Abstract
Cancer survival is largely impacted by the dissemination of cancer cells from the original tumor site to secondary tissues or organs through metastasis. Targets for anti-metastatic therapies have recently become a focus of research, but progress will require a better understanding of the molecular mechanisms driving metastasis. Selenoproteins play important roles in many of the cellular activities underlying metastasis including cell adhesion, matrix degradation and migration, invasion into the blood and extravasation into secondary tissues, and subsequent proliferation into metastatic tumors along with the angiogenesis required for growth. In this review the roles identified for different selenoproteins in these steps and how they may promote or inhibit metastatic cancers is discussed. These roles include selenoenzyme modulation of redox tone and detoxification of reactive oxygen species, calcium homeostasis and unfolded protein responses regulated by endoplasmic reticulum selenoproteins, and the multiple physiological responses influenced by other selenoproteins.
Keywords: Selenium, angiogenesis, calcium flux, cancer, redox, antioxidant, migration
1. Introduction
Cancer treatment approaches have conventionally focused on reducing cancer cell proliferation or ‘shrinking tumors’. However, cancer patients most often die not from primary tumors but from metastasis, which involves the spread of cancer from primary sites to other tissues and accounts for greater than 90% of overall patient mortality (Gupta and Massague, 2006). Shrinking tumors is, of course, clinically important and represents one way to limit metastasis and thereby increase survival, but developing drugs that target other steps of the metastatic process can provide additional weapons to reduce cancer deaths. While such efforts have produced limited results thus far in generating effective treatments (Weber, 2013), researchers should not be deterred from delving deeply into the molecular underpinnings of metastasis so that targets can be identified and therapies pursued. The selenoprotein family is comprised of a diverse group of member proteins that may represent valuable targets for anti-metastatic drugs. Some selenoproteins are well characterized enzymes while others may play non-enzymatic roles (Reeves and Hoffmann, 2009). The cellular activities regulated by selenoproteins involving redox reactions, calcium homeostasis, and stress mitigation suggest that many are functionally integrated into the steps of metastasis, and also suggests that these proteins may be targeted for effective new anti-cancer therapies. This review will describe what is known to date regarding the role of selenoproteins in metastasis.
2. Metastasis
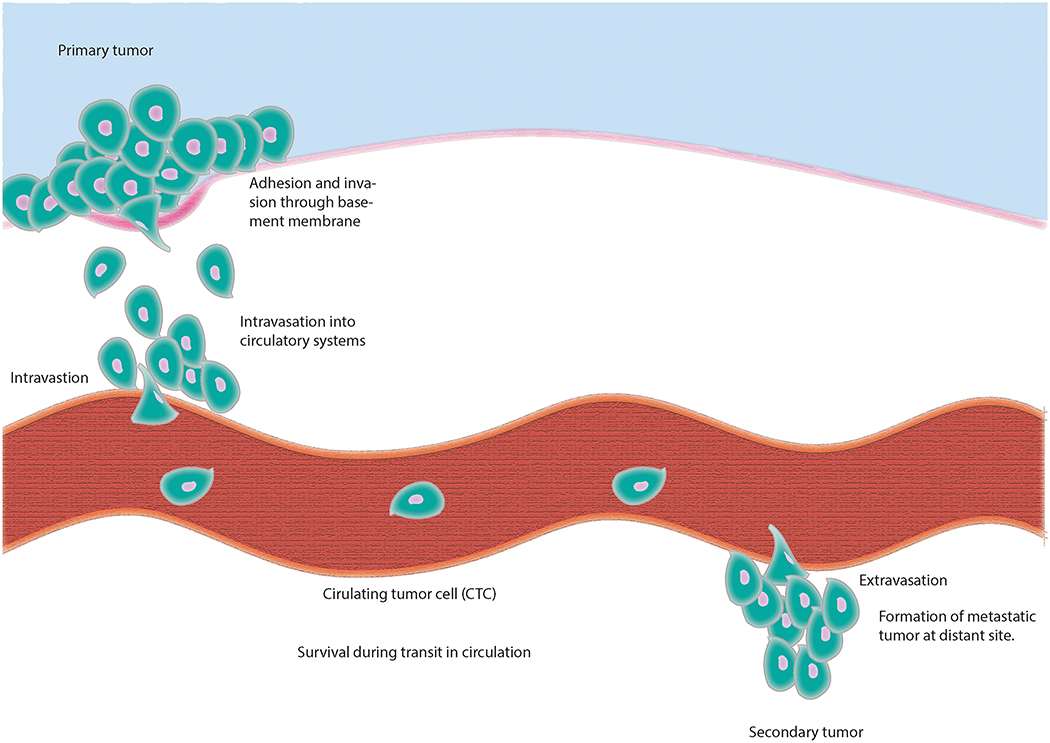
Metastasis is a term to describe a set of biological steps promoting the dissemination of transformed cells from their original tissue to other sites in the body and the subsequent outgrowth of secondary tumors. As shown in Figure 1, the main steps can be partitioned into the following: (1) local invasion of primary tumor cells through adjacent extracellular matrix (ECM), (2) intravasation into blood vessels or entry into lymphatic vessels, (3) survival during transit, (4) arrest at basement membrane of distant organ sites, (5) extravasation into parenchyma or entry into draining lymph nodes, (6) survival in an unfamiliar microenvironment, and (7) reestablishment of programs associated with proliferation including angiogenesis.
Figure 1.
The metastatic process. Metastases are formed via a multi-step process occurring after an initial tumor has been established. During metastasis tumor cells escape their primary site by passage through extracellular matrix and intravasation into blood vessels or entry into lymphatic vessels. They then translocate via systemic circulatory systems protected by platelets (tumor cell embolus, adhesion to basement membrane and extravasation) or they travel to draining lymph nodes. Once in distant tissue they proceed through metastatic deposit, angiogenesis and growth.
2.1. Local invasion
Local invasion involves the entry of cancer cells originating from the defined area of the primary tumor into adjacent normal tissue. An essential component characterizing local invasion is the ability of carcinoma cells to breach the basement membrane (BM), which is a specialized ECM component that organizes epithelial tissue. The organized structure of the BM acts as an inherent barrier to invasiveness that metastatic cancer cells must surmount. In fact, loss of expression of ECM proteins, such as type IV collagen, enhances cancer cell invasiveness in colorectal cancer (Ikeda et al., 2006). In general, active proteolysis performed by matrix metalloproteinases (MMPs) is a major contributor to the loss of the BM barrier, leading to invasion into adjacent tissue. MMPs are also implicated in other steps in the metastatic process, including intravasation and angiogenesis, and may be more relevant in the context of overall metastasis in those later steps (Stamenkovic, 2000). In addition to organizing epithelial tissue, the ECM contains a repository of growth factors that can be liberated via MMP actions, which can then be utilized by the carcinoma cells during invasion (Mott and Werb, 2004).
2.2. Intravasation
Intravasation is characterized by local invasion of the carcinoma cells into the lumina of lymph or blood vessels. At the location of intravasation, tumor cells make contact with endothelial cells via complex interactions involving proteins, lipids and carbohydrates. For example, interactions between a complex of glycosphingolipids (GSLs) and α2β1 integrin receptors was shown to be responsible for metastatic behavior of C4–2B prostate cancer cells (Van Slambrouck et al., 2014). The mechanism of intravasation is also likely to be dependent on the formation of new blood vessels (neoangiogenesis) via the action of vascular endothelial growth factors (VEGF). Compared to normal vasculature, the new blood vessels originating from carcinoma cells are ‘leaky’ and dynamic (Tong et al., 2004). Many mediators driving these early stage vasculature dynamics originate from the cancer cells themselves. For example, cyclooxygenase-2 (COX-2) is expressed in 40% of human breast cancer and mediates production of prostaglandins, thereby promoting breast cancer metastasis to bone (Singh et al., 2007).
