Abstract
One promise of synthetic biology is to provide solutions for biomedical and industrial problems by rational design of added functionality in living systems. Microbes are at the forefront of this biological engineering endeavor due to their general ease of handling and their relevance in many potential applications from fermentation to therapeutics. In recent years, the field has witnessed an explosion of novel regulatory tools, from synthetic orthogonal transcription factors to posttranslational mechanisms for increased control over the behavior of synthetic circuits. Tool development has been paralleled by the discovery of principles that enable increased modularity and the management of host-circuit interactions. Engineered cell-to-cell communication bridges the scales from intracellular to population-level coordination. These developments facilitate the translation of more than a decade of circuit design into applications.
Graphical abstract
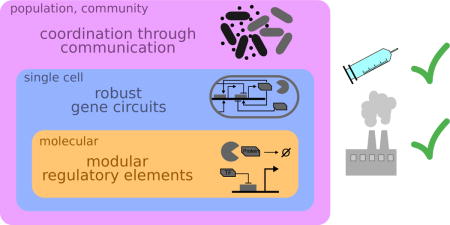
Introduction
Synthetic biology emerged from the demonstration that gene regulatory elements can be combined to resemble classical electrical engineering circuits, resulting in synthetic gene circuits that act as switches and clocks [1,2]. Since its inception, the field has undergone immense expansion and diversification, driven by the construction of more complex networks and by requirements for specific applications. These efforts span all layers of microbiological organization, from molecular regulatory mechanisms to single cells, multi-cellular populations and multi-species consortia. Although these different layers are inherently coupled, the development of distinct tools at each scale contributes to the expanding scope, modularity, orthogonality and interoperability of synthetic constructs. We first review current progress in creating new regulatory elements, particularly those which are complementary to existing tools or provide more fine-grained control. Subsequently, we identify emerging design principles and tools that ease the construction of robust and efficient circuits at the single-cell and the population level, while enabling novel modes of interaction within engineered multi-strain communities. Concurrently, we describe the applications that demonstrate the potential of these strategies for addressing biotechnological and medical problems. We limit our focus to bacteria and yeast as the main model systems for prokaryotic and eukaryotic microbes, respectively (for reviews on the significant progress of mammalian synthetic biology thanks to novel technologies such as CRISPR, see refs, [3,4]).
New regulatory tools
At the core of synthetic biology and its applications lies the ability to control gene expression in response to external stimuli or changes in the intracellular environment. Traditionally, such regulation has been achieved using a small core set of promoters and transcription factors (TFs) such as LacI, TetR, λ cI, luxR and araC, which act as activators or repressors of gene expression. Through interaction with these proteins, expression can be externally modulated by inducers such as chemicals, temperature, and light (Figure 1a, left section). Recently, the flexibility of light induction was considerably improved [5,6], and electric current has been added as another mode of environmental regulation [7]. However, even if accurately characterized [8], the limited number of TFs imposes constraints on the complexity of engineered networks. In yeast, the choice of well-characterized parts is even more limited, although this deficiency is being addressed by creating genetic part libraries and rapid assembly procedures [9,10]. Due to these limitations, there is a constant search for additional, ideally orthogonal, TF-promoter pairs [11], and ways to achieve more predictable expression levels [12] and flexible control at all stages of gene expression (Figure 1a). Since here we focus on novel additions to the regulatory toolbox, we refer the reader to Bradley et al. [13] for an in-depth review.
Figure 1. Versatile regulatory tools for robust gene circuits.
