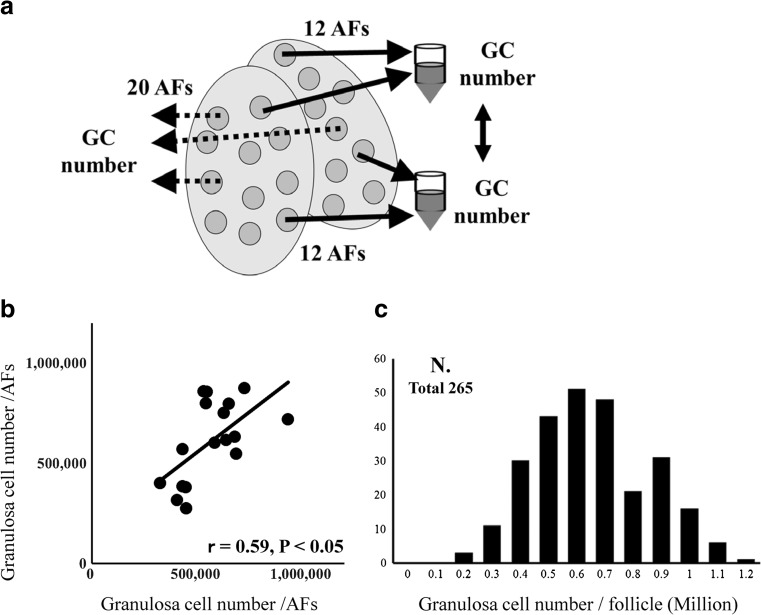
First, we addressed the consistency of GC number within a donor gilt. The number of antral follicles (3–5 mm in diameter) in the ovaries of individual donors ranged from 22 to 48 (average 34.9 ± 1.1, N = 42, preliminary experiment). Twelve AFs were aspirated from each gilt ovary to obtain GCs. The aspiration was repeated to obtain two sets of GCs from each gilt (Fig. 1a). There was a significant correlation between the two GC numbers (17 gilts, r = 0.59, P < 0.05, Fig. 1b), which means that the GC number obtained from at least 12 AFs reflects the characteristics of the donors. Based on this result, we measured the average GC number using 20 AFs. In addition, when the average GCs from 265 gilts were examined, they ranged from 134,616 to 1,251,068 indicating that there were gilts whose follicles were GC-rich and those whose follicles were GC-poor (Fig. 1c). It is worth noting that the number of GCs counted does not include all GCs in follicles, because small GCs can escape aspiration.
Fig. 1.
Examination of the consistency of granulosa cell (GC) number per follicle in a donor. a 12 randomly selected AFs were aspirated (solid arrow) from each gilt ovary to obtain GC. The aspiration was repeated to obtain two sets of GCs from each gilt. Seventeen gilts were used for this comparison. The GC number per follicle was determined using real-time PCR. b There was a significant correlation between the GC number of the two sets (R = 0.59, P < 0.05). Based on this result, we measured the GC number for each gilt using 20 AFs (dotted arrow in a). c Average granulosa cell number in the follicles of donors. Granulosa cells were collected from 20 AFs from each gilt. Two hundred sixty-five gilts were examined. X axis represents GC number and Y axis represents the number of donors
The GC number is related to the ability of FFs to support oocyte development and GC proliferation
Next, the average GC numbers of FF origin were compared with developmental rate (day 7) to the blastocyst stage when the corresponding FFs were added to the maturation medium. Figure 2a shows that there is a significant correlation between these two parameters (10gilts, r = 0.7, P < 0.05), which suggests that the higher the GC number in the follicles, the better the corresponding FF supports subsequent oocyte development. When these FFs were added to a GC culture medium, we also observed a significant correlation between the average GC number in the FF origin and the proliferative activity of the GCs (Fig. 2b, ten gilts, r = 0.7, P < 0.05). Next, we further addressed the hypothesis that there was relationship between GC number in the gilts and ability of FFs to support oocyte development. In addition, supplementation with GC-rich FF (10%) resulted in a significantly higher number of oocytes that developed to the blastocyst stage than supplementation with either GC-poor FF, or no supplementation (control oocytes 7.0%, GC-poor 9.0%, and GC-rich 15.3%, Table 1), whereas the maturation rate of oocytes did not differ among groups. In addition, the number of cumulus cells surrounding the oocytes and their survival rates were significantly greater in COCs cultured with FF irrespective of the GC number compared to cultures without FF (Table 1); a paired Student’s t test showed that there was significant difference between the GC-rich and GC-poor groups (GC-rich 2312, vs. GC-poor, 2043, P = 0.031).