2.3. Survival in vasculature, extravasation and formation of micrometastases
After carcinoma cells have undergone intravasation into blood vessels, they have the ability to disseminate throughout the body and may now be considered circulating tumor cells (CTC). Alternatively, they may enter lymphatic vessels and travel to draining lymph nodes. In order for CTCs to survive in hematogenous circulation they must overcome a number of stresses including absence of anchorage, hemodynamic shear forces, and immune surveillance. In the absence of adhesion to the ECM, epithelial cells normally undergo a type of apoptosis referred to as anoikis (Guo and Giancotti, 2004). Mutations involving genes that regulate apoptosis and/or anoikis that promote survival would presumably enhance metastasis. For example, transfection of the anti-apoptotic protein Bcl-xL into human breast cancer cell line MDA-MB-435 increased their metastatic ability in mice (Fernandez et al., 2000).
Given that the average size of carcinoma cells is 20–30 μm and the luminal diameter of capillaries is approximately 8 μm, the cells must often squeeze through these spaces and CTCs eventually become trapped in various capillary beds. At this point CTCs can penetrate endothelial cells and migrate into new stromal microenvironment distal from the original tumor site (extravasation). Unlike the tumor-associated blood vessels available during intravasation, the vasculature at the site of extravasation is not ‘leaky’ and therefore additional mechanisms must be activated for the CTCs to induce permeability and extravasate. One example is neuropilin-2 (NRP-2), which is a receptor for VEGF and other growth factors that interacts with α5 integrin on endothelial cells to mediate extravasation in zebrafish and murine xenograft models of clear cell renal cell carcinoma (RCC) and pancreatic adenocarcinoma (Cao et al., 2013). Furthermore, angiopoietin-like 4 (Angptl4), epidermal growth factor (EGF) family member Epiregulin (EREG), COX-2, MMP-1 and MMP-2 disrupt endothelial cell-cell junctions in breast cancer cells in the lungs (Gupta et al., 2007). Similar to the primary tumors, the promotion of angiogenesis in secondary tumors influences the final stages of metastasis and affects patient outcome.
3. Dietary selenium and selenium supplementation
Before discussing selenoproteins, one must first consider what is known about selenium metabolism and metastasis. This is because the biological effects of dietary selenium are largely exerted through the incorporation of this element into selenoproteins as selenocysteine, but when it comes to cancer biology the intermediates of selenium metabolism can play significant roles (Jackson and Combs, 2012; Rayman, 2005). The notion that selenium metabolites affect carcinogenesis and tumor progression is supported by the observation that effective doses for anti-cancer effects are substantially greater than those required for maximal selenoprotein expression. In fact, selenium metabolites may in some cases assert effects on signaling pathways in cancer cells that are opposed to the actions of certain redox regulating selenoproteins (Gopalakrishna and Gundimeda, 2001). Thus, it is important to consider the effects of various selenium metabolites separately from the actions of selenoproteins when analyzing how dietary selenium influences cancer biology and metastasis. Extensive reviews have been published describing how different forms of selenium may impact carcinogenesis, tumor proliferation, angiogenesis, and metastasis (Bera et al., 2013; Brigelius-Flohe, 2008; Chen et al., 2013b; Jackson and Combs, 2012), but there are some key issues worthy of discussion herein. First, it is important to note that data pertaining to selenium and metastasis in patients is scarce, and most of what is known has been obtained through mouse models and cell culture studies. Most of these data suggest that the form of selenium ingested and the type of metabolites they generate influence metastasis differently. One example is the finding that dietary supplementation with methylseleninic acid reduced spontaneous metastasis of Lewis lung carcinoma (LLC) in mice while supplementation with selenomethionine was ineffective (Yan and DeMars, 2012). Selenomethionine is more readily metabolized to hydrogen selenide on its way to supporting selenoprotein synthesis, while methylseleninic acid generates the bioactive methylselenol. There are also differences in how these selenocompounds affect redox status and reactive oxygen species, which both can affect the proliferation and metastatic properties of cancer cells. Using the same LLC model, the anti-metastatic effects of methylseleninic acid were abrogated by feeding the mice a high fat diet (Yan and Combs, 2014), suggesting adipose-derived inflammatory mediators can impinge upon the effector mechanisms of selenium metabolites regarding cancer cell movement in vivo. These results suggest that obesity and chronic inflammatory conditions must be taken into consideration when investigating the effects of dietary selenium and metastasis.
In a metastasis model utilizing B16BL6 murine melanoma cells injected i.v. in C57BL/6 mice followed by analyses of tumors formed in the lung, both selenite and selenomethionine supplemented at high levels (2.5 and 5.0 ppm Se) were found to inhibit tumor size in the lungs, with selenite exhibiting a more effective reduction of lung tumor formation (Yan et al., 1999). However, the high levels of selenium supplementation and model used in these studies may not reflect physiological effects of selenium on actual tumor metastasis. This same group found other forms of supplementation such as selenium containing soy protein was effective in reducing pulmonary metastasis of melanoma in mice (Li et al., 2004; Yan et al., 1997). A different model involving mice fed either selenium deficient or adequate diets (0.01 and 0.08 ppm Se, respectively) or diets supplemented with selenite (0.4 ppm Se), selenomethionine (3 ppm Se), or methylesleneninic acid (3 ppm Se) were examined for metastasis using mammary gland inoculation of 4T1.2 murine breast cancer cells into mammary tissue followed by tumor size analyses in secondary organs (Chen et al., 2013a). While no differences were found between selenium deficient and adequate dietary groups, reduction of metastases to some organs was found with selenomethionine or methylesleneninic acid, but selenite appeared to increase tumors in secondary tissues. Since the molar levels of selenium were not equivalent and bioavailability of selenium from each is different, it is difficult to compare these supplemented forms. However, the lack of differences between selenium deficient and adequate diets suggests that subtle, physiologically relevant differences in selenium intake may not have strong effects. Also, the variability in secondary tumor formation in supplemented groups also does not provide conclusive results regarding how selenium metabolites affect metastasis. In another study involving the 4T1.2 breast cancer model, selenium enriched Lactobacillus brevis was found to reduce secondary tumor formation in the liver (Yazdi et al., 2013). The bioavailable levels from different bioactive forms of selenium leading to these results were not determined, but these results are consistent with the general notion that selenium supplementation can affect metastasis.
A number of studies have focused on how selenium compounds affect different steps of the metastatic process such as detachment of cells from the extracellular matrix or tumor tissue, degradation of extracellular matrix and migration, invasion into the blood and extravasation into secondary tissues, and subsequent proliferation into metastatic tumors. For example, in vitro studies using a human fibrosarcoma cell line, HT1080, showed that supplementation with selenite, but not selenite, decreased the invasiveness due to inhibition of cell-matrix interactions and alteration of protease expression involved in matrix degradation (Yoon et al., 2001). Methylseleninic acid and methylselenol also were found to affect the expression profile of matrix metalloproteinases (MMPs) along with their inhibitors in HT1080 cell studies that resulted in decreased invasiveness (Park et al., 2007; Zeng et al., 2006). Another research group examined the effects of methylselenol generated from selenomethionine on cell proliferation, adhesion, and expression of integrins in murine melanoma B16F10 cells, which are metastatic in the lungs of syngeneic C57BL/6J mice (Kim et al., 2007). Treatment with both selenomethionine and the enzyme that converts it to methylselenol, methioninase, led to reduced integrin expression and inhibited melanoma-ECM adhesion. This detachment led to increased apoptosis and the approach used suggests that the anti-cancer effects of selenium supplementation can be enhanced by therapeutic manipulation of factors that regulate certain metabolic steps. Migration of melanoma cells was also decreased by selenite treatment by decreasing IL-18 gene and protein expression, which was utilized by the cells to promote migration (Song et al., 2011).
