Abstract
A 48-year-old woman was infected with a vpr-defective human immunodeficiency virus (HIV)-1 molecular clone. Seroconversion was markedly delayed, and without treatment she had durably suppressed viremia and normal T-cell levels. Neutralizing antibody and CD8+ T-cell immune responses against HIV-1 were unremarkable. Viral sequences confirmed the source but evolved defective nef, suggesting an unknown mechanistic link to vpr. There were subtle qualitative defects in T and B cells. To our knowledge, this is the only case of human infection with a characterized defective HIV-1 molecular clone, which furthermore recapitulated live-attenuated vaccination in macaque models of HIV-1 vaccine research.
Keywords: elite control, HIV-1, vpr
A person who was infected with a vpr-deleted molecular clone of HIV-1 subsequently developed a highly attenuated course of infection.
Rare individuals maintain asymptomatic immune containment of human immunodeficiency virus (HIV)-1 infection, with viremia below the range of detection by commercial polymerase chain reaction (PCR)-based assays and stable blood CD4+ T-cell levels for years or decades without antiretroviral treatment. A major contributing factor is the CD8+ T lymphocyte (CTL) response associated with certain human leukocyte antigen class I (HLA-I) types. However, many persons with viremic containment lack this correlate. Another mechanism is genetic defects in HIV-1, eg, partially defective nef. We describe a unique case of accidental human infection with a molecular clone of HIV-1 containing a genetic deletion of vpr, a gene whose role in pathogenesis is unclear [1], resulting in a highly attenuated clinical course.
METHODS
Subject
Informed consent was provided under an institutional review board-approved protocol, with additional consent for publication.
Human Immunodeficiency Virus-1 Testing
Standard HIV-1 enzyme-linked immunosorbent assay (ELISA), Western blot, and blood T-cell subset enumeration were performed through Clinical Laboratory Improvement Amendments-certified clinical laboratories.
Enzyme-Linked Immune Spot Assays of Human Immunodeficiency Virus-1-Specific CD8+ T Lymphocytes
CD8+ T lymphocyte responses against the whole proteome of HIV-1 were assessed as previously described by gamma interferon enzyme-linked immune spot (ELISpot) assays [2].
Human Immunodeficiency Virus-1-Neutralizing Antibody Assays
Neutralizing antibody responses were performed as previously described using luciferase reporter viruses with pseudotyped HIV-1 envelopes [3].
Human Immunodeficiency Virus-1 Sequencing
Human immunodeficiency virus-1 sequences were recovered from peripheral blood mononuclear cells (PBMCs) as previously described, directly from PBMCs [4] or from ex vivo- expanded CD4+ T-cell deoxyribonucleic acid (DNA) [2].
Flow Cytometric Analyses of Blood Immune Cell Phenotypes
Ficoll-hypaque-purified PBMCs were stained in 100 µL of phosphate-buffered saline containing 1% adult bovine serum for 20 minutes with preconjugated antibody panels at room temperature. A T-cell antibody panel included CD4-PE-Cy7*, CCR7-PerCP*, CD27-PE-Texas Red**, CD45RA-FITC*, HLA-DR-BV650*, CD8-BV605*, CD38-APC-Cy7*, CD3-Pacific Blue**, CD31-PE*, CD25-AF700*, CD28-APC*. A B-cell antibody panel included CD38-PE-Cy7*, CD3-PerCP** and CD16-PerCP** (dump), CD27-PE-Texas Red**, CD24-FITC*, CD10-BV650*, CD20-BV605*, CD19-APC-Cy7**, CD21-Pacific Blue**, IgA-PE** and IgG-PE**, IgD-AF700*, IgM-APC**. An innate immune cell antibody panel included CD123-PE-Cy7*, CD16-PerCP**, CD3-PE-Texas Red** and CD19-PE-Texas Red** (dump), HLA-DR-BV650*, CD80-BV605*, CD56-APC-Cy7*, CD86-Pacific Blue*, CD83-PE*, CD11c-AF700*, CD14-APC**. Each panel also included LIVE/DEAD Fixable Aqua Dead Cell Stain (Invitrogen, Eugene, OR) as a vital dye. All antibodies were obtained from BD BioLegend (San Diego, CA; denoted as *) or Biosciences (San Jose, CA; denoted as **) and were used at saturating conditions. A combination of antimouse immunoglobulin (Ig)G Negative Control Compensation Particles (BD Biosciences) and PBMCs were used to establish compensation settings. Cytometry was performed with a LSR II Flow Cytometer (BD Biosciences), collecting at least 100000 viable cell events. Analysis was done with FlowJo software (Tree Star Inc., Ashland, OR).