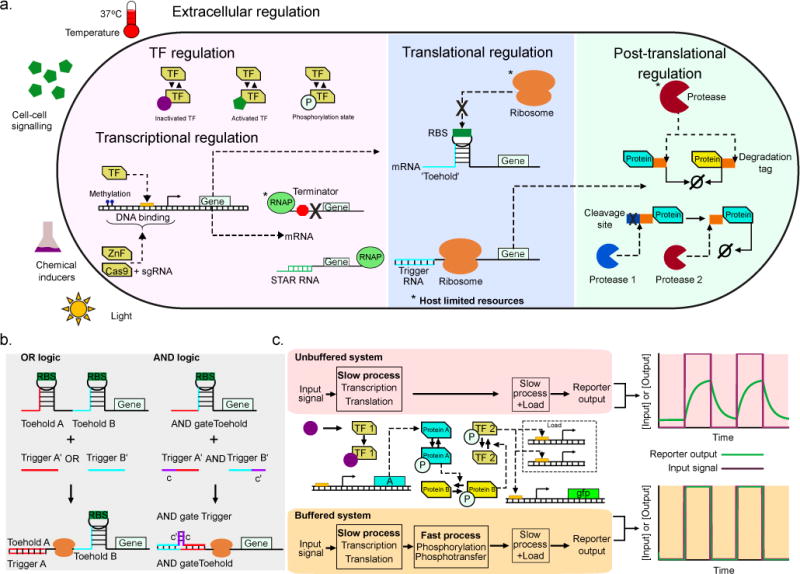
(a) Examples of regulation at various levels of expression. Examples of host resources potentially affected by synthetic constructs (among others) are marked by “*”. Left: Transcriptional regulation. Traditional transcription factors (TFs), modulated by the chemical and physical environment (including light [5,6]) and phosphorylation state; synthetic TFs such as zinc fingers (ZnF) and dCas9 guided by a single guide RNA (sgRNA) [18]; DNA methylation state [22]; and small transcription activating RNAs (STARs) based on the conditional formation of terminator hairpins [26]. Center: Translational regulation through in silico designed, de-novo post-transcriptional “toehold switches” [27]. A specific secondary structure usually prevents access of ribosomes to the ribosome binding site (RBS), which is only exposed by Watson-Crick base pairing of a trigger RNA initiating translation. Right: Post-translational regulation through proteases. Proteins tagged for degradation by host or orthogonal proteases (top) [33,34]; engineered cleavage sites for host-orthogonal proteases where other tags can be conditionally exposed as well (bottom) [35]. (b) Logic gates based on computationally designed, purely post-transcriptional regulators (toehold switches, see panel a) [39]. Two separate switches for the same gene yield OR gate; split trigger sequence between two trigger RNAs yields AND gate. (c) Load driver enabling the rapid load-independent response [50]. In the unbuffered system (top row), a large number of downstream promoters leads to titration of TFs, slowing and reducing the response. In the buffered system (bottom row), a fast intermediate load driver module responding the original TF 1 eventually leads to phosphorylation of TF 2, which is necessary for activating downstream promoters. Due to a large pool of TF 2 and fast load driver kinetics, rapid response of the system is restored, independent of load.
Even simple changes to existing TF-promoter systems, such as the adjustment of receptor protein concentration, can provide accurate control over the dynamic range and sensitivity of inducible expression [14]. More advanced studies, still based on natural TF-promoter pairs, have evolved elements from these systems and combined them in new ways to enable orthogonal control of expression using multi-input bidirectional promoters [15]. Other researchers constructed combinatorial synthetic promoters for different transcriptional repressors from individual core promoters and operators with highly predictable characteristics [16]. In contrast to regulatory elements that are based on naturally occurring TFs, purely synthetic TFs allow transcriptional regulation at almost arbitrary DNA sequences by repurposing of genome-editing tools that have been deprived of their editing function but retain flexible DNA binding specificity. These include transcription activator-like effectors and, more recently, nuclease-deficient CRISPR-Cas9 activated by guide RNAs [17]. Recent advances include methods for the automated design of large sets of transcriptional repressors and synthetic TF-promoter pairs that are orthogonal to the host genome [18,19] and the creation of suitable RNA scaffolds in the guide RNA which recruit effector proteins to a locus of interest [20].
TFs are not the only regulators of transcription. For example, major factors in eukaryotes are epigenetic modifications and chromatin structure, which offer distinct features such as the persistence of an encoded state across generations (reviewed in detail in [21]). Notably, DNA methylation in prokaryotes can be used in a similar way to yield persistent memory [22], apart from the modification of DNA itself ([23] and references therein). A previously overlooked mode of regulation uses transcription from an anti-sense promoter behind a unidirectional transcriptional terminator to achieve > 30-fold repression [24]. From an engineering standpoint, this mechanism could prove valuable as it is independent of the designated promoter for an operon (and its interaction with potential TFs) and can therefore be combined with existing regulatory motifs.