As these studies demonstrate, the relationship between dietary selenium and metastasis is quite complex. The physiology underlying how selenium status affects the stages of tumor development, metastasis, and clinical outcome depends on the type of cancer, the form of selenium ingested and its interaction with several patient factors such as inflammatory conditions and others. The use of pharmacological levels of selenocompounds for therapy appears to show promise based on mouse models and cell studies. However, results from clinical trials and the wide variety of tissues affected by using high doses of selenocompounds to reduce tumor progression and metastasis do not provide a clear picture of how this therapeutic approach may be optimally utilized. This has led many researchers to turn their attention to better understanding roles that individual selenoproteins may play in cancer metastasis. Selenoproteins may serve as more selective targets for controlling certain steps of cancer progression and metastasis. In addition, underlying genetic factors that influence selenoproteins levels or functions may affect cancer in concert with or independent of dietary selenium intake. The remainder of this review will summarize issues regarding how different selenoproteins may be involved with metastasis.
4. Thioredoxin reductases and redox tone
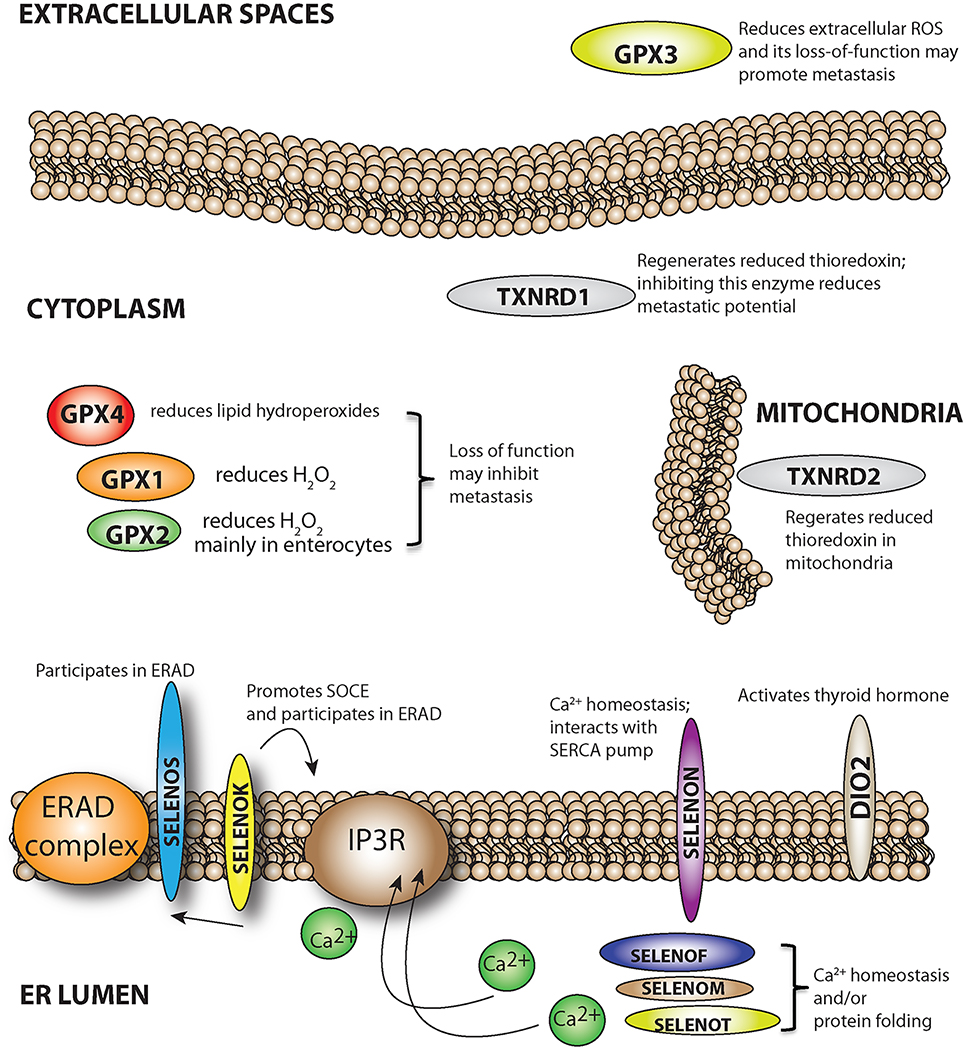
As primary cells of various types transform and develop into aggressive malignant cells, their energy demands and redox tone are altered (Cairns et al., 2011). As discussed above, selenium metabolites may influence redox cycling in different ways, and for the most part the selenoproteins function to tip redox balance toward a reductive state. As described in Chapter 9 of this series and in other published work (Arner, 2009), one selenoprotein subfamily that plays a key role in redox adjustment during tumor progression is the thioredoxin reductases (TXNRDs). TXNRD1 is localized to the cytosol and nucleus and represents the best characterized member of the subfamily in regards to cancer. TXNRD2 is localized to the mitochondria in most cells and TXNRD3 is mainly expressed in sperm during maturation. All three TXNRDs utilize NADPH as an electron donor to regenerate reduced thioredoxin in cells and thus increase reducing capacity (Mustacich and Powis, 2000).
The essential role for TXNRD1 and 2 in development are evident by the fact that deletion of either in mice results in embryonic lethality (Conrad et al., 2004; Jakupoglu et al., 2005). As is the case with many proteins important for development, the TXNRDs are upregulated in many types of cancer. The best example is TXNRD1, which is overexpressed in a variety of malignant cells compared to normal tissue of the same region and with progression toward a metastatic phenotype (Selenius et al., 2010). Microarray and immunohistochemical analyses found that increased TXNRD1 expression correlated with regional lymph node metastasis and clinical stage in 50 patients with oral squamous cell carcinoma (Iwasawa et al., 2011). Mice injected with mouse lung carcinoma cells in which RNAi was used to reduce TXNRD1 expression showed a dramatic reduction in tumor progression and metastasis compared with those mice injected with control cells (Yoo et al., 2006). While much has been uncovered regarding the role of TXNRD1 in steps of carcinogenesis and tumor formation (Schmidt and Arnier, 2016), limited information is available on its specific roles in metastasis. One exception to this is the demonstrated importance of TXNRD1 in angiogenesis during the metastatic process. Reduced expression of TXNRD1 or treatment with reagents that target the TXNRD members exhibit antiangiogenic properties (Xie et al., 2016a). A pro-invasive role for TXNRD1 was shown in breast cancer studies using MDA-MB-231 cells and meta-analysis of 25 gene array studies with 5910 patients showed that thioredoxin-1 and TXNRD1 were both associated with a poor patient prognosis in terms of overall survival, distant metastasis free survival and disease free survival (Bhatia et al., 2016). This is consistent with data showing levels of this enzyme in serum as measured by ELISA correlated with prostate cancer metastasis risk (Zhang et al., 2015) and TXNRD1 levels measured by histological analyses correlating with gallbladder carcinoma invasiveness risk (Nagano et al., 2012). Also, metastasis of salivary adenoid cystic carcinoma correlated with increased thioredoxin and TXNRD1 and a TXNRD1 inhibitor, BBSKE, reduced metastatic potential by limiting EMT in a xenograft mouse model (Jiang et al., 2015). EMT via anchorage independent growth properties of mouse lung carcinoma (LLC1) cells in vitro and in mice injected with these cells was reduced using RNAi techniques targeting TXNRD1 (Yoo, et al., 2006). In fact, EMT has become a focus of TXNRD1 inhibitors as a potential therapeutic strategy for reducing metastasis (Durand and Storz, 2017). TXNRD2 has been less studied, but this selenoenzyme was upregulated in metastatic osteosarcoma and the TXNRD inhibitor, auranofin, was found to reduce metastasis to pulmonary tissues (Topkas et al., 2016). Prostate cancer patients carrying a TXNRD2 single nucleotide polymorphism (rs1005873) had a higher risk of cancer aggressiveness (Xie et al., 2016b), although the functional significance of this polymorphism was not determined. Thus, both TXNRD1 and 2 may not only serve as prognostic biomarkers for metastasis but as therapeutic targets for reducing metastatic capacity for a variety of cancers by limiting multiple steps including EMT, angiogenesis, as well as the redox dependent survival of these cells during the metastatic process.