Histopathology of Rectal Mucosal T Lymphocytes
Rectal biopsies were obtained by flexible sigmoidoscopy, and T-cell subsets were enumerated by immunohistochemistry as previously described [5].
RESULTS
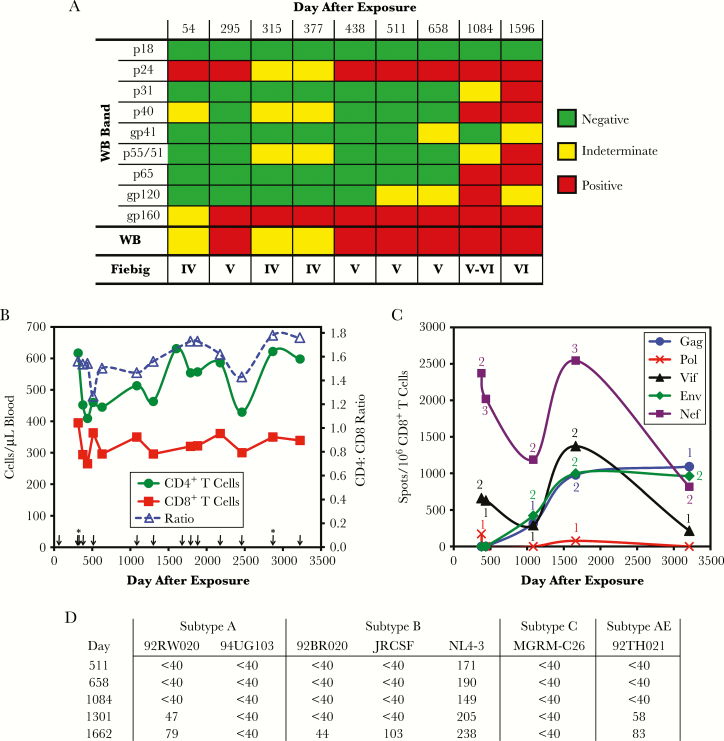
A healthy 48-year-old woman sustained an occupational accidental needlestick exposure to a molecular clone of HIV-1. Multiple past ELISA tests for HIV-1 had been negative, and the most recent was 37 days prior. No antiretroviral medications were administered and she was asymptomatic. Upon initial presentation, ELISA turned positive, but Western blot was indeterminate 54 days postexposure (Figure 1A). All subsequent ELISAs were positive, and Western blot became consistently positive 377–438 days after exposure (Fiebig stage V) [6]. Anti-p31 antibodies were borderline at 1084 days and positive at 1596 days (Fiebig stage VI).
Figure 1.
Immune parameters. (A) Western blot results, including individual bands and overall interpretation are given. The Fiebig stage according to these results is indicated. (B) Peripheral blood T-cell counts are plotted by time after exposure. Arrows indicate plasma human immunodeficiency virus (HIV)-1 ribonucleic acid tests, which all yielded undetectable viremia (<48 copies/mL for tests performed up to day 512, and <20 copies/mL for tests performed after day 512), with the exception of detected viremia below the linear range of quantitation on days 315 and 2868 (indicated by *). (C) Frequencies of CD8+ T-cell responses defined by enzyme-linked immune spot assay using a library of peptides spanning the HIV-1 subtype B sequence whole proteome are plotted. The number labels additionally indicate the number of epitope regions targeted at each time point (recognized 15-mer peptide in isolation or overlap between 2 consecutive overlapping 15-mer peptides). (D) Neutralizing antibody titers are expressed as 1/highest serum dilution to achieve 50% inhibitory concentration of the indicated Env-pseudotyped virions.