RNA itself has (re-)gained significance as an engineering tool for synthetic regulation in recent years [25]. Multiple examples demonstrate the construction of orthogonal de-novo RNA regulators of both transcription [26] and translation [27,28] (Figure 1a, center section) which are activated by specific trigger RNAs or small molecules. Besides the utility of the regulators themselves, the new (computational) methods used to construct them serve to overcome unpredictability and low fold changes of previous riboswitches and riboregulators, and can easily be adapted for arbitrary, application-specific trigger sequences, for example in a diagnostic tool to detect viral RNA [29].
Post-translational regulation represents an additional and orthogonal layer of control on top of existing regulatory interactions, acting on timescales substantially faster than transcriptional regulation (Figure 1a, right section) [30]. Post-translational modifications are at the heart of most cell signaling cascades which usually begin with ligand-repressor interactions at cell membrane and continue through multiple enzymatic processes (e.g. phosphorylation, dephosphorylation) and other protein-protein interactions (e.g. dimerization, active transport), eventually causing specific transcriptional changes. Synthetic biologists are beginning to harness these processes to either fine-tune existing signaling cascades or to develop entirely new ones [31]. For example, Ganesh et al successfully created a synthetic two-component fumarate screening system by fusing the sensor histidine kinase of DcuS with the cytoplasmic catalytic domain of EnvZ in E. coli [32]. Another class of synthetic post-translational regulation exploits active protein degradation with endogenous and orthogonal proteases [33,34]. By creating designated cleavage sites, Fernandez-Rodriguez & Voigt demonstrated that Potyvirus proteases are orthogonal to E. coli host machinery and can be used to activate a protein’s repressor function, and to trigger or prevent protein degradation [35].
Robust gene circuits
The promise of synthetic biology is that the versatile regulatory arsenal developed over the past decade can be used to forward-engineer networks which process information from the environment and produce predictable behavior according to design specifications. While the foundational circuits of synthetic biology implemented fundamental dynamical behaviors, viz. oscillations and switching (bistability), recently, larger-scale systems have been created, including post-translationally coupled genetic oscillators [33], multistable networks [36] and networks of logic gates [37–39]. Detailed characterization of existing transcriptional regulatory parts has enabled the automated assembly of components with well-defined interfaces to achieve a desired digital output function [37], while de-novo regulatory elements have been used to implement recombinase-based comparator circuits which can convert continuous input signals to discrete outputs [38], and complex logic circuits at the post-transcriptional level [39] (Figure 1b). These studies demonstrate how the multitude of regulatory tools enables sophisticated higher-order gene circuits.
As the complexity of regulatory networks increases and the field moves towards applications, a question which is of utmost importance in dynamical systems and control theory is how a desired physiological state can be maintained to ensure robust operation of a synthetic system [40]. One possibility is to implement such regulation in silico by electronically controlling chemical release or light stimulation to bring a readout of expression to a desired level [41–43], circumventing potential intricacies of engineering the underlying biological system. However, many applications would benefit from a biological system that can perform a given task without external intervention and automatically compensate for intracellular and environmental perturbations. A recent example shows that steady production of a toxic compound can be ensured by on-demand protection of the cells via an active feedback [44]. A general framework for host and environment-independent behavior of arbitrary gene circuits was devised by Kushwaha and colleagues, who created a universal bacterial expression resource (UBER) based on host-orthogonal T7 polymerase whose capacity and toxicity is controlled by regulatory feedback loops [45]. The authors demonstrate that the same genetic construct, in this case a synthetic metabolic pathway, can function reliably across species using UBER.
The minimization of interactions with the host and between otherwise unrelated synthetic constructs in the same cell also represents a general desire to modularize synthetic components [46]. Synthetic circuits are particularly prone to distorting the host resource allocation as they are often implemented on multi-copy plasmids—implying high expression levels and resource demand—and converting them to genomically integrated single-copy circuits often requires careful redesign to maintain function [47]. An in-vivo sensor was constructed by Ceroni et al. to assay this burden of synthetic circuits, making it possible to compare alternative designs and choose a circuit that minimizes impact [48]. While it might seem natural to optimize the synthetic circuit itself to avoid undue resource demand, alternatively, cellular resources can be diverted towards the synthetic circuit: Venturelli and colleagues achieved this by artificially lowering the RNA stability of all transcripts except those of the target genes, leading to increased expression or metabolite yield [49]. The intentional alteration of resource allocation might thus be an important future consideration for increased robustness.