5. GPX and ROS regulation
While TXNRDs appear to mainly serve a pro-metastasis role as described above, the relationship between the glutathione peroxidases (GPXs) and metastasis is more dependent on the member of the GPX family being considered. All GPX enzymes utilize glutathione that is oxidized to glutathione disulphide (GSSG) in the process of reducing hydroperoxides (Brigelius-Flohe and Maiorino, 2013). Hydroperoxides are non-radical reactive oxygen species (ROS) generated in cells that in some cases can diffuse to extracellular spaces and possibly back into cells (Finkel, 2003). Hydrogen peroxide (H2O2) and lipid hydroperoxides are reduced by GPX1–3 and 4, respectively, along with other antioxidant systems such as peroxiredoxins and catalase. This detoxification process serves to reduce damage these ROS may cause in cells and the extracellular matrix but also regulates the signaling functions of these secondary messengers as well. The signaling roles hydroperoxides, particularly H2O2, play in non-cancer cells can be exploited during the transformation of primary cells to cancer cells and subsequent development into metastatic tumor cells (Liou and Storz, 2010; Storz, 2005; Wu, 2006). In some cases, GPX loss-of-function may lead to increased ROS that may promote the metastatic potential of tumors. This was clearly demonstrated in studies showing that the treatment of carcinoma cells with H2O2 prior to intravenous injection into mice enhanced metastasis (Kundu et al., 1995). This may reflect a particularly important role for extracellular ROS in promoting metastasis, and the enzyme that regulates levels of H2O2 in the extracellular space is the secreted GPX3 enzyme. In human thyroid cancer, methylation of the GPX3 gene and reduced GPX3 protein levels were correlated with tumor size and lymph node metastasis (Zhao et al., 2015). Wnt signaling was inhibited by overexpressing GPX3 in TPC-1 and FTC133 cells, suggesting that these cells utilize secreted GPX3 to regulate extracellular H2O2 levels and this regulates cellular function. There are other examples of both methylation of the GPX3 gene and reduced GPX3 protein being associated with metastatic potential including studies involving prostate cancer (Yu et al., 2007), hepatocellular carcinoma (Cao et al., 2015), gastric carcinoma (Peng et al., 2012), as well as a cervical cancer study in which downregulation of GPX3 was associated with lymph node metastasis and prognosis (Zhang, et al., 2015).
An elevation in bioavailable extracellular H2O2 may facilitate cancer cells during various stages of the metastatic process, but since this ROS can diffuse through the plasma membrane, it is likely intracellular GPX enzymes, such as GPX1, 2 and 4, are involved as well. In contrast to secreted GPX3, loss-of-function of these intracellular GPXs is in some cases associated with decreased metastatic potential. Perhaps the best example is GPX2, which is highly expressed in gastrointestinal tissues and mostly examined in the context of colorectal cancer due to its upregulation in intestinal adenomas and in colon tumors (Florian et al., 2001). In a study using patient-derived colonosphere cultures, GPX2 silencing caused accumulation of reactive oxygen species, sensitization to H2O2-induced apoptosis, and strongly reduced metastases from primary tumors in the spleen to secondary tumors in the liver (Emmink et al., 2014). These cancer cells rely on GPX2 to keep ROS at optimal levels for metastatic capacity. GPX2 expression may be induced through NF-E2-related factor-2 (Nrf2) or through transcription factors of the signal transducers and activators of transcription (STAT) family (Emmink, et al., 2014). Interestingly, GPX2 loss generated a population of stem-like cancer cells, suggesting that GPX2 and ROS play important roles during both early and late stages of cancer development. How these regulatory pathways operate during progression to metastatic stages of cancer cell development are not completely understood, but may offer potential as targets of controlling tumor progression. This is highlighted by studies showing more aggressive tumors have higher levels of Nrf2 compared to less aggressive tumors and this can upregulate expression of both GPX2 and TXNRD1 for exploitation of their roles in regulating ROS (Brigelius-Flohe et al., 2012; Chen et al., 2010). GPX2 was also found to contribute to metastatic potential in hepatocarcinoma (Suzuki et al., 2013). There may be tissue-specific effects of GPX2 regulating metastasis as demonstrated by findings with non-gastrointestinal or liver cancers such as the urothelial carcinoma in which higher risk of metastasis-free survival has been associated with lower levels of GPX2 transcripts (Chang et al., 2015).
Even though ROS induced damage and apoptosis need to be controlled through the activity of antioxidant enzymes in cancer cells, metastatic cancers still require usable levels of H2O2 to migrate and invade. This was shown in the HT-29 human colon cancer cell line in which stable GPX2 knockdown (not complete knockout) cells exhibited a higher capacity to migrate and invade than the GPX2-expressing controls and this was dependent on cyclooxygenase-2 (COX-2) (Yoon et al., 2002). Thus, GPX2 and H2O2 levels may need to fluctuate depending on the stage of tumor progression for promoting metastasis. So, how does H2O2 promote metastasis? Part of the answer may involve MMPs. As described above, prior to migration of cancer cells to distal sites they undergo EMT to release themselves from the restraint of the basal membrane. During this process, MMPs are upregulated to degrade the proteins that compose the basal membrane and are key for the metastatic process (Duffy et al., 2000; Nelson et al., 2000), and these enzymes are regulated by ROS at several levels. One example is the regulation of the MMP activation state through redox modulation of the cysteines in their catalytic domains (Koch et al., 2009; Rajagopalan et al., 1996). In addition, MMPs can be regulated transcriptionally by ROS via Ets and AP-1 responsive elements present in promoter regions (Nelson et al., 2006). Sustained production of H2O2 was also shown to influence expression of MMPs through activation of the phosphatidylinositol 3-kinase/NF-κB pathway, and this affected cell motility and invasion in HT1080 fibrosarcoma cells (Yoon, et al., 2002). Also, in esophageal cancer cell lines EC109 and EC9706, enhanced GPX1 activity led to higher capacity for invasion and migration through enzymes MMP2 and urokinase type plasminogen activator (uPA), which are crucial for tumor formation and metastasis (Qian et al., 2010). Consistent with these results are data showing a role for selenoprotein expression in macrophages for influencing MMP function through transcriptional regulation of inhibitors of ECM proteolysis (Carlson et al., 2009). These inhibitors affected migration of these cells through ECM and may reflect mechanisms by which selenoproteins control ROS regulated migration of cancer cells during metastasis, although specific roles for GPXs were not addressed in the macrophage study.
