Synthetic biologists seek to develop novel control mechanisms that will enable living cells to be deployed for a wide range of biotechnological and biomedical applications. In many cases, control is established by synthetic biological circuits that take existing biological parts and repurpose them toward new functions in new cellular hosts. Starting from the earliest demonstrations in bacteria more than a decade ago, synthetic biological circuits have been employed for a range of artificial cellular functions, enabling living cells to oscillate, self-destruct, compute logic expressions, and store information. However, as the field has progressed, it has become increasingly difficult to scale up the complexity of existing circuits so that they can take on more advanced functions. Most existing circuits rely heavily on protein regulators that often exhibit idiosyncratic behavior that hampers their integration into larger circuits. Unlike electronic circuit elements that are uniquely connected and buffered, biological components also interact with one another in a complex cellular environment and suffer from extensive crosstalk between components if they are not carefully selected or designed. These challenges point to the pressing need for scalable strategies to increase the complexity of synthetic circuits together with large libraries of compatible parts that can integrate seamlessly into these circuits.
Biological parts crafted from RNA represent a promising source of circuit components. Fortunately, several mechanisms have been elucidated for natural noncoding RNA molecules that play important and diverse roles in regulating cellular function, including splicing and editing RNA, catalyzing biochemical reactions, and regulating gene expression. Such natural mechanisms provide a basis for the development of synthetic RNA-based circuit components. These regulatory I elements can take advantage of predictable Watson–Crick base pairing to control cellular behavior and can harness sophisticated software tools that have been developed for modeling RNA–RNA interactions. Moreover, the sequence diversity and programmability of RNA enable the structural specification of a single RNA part to serve as a blueprint to generate an essentially limitless supply of circuit components having different sequences. In 2004, Isaacs and colleagues demonstrated one of the pioneering works for engineered riboregulators in Escherichia coli.1 These RNA-based regulatory elements were inspired by natural small RNA regulators found in bacteria and enabled protein expression to be activated through the binding of a trans-activating trigger RNA, unlike other works utilizing antisense RNA or ribozymes. This work was soon followed by an assortment of different RNA-based biological parts, including new riboregulators to modulate transcription or translation in response to trigger RNAs, and new engineered riboswitches to convert signals from small molecule or protein ligands into protein outputs. These nature-inspired RNA components, however, could not match the regulatory dynamic range provided by protein components or, because of sequence constraints or design limitations, failed to provide the large libraries of interchangeable elements that are ideal for biological circuit construction.
This changed in 2014 when we reported a new class of riboregulators called toehold switches.2 Unlike previous systems, toehold switches were designed completely from scratch and were devised specifically to eliminate nearly all sequence constraints on the cognate RNA to be detected (Figure 1). They work by concealing the ribosomal binding site (RBS) and start codon required for translation initiation within a designed hairpin, leaving a small single-stranded “toehold” region exposed. This toehold serves as the seed region to initiate interaction with a cognate trigger RNA molecule, which ultimately unwinds the hairpin and enables translation of a downstream open reading frame. The absence of sequence constraints for the toehold switches enabled us to validate dozens of functional RNA-based parts and ensured that multiple toehold switches could operate simultaneously in the cell. Unexpectedly, we found that the toehold switches also provided dynamic range comparable to those of widely used protein-based regulators, increasing the level of gene expression by several hundred-fold after activation by trigger RNAs.
Figure 1.
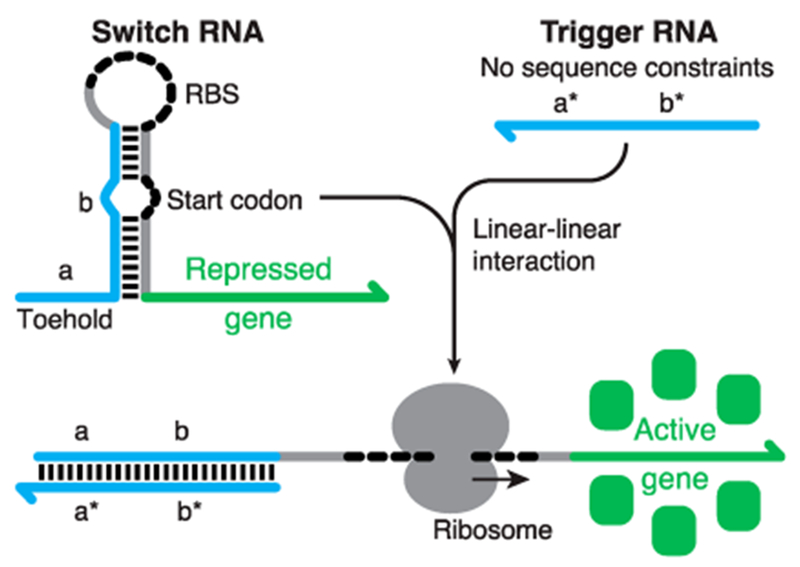
Design of the toehold switch riboregulator. The switch RNA is a functional mRNA whose translation is repressed by sequestering the RBS and start codon within a hairpin secondary structure. A cognate trigger RNA, which can adopt virtually any sequence in domains a* and b*, can initiate binding to the switch RNA through the single-stranded toehold domain and unwind the switch hairpin through hybridization with complementary domains a and b. The resulting trigger–switch complex has an exposed RBS and start codon enabling translation of the output gene.