Fig. 2.
a Relationship between the number of granulosa cells of FF origin and the developmental rate (day 7) to the blastocyst stage (%) of oocytes matured in medium containing the corresponding FF. b Relationship between number of granulosa cells of FF origin and the proliferative activity of granulosa cells cultured in medium containing the corresponding FFs
Table 1.
Effect of supplementation of maturation medium with GC-rich and -poor FFs on developmental ability of oocytes and number of cumulus cells
| Groups | No. of trials | No. of oocytes | Rate of M2 | Rate of blasturation (%) | No. of blastomere | No. of cumulus cells | Survival rate of cumulus cells (%) |
|---|---|---|---|---|---|---|---|
| Control | 5 | 325 | 88.8 ± 4.8a | 7.0 ± 1.5a | 35.4 ± 3.4 | 1310 ± 151a | 71.8 ± 1.7a |
| GC-poor | 5 | 325 | 87.8 ± 1.7a | 9.0 ± 1.5a | 45.7 ± 4.2 | 2043 ± 73b | 84.3 ± 0.7b |
| GC-rich | 5 | 325 | 80.0 ± 2.9a | 15.3 ± 1.3b | 47.4 ± 3.3 | 2312 ± 97b | 86.9 ± 2.1b |
Data were represented as means ± SEM. a–b: < 0.05
GC number relates to the developmental ability of enclosed oocytes
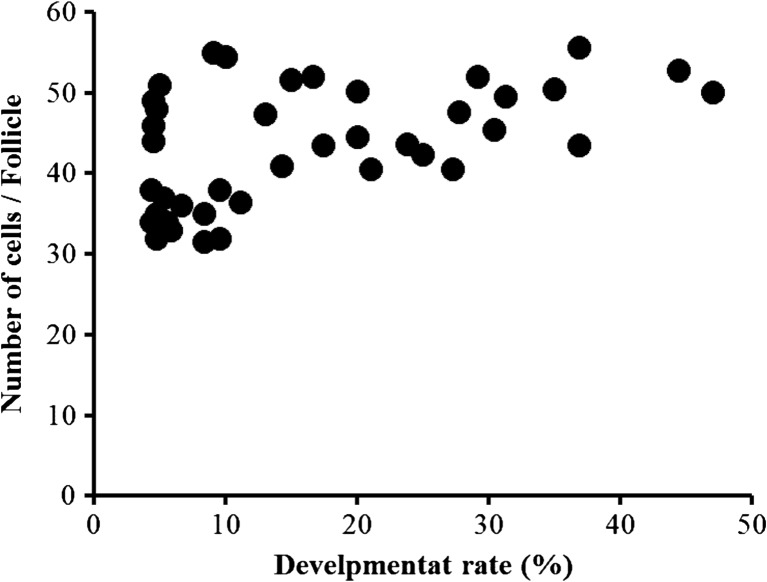
We examine the relationship between the number of GCs and the developmental ability of enclosed oocytes. All middle-sized follicles were aspirated from each donor, and the average GC number was determined for each donor using the same formula described above. All OGCs (25–40 for each gilt) were subjected to IVM, activation, and IVC to determine the developmental rate to the blastocyst for each gilt. The rate of blastulation was significantly related to the average GC number of the corresponding gilts (Fig. 3, N.41, r = 0.51, P < 0.001).
Fig. 3.
Relationship between the number of granulosa cells (GCs) in follicles and the developmental rate (at day 7) to the blastocyst stage (%) of oocytes among donors. Oocytes were collected from all middle size follicles (3–5 mm in diameter) from each donor, and all oocytes (25–40 for each donor) were used to determine the developmental rate to the blastocyst stages. The GC number per follicle was determined using the following formula: GC number per follicle = (copy number of single copy gene / 2) / number of follicles aspirated. GC number and developmental rate to the blastocyst stage were compared among 41 donors
Gene expression analysis reveals that GC-rich conditions are associated with several activation upstream regulators
The molecular mechanisms that underlie the GC number in follicles were investigated using NGS technology. NGS compared the expression levels of 22,862 genes, and 2075 of these genes were significantly differentially expressed between GC-rich and GC-poor groups. Using these differentially expressed genes, an IPA analysis revealed the activation and inhibition of upstream regulators. Tables 2 and 3 show the top 21 activated and the top 10 inhibited upstream regulators. Activated regulators included GATA4, forskolin, RAF1, INHBA, IGF1R, EGF, and estradiol and inhibited regulators included PD98059 and cycloheximide. Furthermore, the IPA analysis showed that the disease and biological functions associated with the differentially expressed genes were related to the reproductive system and development and function, particularly to folliculogenesis and the quantity of germ cells (Table 4).