GPX1 has largely been studied in relation to carcinogenesis, although transcript levels for GPX1 along with other antioxidants have been suggested to have predictive value for metastasis (Giesing et al., 2012; Han et al., 2013; Miar et al., 2015; Min et al., 2012). Increased GPX1 activity, but not expression levels, was linked to increased vascular invasion using a hepatocellular carcinoma cell model (Huang et al., 2012). GPX4 has been found to be decreased in many cancer tissues, and GPX4-overexpressing cancer cells have low COX-2 activity and tumors derived therefrom are smaller than from control cells and do not metastasize (Brigelius-Flohe and Kipp, 2009). Overall, the collective data suggest a dependence on increased extracellular H2O2 for metastasis by decreasing GPX3 secretion, but the cancer cells themselves may upregulate other GPXs in a tissue-specific manner to manage intracellular ROS. In this sense, treatments focused on antioxidant drugs that are restricted to the extracellular spaces may prove more effective than those targeting extra- and intracellular spaces.
6. Selenoproteins involved in ER stress and calcium flux
Seven of the twenty-five members of the selenoprotein family are localized to the ER, including SELENOF, SELENOK, SELENOM, SELENON, SELENOS, SELENOT, and the type-2 iodothyronine deiodinase (Shchedrina et al., 2010). While the latter is an enzyme regulating thyroid hormone metabolism, the other six have been found to be involved in functions such as quality control of protein folding in the ER, retrotranslocation of misfolded proteins from the ER to the cytosol, protein palmitoylation at the ER surface, and regulation of calcium (Ca2+) homeostasis. Ca2+ flux from the ER and ER stress are important for regulating metastasis, which implicates the ER resident selenoproteins involved in these cellular processes.
The importance of ER stress generated through the unfolded protein response in regulating metastasis is well recognized (Hazari et al., 2016). The ER selenoproteins involved in the unfolded protein response within this organelle include SELENOF, SELENOK, and SELENOS (Kelly et al., 2009; Labunskyy et al., 2009; Lee et al., 2015). Polymorphisms in SELENOS have been linked to increased risk for different cancers (Hart et al., 2011; Shibata et al., 2009). SELENOF has been shown to have a pro-tumorigenesis and metastasis role, although exactly how it promotes these activities is unclear (Irons et al., 2010; Tsuji et al., 2015). Altogether, little experimental data are available regarding specific mechanisms regulated by these selenoproteins in relation to metastasis. A similar lack of knowledge pertains to roles for the ER selenoproteins involved in Ca2+ homeostasis and/or flux, which include SELENOK, SELENOM, SELENON, SELENOT (Appenzeller-Herzog and Simmen, 2016; Grumolato et al., 2008; Reeves et al., 2010; Verma et al., 2011). Direct examination of how these family members may influence metastasis through Ca2+ homeostasis or flux from the ER is scant. Ca2+ signaling within normal and cancer cells regulates diverse cellular functions including proliferation, migration, differentiation and cytokine secretion (Berridge et al., 2003). Store operated Ca2+ entry (SOCE) is a mechanism utilized to release Ca2+ from ER stores and activate Ca2+ signaling, and SOCE has been shown to depend on SELENOK in immune cells (Verma, et al., 2011). In particular, SELENOK regulates SOCE by complexing with the DHHC6 enzyme to promote palmitoylation and stable expression of the inositol 1,4,5-triphosphate receptor for efficient SOCE (Fredericks et al., 2014). There is an emerging recognition of SOCE as playing a major role in metastasis (Chen et al., 2016; Umemura et al., 2014; Xia et al., 2016; Yu et al., 2016), suggesting that SELENOK may represent a potential therapeutic target for reducing tumor progression. In fact, ongoing studies in our laboratory support this notion and the fact that SELENOK deficient mice show no major defects suggest that side-effects of targeted SELENOK inhibition for cancer patients may be minimal. The stoichiometry of the SELENOK cofactor and the DHHC6 enzyme may be important for migrating cancer cells in that overexpression of SELENOK was found to inhibit migration of human gastric cancer BGC-823 cells in transwell assays (Ben et al., 2015). This was true for full-length SELENOK and not a C-terminally truncated version, suggesting an overabundance of the selenocysteine containing SELENOK was capable of exerting a dominant negative effect. Overall, there remains a major gap in knowledge regarding how the ER selenoproteins may be involved in metastasis and further investigation is warranted.
7. Other potential roles for selenoproteins in metastasis
Among all selenoproteins, selenoprotein P (SELENOP) is the only one that contains more than one selenocysteine residue per protein with human and rodent SELENOP containing ten selenocysteines per molecule. SELENOP is found in the plasma and mainly secreted by the liver, although many cell types are capable of expressing and secreting this selenoprotein (Schweizer et al., 2016). A main function of SELENOP is transporting selenium to various tissues, although the selenocysteine most proximal to the N-terminus has antioxidant capacity (Burk and Hill, 2015). In this sense, understanding how SELENOP is related to metastasis parallels the discussion above regarding the relationship of dietary selenium to metastasis since SELENOP delivers bioactive selenium to tissues. However, because different cell types synthesize SELENOP at different levels and its synthesis comes at a potential cost to the synthesis of other selenoproteins, there are some additional issues. For the most part, tumors themselves exhibit lower levels of SELENOP compared to healthy tissues at the same location (Rayman, 2005), but neighbor cells may secrete SELENOP to compensate. For example, a recent study in which tumor cell culture media was used to differentiate macrophages into a pro-tumor phenotype (M2) revealed that SELENOP was upregulated 95-fold at the transcript level (Solinas et al., 2010). Colonocytes from late stage colorectal cancer (containing both normal and neoplastic human epithelial cells) had increased SELENOP transcript levels compared to early stage and healthy groups (Yajima et al., 2007). Whether this led to more secreted SELENOP for use by the tumor cells was not determined. However, another study looking directly at prostate cells as they transformed into highly metastatic cancer cells found that SELENOP levels decreased (Calvo et al., 2002). There may be a stronger demand of bioavailable selenium for synthesis of other selenoproteins and, since SELENOP consumes 10-fold more selenium than the other members of the selenoprotein family, this may mean that synthesis of SELENOP is sacrificed. In this sense, the increased SELENOP in M2 macrophages may offset the loss of SELENOP in tumor cells and support metastasis by supplying this in a paracrine manner, although this has not been tested experimentally. This may depend also on the location of cancer as a study of gastric adenocarcinoma found that SELENOP expression was low in cancer tissues compared with control tissues and was related to the degree of gastric adenocarcinoma differentiation but not to tumor node metastasis stage (Wang et al., 2009).
The 15 kDa selenoprotein, selenoprotein F (SELENOF; SEP15), has been found to be upregulated in a wide variety of cancers and reduced in others (Carlson et al., 2016). Regarding metastasis, SELENOF was shown to promote both anchorage-dependent and anchorage-independent growth and formation of experimental metastases of mouse colon carcinoma CT26 cells (Tsuji, et al., 2015). In contrast to CT26 cells, SELENOF-targeted down-regulation in Lewis Lung Carcinoma (LLC1) cells did not affect anchorage-dependent or -independent cell growth (Irons, et al., 2010). Thus, the role of SELENOF in facilitating steps of metastasis may be tissue specific and this requires further investigation.