It is worth noting that, even if sufficiently independent of the host, load effects within a synthetic circuit can also lead to distortions that impede modular design. Taking as an example the undesired titration of repressor when additional downstream “load” (in the form of regulated promoters) is added to a genetic circuit, Mishra et al. showed how an intermediate “load driver” module can be used to mitigate retroactive effects and ensure a swift system response independent of the downstream load [50] (Figure 1c).
Several studies have aimed at deriving general rules that predict how resource demands shape the behavior of synthetic gene circuits and can be managed to avoid undesired cross-talk between components [51–53]. A whole-cell view was taken by Liao et al., who developed a mathematical framework to integrate the biochemical dynamics of synthetic circuits with an explicit model of host physiology [54]. Such predictive models for the bi-directional coupling between synthetic constructs and the host [55] will be essential when seeking to translate sophisticated regulatory networks from ideal lab conditions into applications in natural, fluctuating environments. Nevertheless, proof-of-principle studies have shown that simple synthetic circuits in bacteria can already function robustly in tissue microenvironments to produce a urine marker for liver cancer metastases [56] or record an environmental signal in the mammalian gut [57].
Engineering microbial populations and consortia
While the current library of regulatory components provides powerful tools for rational circuit engineering, these synthetic networks typically act at the level of a single cell. However, microbes naturally live in populations and communities, and hence, there is an increasing effort to exploit and coordinate multicellular behavior. Even without intercellular communication, populations can provide distinct advantages over single cells, simply by virtue of the vast number of their members. At the most basic level, population self-averaging automatically reduces stochastic noise and inherent cell-cell variability in collective response to external stimuli. Furthermore, large amounts of data—such as a sequence of images—can be stored among the individuals of a population using the CRISPR-related Cas1-Cas2 integrase and retrieved later via sequencing [58]. The macroscopic spatial extent of bacterial colonies has been exploited to produce differential behavior in the presence of gradients of environmental inducers, leading to formation of robust patterns of expression and growth [59,60].
An additional layer of self-organization and emergent behavior can be tapped into through the use of cell-to-cell communication, where diffusible signals produced in one cell can alter gene expression in other cells in the population by binding to a receptor protein transcription factor [61]. In many species, natural quorum-sensing (QS) systems feature a positive feedback loop, leading to threshold-like induction of downstream pathways, such as virulence and biofilm formation, at critical population densities [62]. In the context of engineering, they represent a type of analog-to-digital converter with particularly low intrinsic noise levels, as fast diffusion of the QS molecule causes averaging across the population. QS oscillators—requiring steep, nonlinear activation characteristics—have leveraged this robust behavior to implement their core positive feedback loops [33,63–65].
Recent studies have quantitatively characterized and tuned the molecular components of QS systems [66,67], creating a versatile toolbox for cell-to-cell communication with and without crosstalk between different QS signals and receiver modules (Figure 2a). It has been shown that the response characteristics can be further tuned by additional synthetic circuitry around a core QS system [68]. In yeast, the plant hormone auxin combined with synthetic CRISPR-based transcription factors [69] and mating pheromones [70,71] have been used as QS analogs. QS signals effectively create regulatory networks spanning multiple cells serving as a bridge between the intracellular and the population level. Most recently, a QS oscillator used this idea explicitly by synchronously modulating the copy number of a plasmid in the population as a way to globally regulate other downstream intracellular circuits [65]. QS-mediated self-organization has also been used to create scale-invariant ring patterns through collective space sensing [72] and to assemble organic-inorganic microstructured materials made of synthetically expressed curli fibrils that bind to gold nanoparticles [73].
Figure 2. Examples of population level engineering via cell-to-cell communication.