Without antiretroviral treatment, monitoring was notable for normal blood T-cell counts and undetectable viremia (Figure 1B). CD4+ and CD8+ T cells varied approximately 500 and 300 cells/mm3, respectively, yielding normal CD4/CD8 ratios. Plasma viremia by commercial reverse transcription-PCR was generally undetectable (<48 or <20 ribonucleic acid copies/mL, except off-scale low detection on days 315 and 2868). Human leukocyte antigen type was absent B*27 or B*57, 2 alleles most commonly associated with “elite control,” and, in fact, demonstrated B*3501, associated with accelerated disease. Thus, over approximately 10 years, this person, without a known protective genotype, lacked the typical findings of untreated HIV-1 infection.
Immunity against HIV-1 was examined in more detail. Blood CD8+ T cell (CTL) screening (Figure 1C and Supplemental Figure S1) revealed responses against only Gag, Vif, Env, and Nef persisting from days 377 to 3211. However, neutralizing antibody responses were low potency against NL4-3 with minimal heterologous activity over years of untreated infection (Figure 1D).
Human immunodeficiency virus-1 sequences from the subject were compared with the exposure virus (Supplemental Figure S2), a modified NL4-3 with replacement of the vpr reading frame with a reporter gene consisting of murine CD24 containing an influenza hemagglutinin antibody epitope (HSA-HA) [7]. Compared with the exposure virus, structural gene sequences gag, pol, and env from day 377 after exposure were highly matched: 1500 of 1503 (99.8%) for gag, 2999 of 3009 (99.7%) for pol, and 2555 of 2565 (99.6%, identical V3 loop) for env, confirming the source of infection (Supplemental Figure S2A–C). None of the nonsynonymous mutations fell within the regions identified to be targeted by CTLs.
Sequencing of vpr again confirmed the source, containing its modifications of vpr start codon alteration and sequence replacement with the HSA-HA reporter gene (Supplemental Figure S2D). Day 377, 1301, and 3211 sequences all had disrupted reporter. Day 377 and 3211 sequences had defective HSA-HA reading frames due to an early stop mutation and altered start codon, respectively. The day 3211 sequence additionally had 5 nonsynonymous mutations and insertion of 2 nucleotides causing a frameshift. The day 1301 sequence was frameshifted.
Sequences of nef (Supplemental Figure S2E) from days 377 and 1301 were examined at the clonal level. Although NL4-3-HSA-HA is fully Nef competent, unexpectedly all nef reading frames were defective. Nine clones from day 377 all had reading frames truncated by early stop mutations, a subsequent frameshift, and deletion from nucleotides 325 to 414. Among 5 clones from day 1301, 3 had early stop mutations immediately followed by a deletion of 221 nucleotides. Another clone had a substitution of nef nucleotides 161 to 260 with 16 nucleotides of unknown source followed by duplication of HIV-1 genome nucleotides 8752 to 8834 from NL4-3-HSA-HA (overlapping the end of nef and beginning of env), and a final clone similarly had a substitution of nef nucleotides 155 to 260 with 23 nucleotides of unknown source followed by duplication of HIV-1 genome nucleotides 8753 to 8834 from NL4-3-HSA-HA. The latter 2 clones also had premature stops at the ends of the inserted duplicated sequences. Thus, none of the identified clonal sequences encoded intact Nef despite its fully intact reading frame in the infecting virus. However, the presence of different portions of the entire nef reading frame across time points indicated that the full nef sequence must have been present originally.