The three selenoenzymes regulating thyroid hormone activity include iodothyronine deiodinases 1,2, and 3 (DIO1, 2, and 3). DIO1 and 2 convert the prohormone (T4) into active hormone (T3), and DIO3 inactivates T3 (Kohrle et al., 2005). Since thyroid hormone regulates a wide range of cellular processes, including the balance between proliferation and differentiation, it is not surprising that these selenoproteins are involved in cancer (Piekielko-Witkowska and Nauman, 2011). DIO3 in particular is reactivated in human neoplasias through MAPK and sonic hedgehog pathways (Romitti et al., 2016). However, specific roles for the DIO enzymes in metastasis are less clear. Some evidence was presented for a potential role of DIO3 in a study involving papillary thyroid carcinoma showing increased mRNA levels and activity for this enzyme were correlated with lymph node or distant metastasis at diagnosis (Romitti et al., 2012). Data from this study also suggested that levels of DIO2 were altered in the opposite direction, although the role in metastasis was not clearly determined. Much of what is known regarding DIOs and metastasis has been garnered through case studies, and more mechanistic studies interrogating the roles they may play in in disease progression through metastatic processes is required.
SELENOH is an oxidoreductase localized to the nucleolus in a variety of tissues that has been shown to be involved in p53 pathway activation related to both development and carcinogenesis (Cox et al., 2016). There is evidence using the MDA-MB-231 metastatic breast cancer cell line that SELENOH is transcriptionally upregulated by the anti-proliferation and anti-migration protein, delta-lactoferrin (Hoedt et al., 2014). These different studies do not clarify how SELENOH may affect metastasis, but redox directed transcription regulated by SELENOH is likely to both influence and be influenced by the stage of cancer cell progression into aggressive metastatic tumors.
8. Conclusions
There are several cellular activities regulated by selenoproteins related to metastasis and poor outcome in cancer patients. These include modulation of redox tone and detoxification of ROS by the TXNRDs and GPXs, Ca2+ homeostasis and unfolded protein responses regulated by ER selenoproteins, and the multiple physiological responses influenced by other selenoproteins. The tissue-specific expression of different selenoproteins is likely to influence particular roles they play during development of different cancers into metastatic tumors and the establishment of secondary tumors in various tissues. Also, it is worth noting that there are sex-specific mechanisms affecting transcription, alternative splicing, translation and post-translational activity of different selenoproteins (Schomburg, 2016). Thus, the effects exerted by selenoproteins on the steps of metastasis may differ between males and females. Acquiring data from studies in humans has proven difficult, although many clues have been obtained through correlative studies. Animal and cell culture model systems have emerged as a source of novel insights and this is sure to expand in the future as genetic models become more available for the full set of selenoproteins and factors regulating their activity. In fact, the exact biological roles for many selenproteins themselves are only beginning to emerge. More mechanistic insight is a prerequisite for developing anti-metastasis therapies focused on modulating selenoproteins, but progress will continue to be made toward this goal as our understanding of the biological roles of selenoproteins increases.
Figure 2.
Potential roles for selenoproteins in metastasis. Selenoproteins located in different sites within the cell or in extracellular spaces may promote or inhibit malignant cells as they progress through the steps or metastasis.
Acknowledgements
This work was supported by U.S. National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases Grant R01AI089999.
References
- Appenzeller-Herzog C, and Simmen T (2016). Biochem Soc Trans 44, 452–9. [DOI] [PubMed] [Google Scholar]
- Arner ES (2009). Biochim Biophys Acta 1790, 495–526. [DOI] [PubMed] [Google Scholar]
- Ben SB, Peng B, Wang GC, Li C, Gu HF, Jiang H, Meng XL, Lee BJ, and Chen CL (2015). Biochemistry (Mosc) 80, 1344–53. [DOI] [PubMed] [Google Scholar]
- Bera S, De Rosa V, Rachidi W, and Diamond AM (2013). Mutagenesis 28, 127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, and Roderick HL (2003). Nat Rev Mol Cell Biol 4, 517–29. [DOI] [PubMed] [Google Scholar]
- Bhatia M, McGrath KL, Di Trapani G, Charoentong P, Shah F, King MM, Clarke FM, and Tonissen KF (2016). Redox Biol 8, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigelius-Flohe R (2008). Chem Biodivers 5, 389–95. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R, and Kipp A (2009). Biochim Biophys Acta 1790, 1555–68. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R, and Maiorino M (2013). Biochim Biophys Acta 1830, 3289–303. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R, Muller M, Lippmann D, and Kipp AP (2012). Int J Cell Biol 2012, 486147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk RF, and Hill KE (2015). Annu Rev Nutr 35, 109–34. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, and Mak TW (2011). Nat Rev Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, Jorcyk C, and Green JE (2002). Cancer Res 62, 5325–35. [PubMed] [Google Scholar]
- Cao S, Yan B, Lu Y, Zhang G, Li J, Zhai W, Guo W, and Zhang S (2015). Clin Res Hepatol Gastroenterol 39, 198–204. [DOI] [PubMed] [Google Scholar]
- Cao Y, Hoeppner LH, Bach S,E,G, Guo Y, Wang E, Wu J, Cowley MJ, Chang DK, Waddell N, Grimmond SM, Biankin AV, Daly RJ, Zhang X, and Mukhopadhyay D (2013). Cancer Res 73, 4579–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA, Hartman JM, and Tsuji PA (2016). In “Selenium: Its Molecular Biology and Role in Human Health.” (U. a. T. D. L. S. Hatfield PAand Gladyshev VN, ed.), Vol. 4, pp. 235–243. Springer, New York. [Google Scholar]
- Carlson BA, Yoo MH, Sano Y, Sengupta A, Kim JY, Irons R, Gladyshev VN, Hatfield DL, and Park JM (2009). BMC Immunol 10, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang IW, Lin VC, Hung CH, Wang HP, Lin YY, Wu WJ, Huang CN, Li CC, Li WM, Wu JY, and Li CF (2015). World J Urol 33, 1777–89. [DOI] [PubMed] [Google Scholar]