Table 2.
Upstream regulators of differential expressed genes between granulosa cell rich and poor follicles
| Upstream regulator | Molecule type | Predicted activation state | Activation z score | P value of overlap | Target molecules in dataset |
|---|---|---|---|---|---|
| Estrogen | Chemical drug | 1.57 | 9.60.E−04 | COL3A1, CYBB, CYP19A1, FLT1, IGFBP4, IG | |
| FBP5, PRLR | |||||
| Beta-estradiol | Chemical-endogenous mammalian | 1.57 | 2.91E−04 | A2M, ACTA1, CCND2, CHI3L1, COL1A2, CO | |
| L3A1, CTSL, CYBB, CYP19A1, FGF1, IGFBP4 | |||||
| IGFBP5, INHA, LPCAT2, LRP8, LYZ, MMP1 | |||||
| PPP2R2C, PRLR, PTPRN, SLCO2B1, SYP | |||||
| Bucladesine | Chemical toxicant | 1.60 | 7.64.E−03 | ASS1, CR2, CYP19A1, IGFBP5, MMP1, SYP | |
| TLR4 | Transmembrane receptor | 1.62 | 9.89.E−04 | ACTA1, BAMBI, CCND2, DLL4, GPR34, MA | |
| RCO, MMP1, TCF4 | |||||
| HRAS | Enzyme | 1.66 | 4.00.E−04 | A2M, ACTA1, ATP1A3, CCND2, COL1A2, CO | |
| L3A1, IGFBP4, IGFBP5, LYZ, MMP1, SORBS1 | |||||
| TGFB3 | Growth factor | 1.68 | 3.15.E−04 | ACTA1, COL1A2, COL3A1, CYP19A1, MMP1 | |
| FGF2 | Growth factor | 1.82 | 3.14.E−05 | CCND2, COL1A2, COL3A1, CTSL, DLL4, FLT1 | |
| GBP1, IGFBP4, IGFBP5, MMP1 | |||||
| A2M, BAMBI, CCND2, CD83, COL1A2, CYB | |||||
| NFkB (complex) | Complex | 1.84 | 1.49.E−04 | B, FGF1, GFPT2, GPR34, IL13RA2, MMP1, MT3 | |
| EGF | Growth factor | 1.85 | 4.75.E−04 | CCND2, COL1A2, COL3A, FLT1, IGFBP4, IG | |
| FBP5, MMP1, MT3, PTPRN, SYP | |||||
| AHR | Ligand-dependent nuclear receptor | 1.86 | 1.18.E−01 | A2M, COL1A2, COL3A1, CYP19A1 | |
| RELA | Transcription regulator | 1.91 | 1.81.E−03 | A2M, CHI3L1, COL1A2, CR2, CYBB, CYP19A1, GBP1, MMP1 | |
| IGF1R | Transmembrane receptor | 1.94 | 3.19.E−03 | ACTA1, CD5L, CD93, COL3A1, IGFBP4, IGFBP5 | |
| ITGB1 | Transmembrane receptor | 1.97 | 1.36.E−03 | ACTA1, COL1A2, MMP1, TCF4 | |
| TCF3 | Transcription regulator | 1.98 | 1.01.E−02 | ACACB, CCND2, CR2, ZP2, ZP3, ZP4 | |
| MAPL1 | Kinase | Activated | 2.00 | 1.21.E−01 | GBP1, IGFBP7, MMP1, MX2 |
| 5-O-Mycolyl-beta-araf-(1- > 1′)-glycerol | Chemical-endogenous non-mammalian | Activated | 2.00 | 1.25.E−03 | CD83, CX3CR1MCYP19A1, MMP1 |
| INHBA | Growth factor | Activated | 2.19 | 4.13.E−03 | BAMBI, CCND2, CYP19A1, INHA, PRLR |
| Mifepristone | Chemical drug | Activated | 2.20 | 2.10.E−02 | CHI3L1, CYBB, IL13RA2, MMP1, NOX4 |
| RAF1 | Kinase | Activated | 2.20 | 6.52.E−03 | ATP6V1B1, CTSL, MMP1, MT3, TUBA4A |
| Forskolin | Chemical toxicant | Activated | 2.22 | 1.17.E−02 | ACTA1, CCND2, COL1A2, CYP19A1, GBP1, I |
| GFBP5, INHA, PTPRN, TUBA4A | |||||
| GATA4 | Transcription regulator | Activated | 2.23 | 2.21.E−03 | ACTA1, COL1A2, COL3A1, CYP19A1, INHA |
Table 3.