Despite having a large library of high-performance toehold switch parts, we still lacked a scalable strategy for using these components in biological circuits. Our solution was to generate RNA-based computing or ribocomputing devices by converting all the major circuit operations into RNA–RNA interactions that could be readily detected by toehold switches3 (Figure 2). For instance, to evaluate an AND logic expression, which requires all inputs present to be evaluated as true, we designed input RNA molecules that could hybridize and form a trigger sequence capable of activating a cognate toehold switch. For NOT logic, we employed deactivating transcripts complementary to trigger RNAs to prevent them from binding to toehold switches. These interactions between input RNAs were assessed using an extended, computationally designed RNA molecule termed a gate RNA. The gate RNA featured multiple toehold switch hairpins upstream of the output gene and encoded the information-processing tasks of the circuit. Ribocomputing devices avoid many of the pitfalls of previous systems. Because computations are performed in a single layer within a single gate transcript, timing delays do not occur and both circuit size and diffusive signal losses are reduced dramatically. Furthermore, we can exploit the programmability of RNA–RNA interactions to generate new circuit elements avoiding part limitations that have hampered previous approaches. Harnessing the capabilities described above, we demonstrated sophisticated multi-input computations within E. coli, including six-input OR and four-input AND. Furthermore, we successfully scaled up circuit complexity by computing the 12-input expression (A1 AND A2 AND NOT A1*) OR (B1 AND B2 AND NOT B2*) OR (C1 AND C2) OR (D1 AND D2) OR (E1 AND E2), which represents one of the most complex logic expressions evaluated to date in vivo.
Figure 2.

Ribocomputing device architecture provides a scalable method for synthetic biological circuit construction using predictable and programmable RNA–RNA interactions. Input RNAs to the circuit interact with one another in a cooperative or inhibitory manner to perform AND or NOT logic, respectively. A gate RNA, which contains multiple toehold switch sensor modules upstream of the output gene, detects the input RNAs and converts their signals into the translation of an output protein.
Our work on ribocomputing devices has several important implications. First, the gate RNA architecture demonstrates that co-localization of sensors and output modules can dramatically improve molecular computation. This strategy can be employed in vitro for nucleic acid nanotechnology and inexpensive paper-based diagnostics. Second, ribocomputing devices require only Watson–Crick base pairing to function. Consequently, similar circuits can be employed broadly in other bacterial hosts and potentially even eukaryotes. Third, ribocomputing circuits have very favorable scalability, particularly in parallel implementations, enabling them to evaluate much more challenging computations going forward. Despite their advantages, ribocomputing devices have limitations in their current form. The production of input RNAs relies on other transcriptional regulatory elements and typically requires strong promoters such as the T7 RNA polymerase promoter to achieve high intracellular input RNA concentrations. For broader applicability, incorporation of ribocomputing devices into sophisticated layered circuits that seamlessly integrate with protein-based circuits is the next challenge. Overall, the development of ribocomputing devices along with parallel advances in synthetic biological circuit design,4 RNA-based transcriptional regulation,5 and CRISPR-based technologies provide us with a wealth of new tools with which to craft biological circuits. For instance, integration of multi-input mRNA-sensing ribocomputing circuits with the SNP selectivity of CRISPR technology in paper-based synthetic biology systems could improve the robustness and reliability of these diagnostic tools when they are deployed in the field. In the coming years, we expect the next-generation biological circuits that take advantage of these tools will enable much more sophisticated cellular and extracellular functions to be achieved and help realize the full potential of synthetic biology.
Acknowledgments
Funding
This work was supported by National Institutes of Health (NIH) Director’s New Innovator and Transformative Research Awards (1DP2OD007292 and 1R01EB018659), an ONR Young Investigator Program Award (N000141110914), NIH Grants N000141010827, N000141310593, N000141410610, and N000141612410, National Science Foundation (NSF) CAREER and Expedition in Computing Awards (CCF1054898 and CCF1317291), NSF Grants CCF1162459 and ERASynBio 1540214, and Wyss Institute Molecular Robotics Initiative support to P.Y. and an NIH Director’s New Innovator Award (1DP2GM126892), a DARPA Young Faculty Award (D17AP00026), an Arizona Biomedical Research Commission New Investigator Award (ADHS16-162400), an Alfred P. Sloan Research Fellowship (FG-2017-9108), Gates Foundation funds (OPP1160667), and Arizona State University funds to A.A.G. J.K. acknowledges a Wyss Institute Director’s Cross-Platform Fellowship.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Isaacs FJ, Dwyer DJ, Ding CM, Pervouchine DD, Cantor CR, and Collins JJ (2004) Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol 22, 841–847. [DOI] [PubMed] [Google Scholar]
- (2).Green AA, Silver PA, Collins JJ, and Yin P (2014) Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell 159, 925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Green AA, Kim J, Ma D, Silver PA, Collins JJ, and Yin P (2017) Complex cellular logic computation using ribocomputing devices. Nature 548, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Nielsen AAK, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, and Voigt CA (2016) Genetic circuit design automation. Science 352, aac7341. [DOI] [PubMed] [Google Scholar]
- (5).Chappell J, Takahashi MK, and Lucks JB (2015) Creating small transcription activating RNAs. Nat. Chem. Biol 11, 214–220. [DOI] [PubMed] [Google Scholar]