- Chen N, Yi X, Abushahin N, Pang S, Zhang D, Kong B, and Zheng W (2010). Int J Clin Exp Pathol 4, 85–96. [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Prabhu KS, Das A, and Mastro AM (2013a). Int J Cancer 133, 2054–64. [DOI] [PubMed] [Google Scholar]
- Chen YC, Prabhu KS, and Mastro AM (2013b). Nutrients 5, 1149–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Hsu KF, and Shen MR (2016). Biochim Biophys Acta 1863, 1427–35. [DOI] [PubMed] [Google Scholar]
- Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, and Brielmeier M (2004). Mol Cell Biol 24, 9414–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AG, Tsomides A, Kim AJ, Saunders D, Hwang KL, Evason KJ, Heidel J, Brown KK, Yuan M, Lien EC, Lee BC, Nissim S, Dickinson B, Chhangawala S, Chang CJ, Asara JM, Houvras Y, Gladyshev VN, and Goessling W (2016). Proc Natl Acad Sci U S A 113, E5562–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, Maguire TM, Hill A, McDermott E, and O’Higgins N (2000). Breast Cancer Res 2, 252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand N, and Storz P (2017). Expert Rev Anticancer Ther 17, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmink BL, Laoukili J, Kipp AP, Koster J, Govaert KM, Fatrai S, Verheem A, Steller EJ, Brigelius-Flohe R, Jimenez CR, Borel Rinkes IH, and Kranenburg O (2014). Cancer Res 74, 6717–30. [DOI] [PubMed] [Google Scholar]
- Fernandez Y, Espana L, Manas S, Fabra A, and Sierra A (2000). Cell Death Differ 7, 350–9. [DOI] [PubMed] [Google Scholar]
- Finkel T (2003). Curr Opin Cell Biol 15, 247–54. [DOI] [PubMed] [Google Scholar]
- Florian S, Wingler K, Schmehl K, Jacobasch G, Kreuzer OJ, Meyerhof W, and Brigelius-Flohe R (2001). Free Radic Res 35, 655–63. [DOI] [PubMed] [Google Scholar]
- Fredericks GJ, Hoffmann FW, Rose AH, Osterheld HJ, Hess FM, Mercier F, and Hoffmann PR (2014). Proc Natl Acad Sci U S A 111, 16478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesing M, Driesel G, Molitor D, and Suchy B (2012). BJU Int 110, E1202–11. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna R, and Gundimeda U (2001). Nutr Cancer 40, 55–63. [DOI] [PubMed] [Google Scholar]
- Grumolato L, Ghzili H, Montero-Hadjadje M, Gasman S, Lesage J, Tanguy Y, Galas L, Ait-Ali D, Leprince J, Guerineau NC, Elkahloun AG, Fournier A, Vieau D, Vaudry H, and Anouar Y (2008). FASEB J 22, 1756–68. [DOI] [PubMed] [Google Scholar]
- Guo W, and Giancotti FG (2004). Nat Rev Mol Cell Biol 5, 816–26. [DOI] [PubMed] [Google Scholar]
- Gupta GP, and Massague J (2006). Cell 127, 679–95. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, and Massague J (2007). Nature 446, 765–70. [DOI] [PubMed] [Google Scholar]
- Han JJ, Xie de R, Wang LL, Liu YQ, Wu GF, Sun Q, Chen YX, Wei Y, Huang ZQ, and Li HG (2013). Gastroenterol Res Pract 2013, 380193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart K, Landvik NE, Lind H, Skaug V, Haugen A, and Zienolddiny S (2011). Lung Cancer 71, 123–9. [DOI] [PubMed] [Google Scholar]
- Hazari YM, Bashir A, Haq EU, and Fazili KM (2016). Tumour Biol 37, 14381–14390. [DOI] [PubMed] [Google Scholar]
- Hoedt E, Chaoui K, Huvent I, Mariller C, Monsarrat B, Burlet-Schiltz O, and Pierce A (2014). PLoS One 9, e104563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Ding G, Gu C, Zhou J, Kuang M, Ji Y, He Y, Kondo T, and Fan J (2012). Clin Cancer Res 18, 3042–53. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Iyama K, Ishikawa N, Egami H, Nakao M, Sado Y, Ninomiya Y, and Baba H (2006). Am J Pathol 168, 856–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons R, Tsuji PA, Carlson BA, Ouyang P, Yoo MH, Xu XM, Hatfield DL, Gladyshev VN, and Davis CD (2010). Cancer Prev Res (Phila) 3, 630–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasawa S, Yamano Y, Takiguchi Y, Tanzawa H, Tatsumi K, and Uzawa K (2011). Oncol Rep 25, 637–44. [DOI] [PubMed] [Google Scholar]
- Jackson MI, and Combs JGF (2012). “Selenium as a cancer preventive agent.” Springer, New York. [Google Scholar]
- Jakupoglu C, Przemeck GK, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, and Conrad M (2005). Mol Cell Biol 25, 1980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Feng X, Zheng L, Li SL, Ge XY, and Zhang JG (2015). Oncotarget 6, 25506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly E, Greene CM, Carroll TP, McElvaney NG, and O’Neill SJ (2009). J Biol Chem 284, 16891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Oh JH, Park JM, and Chung AS (2007). J Cell Physiol 212, 386–400. [DOI] [PubMed] [Google Scholar]
- Koch S, Volkmar CM, Kolb-Bachofen V, Korth HG, Kirsch M, Horn AH, Sticht H, Pallua N, and Suschek CV (2009). J Mol Med (Berl) 87, 261–72. [DOI] [PubMed] [Google Scholar]
- Kohrle J, Jakob F, Contempre B, and Dumont JE (2005). Endocr Rev 26, 944–84. [DOI] [PubMed] [Google Scholar]
- Kundu N, Zhang S, and Fulton AM (1995). Clin Exp Metastasis 13, 16–22. [DOI] [PubMed] [Google Scholar]
- Labunskyy VM, Yoo MH, Hatfield DL, and Gladyshev VN (2009). Biochemistry 48, 8458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Park KJ, Jang JK, Jeon YH, Ko KY, Kwon JH, Lee SR, and Kim IY (2015). J Biol Chem 290, 29941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Graef GL, Yee JA, and Yan L (2004). J Nutr 134, 1536–40. [DOI] [PubMed] [Google Scholar]
- Liou GY, and Storz P (2010). Free Radic Res 44, 479–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miar A, Hevia D, Munoz-Cimadevilla H, Astudillo A, Velasco J, Sainz RM, and Mayo JC (2015). Free Radic Biol Med 85, 45–55. [DOI] [PubMed] [Google Scholar]
- Min SY, Kim HS, Jung EJ, Jung EJ, Jee CD, and Kim WH (2012). Anticancer Res 32, 3169–75. [PubMed] [Google Scholar]
- Mott JD, and Werb Z (2004). Curr Opin Cell Biol 16, 558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustacich D, and Powis G (2000). Biochem J 346 Pt 1, 1–8. [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Hatakeyama K, Kai M, Nakamura H, Yodoi J, Asada Y, and Chijiiwa K (2012). HPB (Oxford) 14, 573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AR, Fingleton B, Rothenberg ML, and Matrisian LM (2000). J Clin Oncol 18, 1135–49. [DOI] [PubMed] [Google Scholar]
- Nelson KK, Subbaram S, Connor KM, Dasgupta J, Ha XF, Meng TC, Tonks NK, and Melendez JA (2006). J Biol Chem 281, 14100–10. [DOI] [PubMed] [Google Scholar]
- Park JM, Kim A, Oh JH, and Chung AS (2007). Carcinogenesis 28, 837–47. [DOI] [PubMed] [Google Scholar]
- Peng DF, Hu TL, Schneider BG, Chen Z, Xu ZK, and El-Rifai W (2012). PLoS One 7, e46214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekielko-Witkowska A, and Nauman A (2011). J Endocrinol Invest 34, 716–28. [DOI] [PubMed] [Google Scholar]