Upstream inhibition regulators of differential expressed genes between granulosa cell-rich and-poor follicles
| Upstream regulator | Molecule type | Predicted activation state | Activation z score | P value of overlap | Target molecules in dataset |
|---|---|---|---|---|---|
| PD98059 | Chemical-kinase inhibitor | Inhibited | − 2.63 | 3.35.E−03 | ATP6V1B1, CCND2, CD83, COL1A2, C |
| OL3A1, CTSL, MMP1, MT3, TUB4A | |||||
| Cycloheximide | Chemical reagent | Inhibited | − 2.49 | 4.10.E−03 | ASS1, CYP19A1, ERG, GBP1, IGFBP4, IG |
| FBP5, MMP1 | |||||
| Cholesterol | Chemical-endogenous mammalian | Inhibited | − 2.43 | 1.14.E−03 | CD5L, COL3A1, CYBB, FLT1, LYZ, MMP1 |
| Tgf beta | Group | Inhibited | − 2.41 | 2.14.E−03 | BAMBI, CCND2, COL1A2, IGFBP5, MMP1, NOX4 |
| Doxorubicin | Chemical drug | Inhibited | − 2.22 | 6.90.E−02 | A2M, COL3A1, CYBB, IGFBP7, NOX4 |
| Actinomycin D | Chemical drug | Inhibited | − 2.12 | 9.73.E−03 | ASS1, COL1A2, IGFBP4, IGFBP5, MAT1A |
| H89 | Chemical-kinase inhibitor | − 1.98 | 6.97.E−03 | CD83, CYP19A1, IGFBP5, MMP1 | |
| Cisplatin | Chemical drug | − 1.96 | 3.15.E−01 | A2M, FLT1, LYZ, MAT1A, NOX4 | |
| N-Nitro-L-arginine methyl | Chemical drug | − 1.95 | 1.33.E−04 | ACTA1, CD83, COL1A2, COL3A1, CYBB | |
| NFKBIA | Transcription regulator | − 1.94 | 1.05.E−02 | A2M, ATP6V1B1, CCND2, CHI3L1, CO L1A2, COL3A1, MMP1 |
Table 4.
Disease and bio-function predicted by IPA analysis
| Categories | Functions | Diseases or functions annotation | P value | Predicted activation state | Activation z score | Molecules |
|---|---|---|---|---|---|---|
| Organ development | CYP19A1, PRLR, I | |||||
| Reproductive system | Growth | Growth of genital organ | 0.0000968 | 1.919 | GFBP7, CCND2, B | |
| Development and function | MP15, INHA | |||||
| Organ morphology | ||||||
| Reproductive system | ZP3, PABPC1L, Z | |||||
| Development and function | Quantity | Quantity of oocytes | 0.0000924 | 1.969 | P2, BMP15 | |
| Tissue morphology | ||||||
| Embryonic development | ||||||
| Organ development | ZP3, CYP19A1, A | |||||
| Organismal development | Development | Development of ovary | 0.00000495 | 1.99 | MHR2, CCND2, P | |
| reproductive system | TPRN, ZP2, BMP1 | |||||
| Development and function | 5, INHA | |||||
| Tissue development | ||||||
| Embryonic development | ||||||
| Organ development | ZP3, CYP19A1, C | |||||
| Organismal development | Folliculogenesis | Folliculogenesis | 0.0000106 | 1.99 | CND2, PTPRN, ZP | |
| Reproductive system | 2, BMP15, INHA | |||||
| Development and function | ||||||
| Tissue development | ||||||
| Organ morphology | ||||||
| Reproductive system | ZP3, CYP19A1, P | |||||
| Development and function | Quantity | Quantity of germ cells | 0.0000997 | Increased | 2.409 | ABPC1L, ZP2, BM |
| Tissue morphology | P15, INHA, CHD5 |
Discussion
The results from the present study showed that GCs are a simple and useful marker of follicle status and oocyte quality, that FF was a decisive factor for determining GC number in follicles and the subsequent developmental ability of oocytes, and that it uncovers the molecular mechanism that underlies differential GC numbers in follicle.