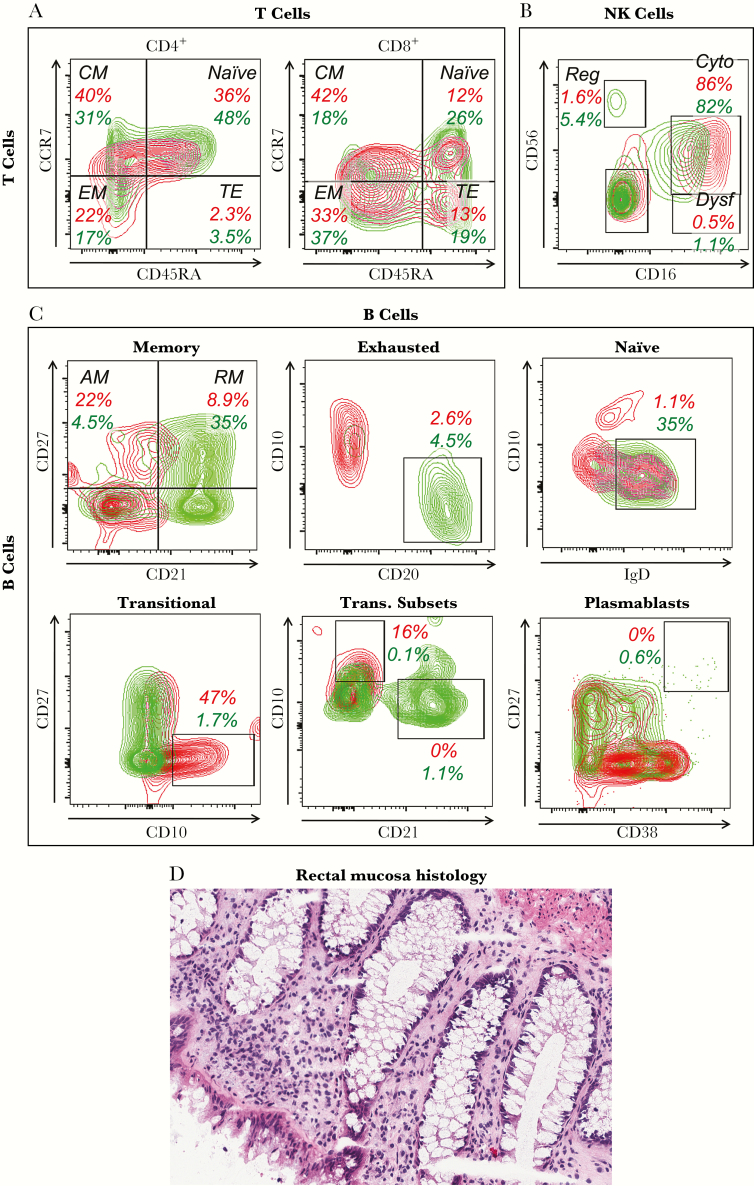
Immunologic consequences were further explored by flow cytometry on T cells (Figure 2A). Unlike typical untreated HIV-1 infection, CD4+ T-cell memory subset percentages were minimally skewed, showing mildly reduced naive and mildly increased effector memory subsets. However, CD8+ T-cell memory subset percentages were more skewed, showing reduced naive, increased central memory, and mildly increased effector memory subsets. CD8+ T cell coexpression of activation markers HLA-DR and CD38 was elevated at 6.9% (data not shown), compared with a typical percentage in uninfected persons of 2.7% ± 1.4% [8].
Figure 2.
Immune cell phenotyping. (A–C) Peripheral blood mononuclear cells obtained on day 3324 after exposure were analyzed by flow cytometry of singlet viable lymphocytes (by forward and side scatter and dye exclusion). The red plots and numbers represent the subject of this study, and the green plots and numbers represent a typical human immunodeficiency virus-1-uninfected control. (A) Differentiation status of blood CD4+ and CD8+ T cells as defined by CD45RA and CCR7 after gating on CD3+ cells. Percentages across all CD4+ and CD8+ T cells are given. (B) Natural killer (NK) cell subsets defined by CD16 and CD56 after gating on CD3−/CD19−/HLA DR−/CD11c− cells. Percentages across all NK cells are given. (C) B-cell subsets are delineated as subpopulations of total B cells defined as CD3−/CD16−/CD19+. Percentages across all B cells are given. Memory B cells are defined using CD21 and CD27. Exhausted B cells are defined by subgating on the CD27−/CD21− B cells from (D), as CD20+/CD10− cells. Naive B cells are defined by subgating on the CD27−/CD21+ B cells from (D), as IgD+/CD10− cells. Transitional-immature B cells are defined as CD10+/CD27−. The transitional B cells in (G) are further subclassified as less immature expressing CD21high/CD10low versus more immature expressing CD21low/CD10high. Plasmablasts are defined as CD27high/CD38high. (D) Rectal mucosal histology (hematoxylin and eosin staining) is shown. Abbreviations: AM, activated memory (CD27+/CD21−); CM, central memory; Cyto, cytotoxic; Dysf, dysfunctional; EM, effector memory; Reg, regulatory; RM, resting memory (CD27+/CD21+); TE, terminal effector.