- Qian Q, Wang Q, Zhan P, Peng L, Wei SZ, Shi Y, and Song Y (2010). Cancer Invest 28, 661–9. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Meng XP, Ramasamy S, Harrison DG, and Galis ZS (1996). J Clin Invest 98, 2572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman MP (2005). Proc Nutr Soc 64, 527–42. [DOI] [PubMed] [Google Scholar]
- Reeves MA, Bellinger FP, and Berry MJ (2010). Antioxid Redox Signal 12, 809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MA, and Hoffmann PR (2009). Cell Mol Life Sci 66, 2457–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romitti M, Wajner SM, Ceolin L, Ferreira CV, Ribeiro RV, Rohenkohl HC, Weber Sde S, Lopez PL, Fuziwara CS, Kimura ET, and Maia AL (2016). Endocr Relat Cancer 23, 135–46. [DOI] [PubMed] [Google Scholar]
- Romitti M, Wajner SM, Zennig N, Goemann IM, Bueno AL, Meyer EL, and Maia AL (2012). Thyroid 22, 897–904. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, and Arnier ESJ (2016). In “Selenium: Its Molecular Biology and Role in Human Health” (D. L. a. S. Hatfield U and Tsuji PA and Gladyshev VN, ed.), Vol. 4, pp. 199–210. Springer, New York. [Google Scholar]
- Schomburg L (2016). In “Selenium: Its Molecular Biology and Role in Human Health” (Hatfield DL, Schweizer U, Tsuji PA, and Gladyshev VN, eds.), pp. 377–388. Springer, New York. [Google Scholar]
- Schweizer U, Schomburg L, and Kohrle J (2016). In “Selenium: Its Molecular Biology and Role in Human Health” (U. a. T. D. L. S. Hatfield PA and Gladyshev VN, ed.), Vol. 4, pp. 261–274. Springer, New York. [Google Scholar]
- Selenius M, Rundlof AK, Olm E, Fernandes AP, and Bjornstedt M (2010). Antioxid Redox Signal 12, 867–80. [DOI] [PubMed] [Google Scholar]
- Shchedrina VA, Zhang Y, Labunskyy VM, Hatfield DL, and Gladyshev VN (2010). Antioxid Redox Signal 12, 839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Arisawa T, Tahara T, Ohkubo M, Yoshioka D, Maruyama N, Fujita H, Kamiya Y, Nakamura M, Nagasaka M, Iwata M, Takahama K, Watanabe M, and Hirata I (2009). BMC Gastroenterol 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Berry JA, Shoher A, Ayers GD, Wei C, and Lucci A (2007). Oncogene 26, 3789–96. [DOI] [PubMed] [Google Scholar]
- Solinas G, Schiarea S, Liguori M, Fabbri M, Pesce S, Zammataro L, Pasqualini F, Nebuloni M, Chiabrando C, Mantovani A, and Allavena P (2010). J Immunol 185, 642–52. [DOI] [PubMed] [Google Scholar]
- Song H, Kim J, Lee HK, Park HJ, Nam J, Park GB, Kim YS, Cho D, and Hur DY (2011). Int Immunopharmacol 11, 2208–13. [DOI] [PubMed] [Google Scholar]
- Stamenkovic I (2000). Semin Cancer Biol 10, 415–33. [DOI] [PubMed] [Google Scholar]
- Storz P (2005). Front Biosci 10, 1881–96. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Pitchakarn P, Ogawa K, Naiki-Ito A, Chewonarin T, Punfa W, Asamoto M, Shirai T, and Takahashi S (2013). Toxicology 311, 115–23. [DOI] [PubMed] [Google Scholar]
- Tong RT, Boucher Y, Kozin SV, Winkler F, Hicklin DJ, and Jain RK (2004). Cancer Res 64, 3731–6. [DOI] [PubMed] [Google Scholar]
- Topkas E, Cai N, Cumming A, Hazar-Rethinam M, Gannon OM, Burgess M, Saunders NA, and Endo-Munoz L (2016). Oncotarget 7, 831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji PA, Carlson BA, Yoo MH, Naranjo-Suarez S, Xu XM, He Y, Asaki E, Seifried HE, Reinhold WC, Davis CD, Gladyshev VN, and Hatfield DL (2015). PLoS One 10, e0124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura M, Baljinnyam E, Feske S, De Lorenzo MS, Xie LH, Feng X, Oda K, Makino A, Fujita T, Yokoyama U, Iwatsubo M, Chen S, Goydos JS, Ishikawa Y, and Iwatsubo K (2014). PLoS One 9, e89292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Slambrouck S, Groux-Degroote S, Krzewinski-Recchi MA, Cazet A, Delannoy P, and Steelant WF (2014). Biosci Rep 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, Nguyen-Wu E, Hashimoto AS, and Hoffmann PR (2011). J Immunol 186, 2127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Gong L, Dong R, Qiao Q, He XL, Chu YK, Du XL, Yang Y, Zang L, Nan J, Lin C, and Lu JG (2009). J Int Med Res 37, 169–74. [DOI] [PubMed] [Google Scholar]
- Weber GF (2013). Cancer Lett 328, 207–11. [DOI] [PubMed] [Google Scholar]
- Wu WS (2006). Cancer Metastasis Rev 25, 695–705. [DOI] [PubMed] [Google Scholar]
- Xia J, Wang H, Huang H, Sun L, Dong S, Huang N, Shi M, Bin J, Liao Y, and Liao W (2016). Cancer Lett 381, 31–40. [DOI] [PubMed] [Google Scholar]
- Xie L, Luo Z, Zhao Z, and Chen T (2016a). J Med Chem. [DOI] [PubMed] [Google Scholar]
- Xie W, Yang M, Chan J, Sun T, Mucci LA, Penney KL, Lee GS, and Kantoff PW (2016b). Prostate 76, 691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima S, Ishii M, Matsushita H, Aoyagi K, Yoshimatsu K, Kaneko H, Yamamoto N, Teramoto T, Yoshida T, Matsumura Y, and Sasaki H (2007). Int J Oncol 31, 1029–37. [PubMed] [Google Scholar]
- Yan L, and Combs GF Jr. (2014). Carcinogenesis 35, 2308–13. [DOI] [PubMed] [Google Scholar]
- Yan L, and DeMars LC (2012). Int J Cancer 131, 1260–6. [DOI] [PubMed] [Google Scholar]
- Yan L, Yee JA, Li D, McGuire MH, and Graef GL (1999). Anticancer Res 19, 1337–42. [PubMed] [Google Scholar]
- Yan L, Yee JA, McGuire MH, and Graef GL (1997). Nutr Cancer 28, 165–9. [DOI] [PubMed] [Google Scholar]
- Yazdi MH, Mahdavi M, Setayesh N, Esfandyar M, and Shahverdi AR (2013). Daru 21, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo MH, Xu XM, Carlson BA, Gladyshev VN, and Hatfield DL (2006). J Biol Chem 281, 13005–8. [DOI] [PubMed] [Google Scholar]
- Yoon SO, Kim MM, and Chung AS (2001). J Biol Chem 276, 20085–92. [DOI] [PubMed] [Google Scholar]
- Yoon SO, Park SJ, Yoon SY, Yun CH, and Chung AS (2002). J Biol Chem 277, 30271–82. [DOI] [PubMed] [Google Scholar]
- Yu C, Tang W, Wang Y, Shen Q, Wang B, Cai C, Meng X, and Zou F (2016). Cancer Lett 376, 268–77. [DOI] [PubMed] [Google Scholar]
- Yu YP, Yu G, Tseng G, Cieply K, Nelson J, Defrances M, Zarnegar R, Michalopoulos G, and Luo JH (2007). Cancer Res 67, 8043–50. [DOI] [PubMed] [Google Scholar]
- Zeng H, Briske-Anderson M, Idso JP, and Hunt CD (2006). J Nutr 136, 1528–32. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zheng X, and Wang X (2015). Am J Cancer Res 5, 2788–98. [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Li J, Li X, Han C, Zhang Y, Zheng L, and Guo M (2015). Curr Protein Pept Sci 16, 316–21. [DOI] [PubMed] [Google Scholar]