The present study found a consistency in the number of GCs (relative total GC number) within the ovaries, such that the average GC number determined by only 12 AFs was very similar with that determined by another group of 12 AFs in the same ovaries. This result suggests that the number of GCs in follicles is a characteristic of the donors, although differences were observed among individual follicles and ovaries. The number of GC may therefore reflect the physiological condition of the ovaries and/or the donors.
It has been reported in both pigs and humans that supplementation of maturation medium with FF improves oocyte maturation, fertilization, and subsequent embryo development [19, 20]. The present study also shows the beneficial effect of FFs on cumulus cell proliferation and survival rate. Furthermore, the study also showed that the origin of the FF profoundly affects oocyte developmental competence and GC proliferation. This is the first report to identify a close correlation between the number of GCs in a follicle, and the ability of the corresponding FF to support the acquisition of oocyte developmental ability and GC proliferation. FF is the sole environment to which GCs and oocytes are exposed, and the data indicate that the characteristics of the FF determine follicular development and oocytes quality. This hypothesis was confirmed in another experiment using FF categorized based on the number of GCs it was derived from, namely GC-rich FF and GC-poor FF. Supplementation of maturation medium with GC-rich FF improved the developmental ability of oocytes, as well as increasing the cumulus cell number surrounding the oocytes. Moreover, the present study showed that high GC number in follicles associated with high developmental ability of enclosed oocytes (Fig. 3). These results led us to conclude that the GC number is a maker of high quality follicles, and that FF contains factors that are decisive for GC proliferation and oocyte developmental competence, even though the FFs were aspirated from the same-sized follicles. These data indicate that although follicles were similar in size and morphology, in pigs, the ability of follicles had already been determined, and the quality of the oocytes had already deteriorated in the FF. However, providing these oocytes with appropriate IVM conditions, their developmental ability can be improved compared to those matured under their intrinsic follicular conditions.
FF contains a myriad of factors with complex interactions; therefore, although an investigation of the effect of individual factors on follicular development and oocyte quality is important, it will be laborious works and possibly hard to integrate due to differential experimental conditions. The intent of the present study was to grasp an overview of the molecular background underlying good (GC-rich) and bad (GC-poor) follicles using RNA-seq. As a result, we identified thousands of genes that were differentially expressed between the two follicular conditions. We first noted the fact that genes associated with glycolysis were highly expressed in GCs from GC-rich follicles compared to those from GC-poor follicles (Fig. 4). This finding is interesting because the upregulation of glycolysis in GCs is a predominant event that occurs during porcine follicular development [21]. With respect to the expression of individual genes, we used an IPA analysis to understand the interactions between these differentially expressed genes. The top bio-functions of these differentially expressed genes were associated with the quantity of germ cells, folliculogenesis, and