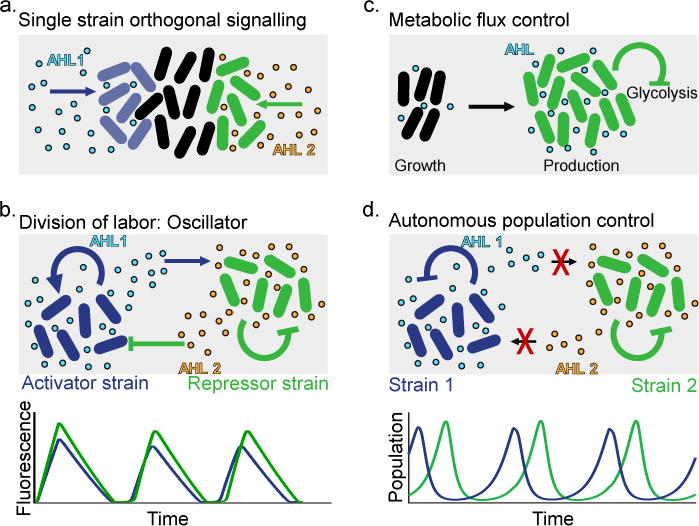
(a) Orthogonal spatial response to two different quorum sensing (QS) signals in a double-receiver strain, achieved by optimizing promoter sequence and receiver protein levels [67] (sender cells for AHL 1 and AHL 2 not shown). (b) Division of labor between two strains, each implementing part of a classical oscillator circuit [63]. The activator strain contains a positive feedback loop involving AHL 1, whereas degradation of QS molecules is triggered in both strains by AHL 2, which is produced by the repressor strain in response to AHL 1. These positive and negative feedback loops spanning both strains lead to emergent oscillations. (c) QS circuit suppressing glycolysis at a critical cell density [79]. Trade-off optimization between biomass production, overflow metabolism and flux through a heterologous pathway maximizes product yield. (d) Population control with tunable dynamics via QS-triggered lysis [64,81]. Self-limitation of individual strains stabilizes co-cultures (oscillatory regime shown), with no communication between strains required.
The ability of QS to link gene circuits across the confines of single cells also makes it possible to split a large-scale network or pathway into components implemented in distinct subpopulations, which can lead to increased modularity, reduced burden on individual cells and the separation of chemically incompatible functions [74]. A prototypical example for applying this division-of-labor strategy to a classical oscillator circuit was implemented by Chen and colleagues using two QS signals in a co-culture of separate “activator” and “repressor” strains which showed emergent oscillations [63] (Figure 2b). Similarly, two yeast strains repressing each other’s pheromone production yielded a bistable system upon co-culture, in analogy to the classical toggle switch design [71]. If the functional separation is accompanied by a spatial segregation, diffusible signals can also broadcast information from sender to receiver cells over larger distances [67,75] and be engineered to induce the formation of stripe and spot patterns in expression and growth [60].
Division of labor can have similar advantages for commercially relevant fermentation processes, whose design is increasingly inspired by synthetic biology approaches [76]. When metabolic pathways are difficult to reconstitute in single cells, compartmentalization into subtasks allows the choice of optimal host strains for each task. For example, Zhou and colleagues used E. coli and S. cerevisiae to synergistically produce a paclitaxel precursor [77]. Such strategies can also significantly increase production yields by avoiding the overproduction of deleterious intermediates [78]. While these systems are inherently pathway-specific, other pathway-independent QS circuits can be used more generally to monitor cell density and initiate regulatory programs to redirect metabolic fluxes and avoid inefficient overflow metabolism (Figure 2c) [79].
Besides metabolism and regulation, population-level circuit design also offers the opportunity to synthetically shape and control population growth itself. In some cases, this might even be a necessity in terms of ecological stability, if the division-of-labor strategy does not implicitly ensure the long-term coexistence of the strains through metabolic interdependencies or cross-feeding [80]. A new strategy to stabilize competitive co-cultures which is not based on traditional ecological principles is through use of orthogonal QS systems: In each strain, an intracellular lysis circuit is activated at a critical cell density, which renders the population self-limiting and thus prevents it from taking over the co-culture (Figure 2d) [81]. A population control circuit was recently implemented to release, upon lysis, therapeutic toxins inside a tumor environment for bacterial cancer therapy [64], demonstrating that synthetic control over growth dynamics can have a functional role beyond studies of bacterial physiology [82] and pattern formation [60]. A complementary system was constructed by Huang and colleagues, who engineered a QS-based survival circuit that produces an antibiotic resistance protein at high enough cell density, which is only reached inside specific microbial “swarmbot” capsules [83]. This effectively confined the engineered strain to its synthetic environment, preventing unintended proliferation.