In contrast, the profile of blood NK cells appeared relatively unperturbed. Functional subsets (Figure 2B) showed relatively normal proportions of cytotoxic and dysfunctional cells, lacking the large expansion of dysfunctional NK cells seen in untreated infection.
B-cell subset examination (Figure 2C) revealed mixed perturbations among those documented in HIV-1 infection. As in prior studies, there were increased activated and decreased resting memory B cells, reduced naive B cells, and increased immature transitional B cells. However, there were not typical changes of increased B-cell exhaustion or plasmablasts usually seen in untreated HIV-1 infection.
Because CD4+ T-cell depletion is far more pronounced in the gut mucosal compartment than blood in typical HIV-1 infection, immunohistochemistry of rectal biopsies (Figure 2D) was examined. The histology was grossly normal, and immunohistochemistry for CD4+ and CD8+ T cells (data not shown ) revealed densities of 20597 ± 1948 and 26939 ± 7237 cells/mm3 tissue, respectively, comparable to previously observed healthy control normal ranges of 21557 ± 332 and 17090 ± 1206 cells/mm3, respectively [5]. Thus, the typical severe CD4+ T-cell deletion in the gut compartment in untreated infection was absent in this person.
Discussion
To our knowledge, this is the only example of a person infected by a defined molecular HIV-1 clone. This virus is a CXCR4-tropic chimeric laboratory strain generated from 2 primary isolate clones [9], further modified to have a reporter by deletion and substitution of the Vpr reading frame but otherwise complete [7]. A version of this virus with the same Vpr replacement with the HSA reporter without the HA epitope has normal pathogenicity in an in vivo-humanized mouse model [10]. Unmodified NL4-3 is a prototypical fully competent strain of HIV-1; it is interesting that this Vpr-deleted version yielded such attenuated infection, and delayed HIV-1 seroresponse demonstrated by reaching Fiebig stage VI between days 1084 to 1596, compared with ~100 days typically [6]. Moreover, the virus did not appear to evolve CCR5 tropism, despite the association of CXCR4-tropism with greater pathogenicity.
It is possible that host factors determined this outcome, but it seems less likely. “Elite control” of HIV-1 infection is rare, and she had no typical protective HLA type, although this is true for approximately 40% of “controllers.” The CTL responses were not anti-Gag dominant (associated with better control), although ELISpot is not a reliable correlate. In addition, immune-driven viral CTL epitope sequence evolution was not observed, but archived proviral sequences could underrepresent such evolution. Reporter protein sequences were ablated in vivo, and thus antireporter immunity is unlikely to have played a role. Finally, broadly neutralizing antibody activity was not observed, and neutralizing activity against autologous NL4-3 was modest, indicating insignificant contribution by humoral immunity.
It seems likely that attenuation was due to virologic factor(s), with particularly deletion of vpr. Vpr has a myriad of reported in vitro cellular effects, whose contributions to pathogenesis remain unclear [1]. However, it is important in vivo; deletion of vpr and vpx in simian immunodeficiency virus (SIV) results in a highly attenuated infection of macaques. Also in macaques, there is strong selective pressure to maintain the vpr open reading frame in vivo, also noted anecdotally in a person who was accidentally infected with the HIV-1 IIIB swarm containing mostly frame-shifted vpr, in whom intact Vpr was selected [11].
Viral Nef was functionally deleted in vivo, although intact in the infecting virus. Despite large deletions in nef, they were in non-overlapping regions at different times, and thus the entire gene must have been present at the time of infection (and archived). Similar to vpr, macaque studies demonstrated strong selective pressure for maintenance of SIV nef. Therefore, it is very unusual that Nef was not maintained in this person. When nef is fully deleted from SIV, infection of macaques is markedly attenuated and they are protected from subsequent challenge with wild-type SIV [12], which has been a model for a successful vaccine. Data in humans also suggests that defective Nef attenuates infection [13]; thus, the loss of Nef likely contributed to the markedly attenuated course of infection in this person.