ovary development (Table 4), indicating that the present comparison between GC-rich and -poor follicles reflects good follicle and oocyte development. The upstream regulators identified include some key genes that have previously been reported to be linked to follicular status and development. Forskolin is a well-known activator of cAMP and hence of PKA; PKA is downstream of both IGF1 and FSH, which are also key regulators of steroidogenesis (Kegg pathway; http://www.kegg.jp/kegg-bin/show_pathway?hsa04913+1588). In our IPA analysis, the IGF1R was listed as being, a significant upstream regulator, having a z score of 1.94 (Table 2). Furthermore, inhibin is a well-known response factor to FSH, and the NGS analysis revealed that the expression levels of INHA and INHBA were significantly higher in GC-rich groups than in GC-poor counterparts (2.0- and 1.9-fold difference, P < 0.001 and P < 0.008, respectively). In addition, INHBA was also found to be an activation upstream regulator in GC-rich groups (Table 2). GATA4 is a cAMP/PKA effector in gonadal cells [22]; Monga et al. [23] have elegantly shown that phosphorylated GATA4 binds to the promotor of CYP19A1 and that siRNA-mediated downregulation of GATA4 leads to a reduction in CYP19A1 expression levels and estradiol secretion in buffalo ovaries. Our NGS analysis showed that CYP19A1 expression levels were significantly higher in GC-rich compared to GC-poor follicles (2.5-fold, P < 0.01). Estradiol is an important stimulator of the development of porcine and bovine oocyte GCs in vitro [24, 25]. Taken together, in response to IGF and/or FSH activation of cAMP-PKA and GATA4-estradiol appears to be the predominant condition that is important for proper follicle development (Fig. 4). EGF has been reported to be a key factor in oocyte development, and the acquisition of EGF responsiveness by cumulus cells is a fundamental event in oocyte developmental competence [26]. Our IPA analysis showed that both EGF and EGFR are significant upstream regulators having positive z scores (1.85 and 1.31, respectively). Furthermore, RAF1 and MAPK1 were found to be significantly activated regulators in GC-rich groups (Table 2). The EGF-RAF1-MAPK1 pathway is well-known to be part of the signal transduction cascade that leads to cellular proliferation. This notion is supported by a fact that PD98059, an inhibitor of MEK, was identified as a significant inhibitory regulator (Table 3). Furthermore, the high proliferation in the GC-rich group mirrors a need for high protein synthesis, which is indicated by the identification of cycloheximide as an inhibitory upstream regulator. From these data, the response to EGF followed by activation of the RAF1-MAPK1 signaling profoundly affects follicle fate and developmental ability of oocytes (Fig. 5). Other factors that have previously been reported to be involved in follicle development are FGF2 and ITGB1, which are predicted to be activated upstream regulators (Table 2). FGF2 is a master inhibitor of follicular atresia and improves EAF development in sheep [27]. ITGB1 (integrin beta 1) expression levels in GCs have been shown to be changed in follicular development, increasing from early antral to large antral follicles [21].
Fig. 4.
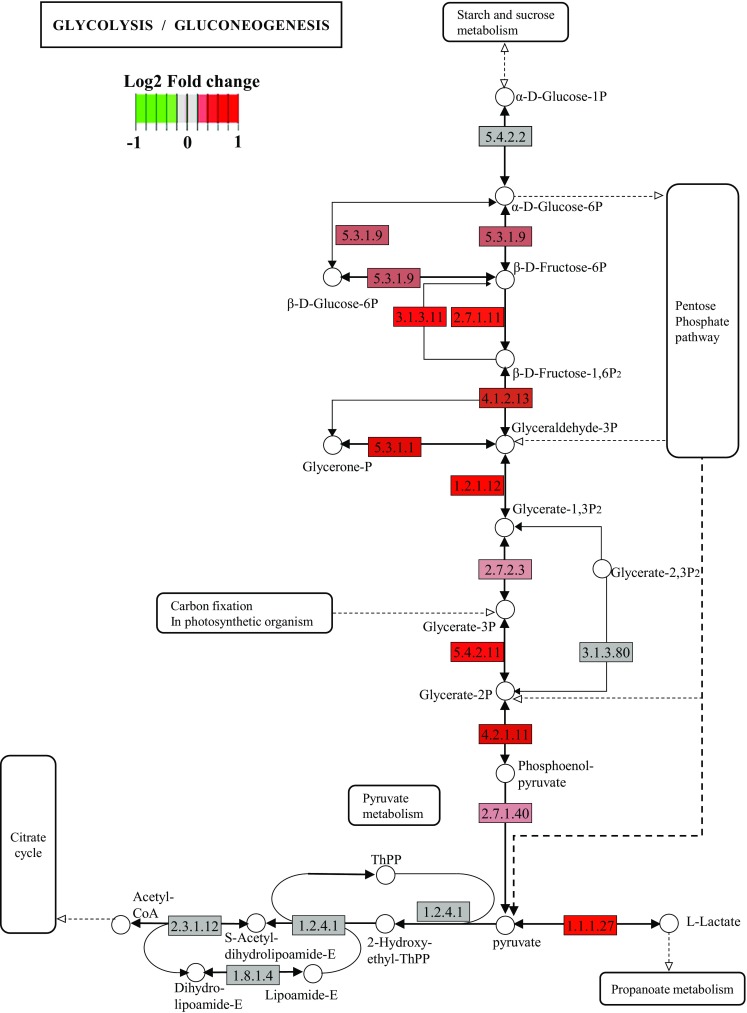
Gene expression profiles of glycolysis associated genes (log2 fold change). The pathway was retrieved from ssc00010 (http://www.genome.jp/kegg-bin/show_pathway?org_name=ssc&mapno=00010&mapscale=&show_description=show). Genes associated with glycolysis were highly expressed in granulosa cell in GC-rich follicles
Fig. 5.
Hypothesis of the factors in follicular fluid the determine granulosa cell number and oocyte developmental competences. In response to IGF1, FSH, and EGF, proper activation of RAF-MAPK and cAMP-PKA followed by estradiol production is an important molecular background for granulosa cell proliferation and successful follicle development
In conclusion, we have found that GC number is a useful marker of follicular development and the characteristics of FFs are closely associated with GC number in follicles and oocyte developmental ability. We have identified several pathways that appear to be involved in the underlying molecular mechanism behind the GC rich condition, as well as the high developmental ability of oocytes.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Clarke HJ. Regulation of germ cell development by intercellular signaling in the mammalian ovarian follicle. Wiley Interdiscip Rev Dev Biol. 2017;7:e294. doi: 10.1002/wdev.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bui HT, Van Thuan N, Kishigami S, Wakayama S, Hikichi T, Ohta H, Mizutani E, Yamaoka E, Wakayama T, Miyano T. Regulation of chromatin and chromosome morphology by histone H3 modifications in pig oocytes. Reproduction. 2007;133:371–382. doi: 10.1530/REP-06-0099. [DOI] [PubMed] [Google Scholar]
- 3.De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292:1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Jeong WJ, Cho SJ, Lee HS, Deb GK, Lee YS, Kwon TH, Kong IK. Effect of cytoplasmic lipid content on in vitro developmental efficiency of bovine IVP embryos. Theriogenology. 2009;72:584–589. doi: 10.1016/j.theriogenology.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Nagano M, Katagiri S, Takahashi Y. Relationship between bovine oocyte morphology and in vitro developmental potential. Zygote. 2006;14:53–61. doi: 10.1017/S0967199406003510. [DOI] [PubMed] [Google Scholar]
- 6.Manosalva I, González A. Aging alters histone H4 acetylation and CDC2A in mouse germinal vesicle stage oocytes. Biol Reprod. 2009;81:1164–1171. doi: 10.1095/biolreprod.109.078386. [DOI] [PubMed] [Google Scholar]
- 7.Niu Y, Wang C, Xiong Q, Yang X, Chi D, Li P, Liu H, Li J, Huang R. Distribution and content of lipid droplets and mitochondria in pig parthenogenetically activated embryos after delipation. Theriogenology. 2015;83:131–138. doi: 10.1016/j.theriogenology.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Tamassia M, Nuttinck F, May-Panloup P, Reynier P, Heyman Y, Charpigny G, Stojkovic M, Hiendleder S, Renard JP, Chastant-Maillard S. In vitro embryo production efficiency in cattle and its association with oocyte adenosine triphosphate content, quantity of mitochondrial DNA, and mitochondrial DNA haplogroup. Biol Reprod. 2004;71:697–704. doi: 10.1095/biolreprod.103.026104. [DOI] [PubMed] [Google Scholar]
- 9.Munakata Y, Ichinose T, Ogawa K, Itami N, Tasaki H, Shirasuna K, Kuwayama T, Iwata H. Relationship between the number of cells surrounding oocytes and energy states of oocytes. Theriogenology. 2016;86:1789–1798. doi: 10.1016/j.theriogenology.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 10.Sugiyama M, Sumiya M, Shirasuna K, Kuwayama T, Iwata H. Addition of granulosa cell mass to the culture medium of oocytes derived from early antral follicles increases oocyte growth, ATP content, and acetylation of H4K12. Zygote. 2016;24:848–856. doi: 10.1017/S0967199416000198. [DOI] [PubMed] [Google Scholar]
- 11.Munakata Y, Kawahara-Miki R, Shirasuna K, Kuwayama T, Iwata H. Polyacrylamide gel as a culture substrate improves in vitro oocyte growth from porcine early antral follicles. Mol Reprod Dev. 2017;84:44–54. doi: 10.1002/mrd.22758. [DOI] [PubMed] [Google Scholar]
- 12.Bijttebier J, Van Soom A, Meyer E, Mateusen B, Maes D. Preovulatory follicular fluid during in vitro maturation decreases polyspermic fertilization of cumulus-intact porcine oocytes in vitro maturation of porcine oocytes. Theriogenology. 2008;70:715–724. doi: 10.1016/j.theriogenology.2008.04.046. [DOI] [PubMed] [Google Scholar]