The relative roles of Vpr and Nef deletion in this case are unclear. It appears that deletion of vpr may have resulted in a disadvantage where maintaining Nef as an immune target outweighed its advantages in viral replication, immune evasion, and other functions. There is no obvious mechanism, because Nef functions have generally been confirmed independently of Vpr in studies of Nef alone or within vpr-deleted reporter viruses [14]. One potential connection is that virion-associated Vpr may help drive earlier translation of viral proteins including Nef from unintegrated proviral DNA [15], although how this would shift the benefit of Nef expression is unknown.
Conclusions
In conclusion, this is the only known case of human infection with a live-attenuated molecular clone of HIV-1. The outcome recapitulates “elite control” and live-attenuated SIV infection of macaques, considered an experimental standard for defining protection in HIV-1 vaccine research, and reveals a yet-undefined mechanistic link of Vpr to Nef in the pathogenesis of infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Abena Kwaa and Dr. Rebecca Veenhuis for technical assistance. We are grateful to the subject of the study for providing samples and permission to report her case.
Financial support. This work was funded by an AIDS Healthcare Foundation research grant (to O. O. Y.), National Institutes of Health (NIH) grants AI043203 and AI051970 (to O. O. Y.), and NIH grants P30 AI094189 and AI120024 (to J. N. B.). Funding was also provided by the UCLA Center for AIDS Research (NIH AI028697), James B. Pendleton Trust, and the McCarthy Foundation. Recombinant human interleukin-2 was provided by the NIH AIDS Reagent Repository.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Guenzel CA, Hérate C, Benichou S. HIV-1 Vpr-a still “enigmatic multitasker”. Front Microbiol 2014; 5:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balamurugan A, Lewis MJ, Kitchen CM, et al. . Primary human immunodeficiency virus type 1 (HIV-1) infection during HIV-1 Gag vaccination. J Virol 2008; 82:2784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simek MD, Rida W, Priddy FH, et al. . Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 2009; 83:7337–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blankson JN, Bailey JR, Thayil S, et al. . Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol 2007; 81:2508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Preza GC, Yang OO, Elliott J, Anton PA, Ochoa MT. T lymphocyte density and distribution in human colorectal mucosa, and inefficiency of current cell isolation protocols. PLoS One 2015; 10:e0122723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fiebig EW, Wright DJ, Rawal BD, et al. . Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 2003; 17:1871–9. [DOI] [PubMed] [Google Scholar]
- 7. Ali A, Yang OO. A novel small reporter gene and HIV-1 fitness assay. J Virol Methods 2006; 133:41–7. [DOI] [PubMed] [Google Scholar]
- 8. Yang OO, Boscardin WJ, Matud J, et al. . Immunologic profile of highly exposed yet HIV type 1-seronegative men. AIDS Res Hum Retroviruses 2002; 18:1051–65. [DOI] [PubMed] [Google Scholar]
- 9. Adachi A, Gendelman HE, Koenig S, et al. . Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 1986; 59:284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jamieson BD, Zack JA. In vivo pathogenesis of a human immunodeficiency virus type 1 reporter virus. J Virol 1998; 72:6520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goh WC, Rogel ME, Kinsey CM, et al. . HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med 1998; 4:65–71. [DOI] [PubMed] [Google Scholar]
- 12. Daniel MD, Kirchhoff F, Czajak SC, Sehgal PK, Desrosiers RC. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 1992; 258:1938–41. [DOI] [PubMed] [Google Scholar]
- 13. Dyer WB, Geczy AF, Kent SJ, et al. . Lymphoproliferative immune function in the Sydney Blood Bank Cohort, infected with natural nef/long terminal repeat mutants, and in other long-term survivors of transfusion-acquired HIV-1 infection. AIDS 1997; 11:1565–74. [DOI] [PubMed] [Google Scholar]
- 14. Ali A, Jamieson BD, Yang OO. Half-genome human immunodeficiency virus type 1 constructs for rapid production of reporter viruses. J Virol Methods 2003; 110:137–42. [DOI] [PubMed] [Google Scholar]
- 15. Poon B, Chen IS. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J Virol 2003; 77:3962–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.