- 13.Takeo S, Kimura K, Shirasuna K, Kuwayama T, Iwata H. Age-associated deterioration in follicular fluid induces a decline in bovine oocyte quality. Reprod Fertil Dev. 2017;29:759–767. doi: 10.1071/RD15228. [DOI] [PubMed] [Google Scholar]
- 14.Sutton-McDowall ML, Wu LL, Purdey M, Abell AD, Goldys EM, MacMillan KL, Thompson JG, Robker RL. Nonesterified fatty acid-induced endoplasmic reticulum stress in cattle cumulus oocyte complexes alters cell metabolism and developmental competence. Biol Reprod. 2016;94:23. doi: 10.1095/biolreprod.115.131862. [DOI] [PubMed] [Google Scholar]
- 15.Yoshioka K, Suzuki C, Onishi A. Defined system for in vitro production of porcine embryos using a single basic medium. J Reprod Dev. 2008;54:208–213. doi: 10.1262/jrd.20001. [DOI] [PubMed] [Google Scholar]
- 16.Yoshioka K, Suzuki C, Tanaka A, Anas IM, Iwamura S. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod. 2002;66:112–119. doi: 10.1095/biolreprod66.1.112. [DOI] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 19.Ducolomb Y, González-Márquez H, Fierro R, Jiménez I, Casas E, Flores D, Bonilla E, Salazar Z, Betancourt M. Effect of porcine follicular fluid proteins and peptides on oocyte maturation and their subsequent effect on in vitro fertilization. Theriogenology. 2013;79:896–904. doi: 10.1016/j.theriogenology.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Tan J, Zou Y, Wu XW, Tian LF, Su Q, He JX, Huang ZH, Zhao Y, Wu LP, Wu QF. Increased SCF in follicular fluid and granulosa cells positively correlates with oocyte maturation, fertilization, and embryo quality in humans. Reprod Sci. 2017;24:1544–1550. doi: 10.1177/1933719117697125. [DOI] [PubMed] [Google Scholar]
- 21.Munakata Y, Kawahara-Miki R, Shiratsuki S, Tasaki H, Itami N, Shirasuna K, Kuwayama T, Iwata H. Gene expression patterns in granulosa cells and oocytes at various stages of follicle development as well as in in vitro grown oocyte-and-granulosa cell complexes. J Reprod Dev. 2016;62:359–366. doi: 10.1262/jrd.2016-022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tremblay JJ, Viger RS. Transcription factor GATA-4 is activated by phosphorylation of serine 261 via the cAMP/protein kinase a signaling pathway in gonadal cells. J Biol Chem. 2003;278:22128–22135. doi: 10.1074/jbc.M213149200. [DOI] [PubMed] [Google Scholar]
- 23.Monga R, Ghai S, Datta TK, Singh D. Involvement of transcription factor GATA-4 in regulation of CYP19 gene during folliculogenesis and luteinization in buffalo ovary. J Steroid Biochem Mol Biol. 2012;130:45–56. doi: 10.1016/j.jsbmb.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Tasaki H, Iwata H, Sato D, Monji Y, Kuwayama T. Estradiol has a major role in antrum formation of porcine preantral follicles cultured in vitro. Theriogenology. 2013;79:809–814. doi: 10.1016/j.theriogenology.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Endo M, Kawahara-Miki R, Cao F, Kimura K, Kuwayama T, Monji Y, Iwata H. Estradiol supports in vitro development of bovine early antral follicles. Reproduction. 2013;145:85–96. doi: 10.1530/REP-12-0319. [DOI] [PubMed] [Google Scholar]
- 26.Ritter LJ, Sugimura S, Gilchrist RB. Oocyte induction of EGF responsiveness in somatic cells is associated with the acquisition of porcine oocyte developmental competence. Endocrinology. 2015;156:2299–2312. doi: 10.1210/en.2014-1884. [DOI] [PubMed] [Google Scholar]
- 27.Santos JM, Menezes VG, Barberino RS, Macedo TJ, Lins TL, Gouveia BB, Barros VR, Santos LP, Gonçalves RJ, Matos MH. Immunohistochemical localization of fibroblast growth factor-2 in the sheep ovary and its effects on pre-antral follicle apoptosis and development in vitro. Reprod Domest Anim. 2014;49:522–528. doi: 10.1111/rda.12322. [DOI] [PubMed] [Google Scholar]