Such artificial confinement of engineered organisms can also be viewed in the general context of safe-guarding strategies needed to prevent the inadvertent spread of synthetic microbes and genes into the environment. Population control circuits can help solve this problem, combined with enhanced genetic stability [84,85] which ensures long-term viability of the engineered function as well as the safeguards themselves. However, it should be noted that there are many other ways of achieving such biocontainment. Novel examples include sophisticated, stabilized circuits which actively kill cells or repress essential genes unless specific environmental signals are present [86–88], auxotrophies to non-standard amino acids [89] and conditionally stable plasmids to prevent spreading of genes to other hosts [90]. For further information, we refer the reader to two recent reviews [74,91].
Conclusion
Synthetic biology relies on a versatile and ever-expanding set of engineering tools, many of which are inherently modular due to their action at different stages of gene expression. Combined with the engineered orthogonality of many novel regulatory elements and design principles which ensure the maintenance of cellular baseline conditions, synthetic constructs become ever more robust and fitter for applications. Given the importance of dynamic gene expression in nature and the possibilities created by the increased modularity and robustness, we anticipate that the field will also see more frequent examples of systems exploiting intrinsic dynamics as opposed to stable steady-states. While there is a limit to the complexity and capacity of a single cell, this development will be facilitated by synthetic control over populations and the composition of synthetic consortia, which open up new perspectives for sophisticated and, with the appropriate precautions, safe biotechnological applications.
Highlights.
Synthetic biology approaches enable microbial systems engineering
Versatile tools rapidly expand regulatory engineering capabilities
Modular components and management of load effects facilitate robust gene circuits
Ecology-level control opens up new possibilities for applications
Acknowledgments
This work was supported by the National Institutes of Health [R01-GM069811]; the Office of Naval Research [N00014-16-1-2093]; and the Human Frontiers Science Program [LT000840/2014-C] (to P.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
- 1.Collins JJ, Gardner TS, Cantor CR. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 2.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 3.Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat Rev Mol Cell Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black JB, Perez-Pinera P, Gersbach CA. Mammalian Synthetic Biology: Engineering Biological Systems. Annu Rev Biomed Eng. 2017;19:249–277. doi: 10.1146/annurev-bioeng-071516-044649. [DOI] [PubMed] [Google Scholar]
- 5.Jayaraman P, Devarajan K, Chua TK, Zhang H, Gunawan E, Poh CL. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Res. 2016;44:6994–7005. doi: 10.1093/nar/gkw548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Rodriguez J, Moser F, Song M, Voigt CA. Engineering RGB color vision into Escherichia coli. Nat Chem Biol. 2017;13:706–708. doi: 10.1038/nchembio.2390. [DOI] [PubMed] [Google Scholar]
- 7.Tschirhart T, Kim E, McKay R, Ueda H, Wu H-C, Pottash AE, Zargar A, Negrete A, Shiloach J, Payne GF, et al. Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling. Nat Commun. 2017;8:14030. doi: 10.1038/ncomms14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudge TJ, Brown JR, Federici F, Dalchau N, Phillips A, Ajioka JW, Haseloff J. Characterization of Intrinsic Properties of Promoters. ACS Synth Biol. 2016;5:89–98. doi: 10.1021/acssynbio.5b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee ME, DeLoache WC, Cervantes B, Dueber JE. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth Biol. 2015;4:975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- 10.Reider Apel A, D’Espaux L, Wehrs M, Sachs D, Li RA, Tong GJ, Garber M, Nnadi O, Zhuang W, Hillson NJ, et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:496–508. doi: 10.1093/nar/gkw1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanton BC, Nielsen AAK, Tamsir A, Clancy K, Peterson T, Voigt CA. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai Q-A, Tran AB, Paull M, Keasling JD, Arkin AP, et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods. 2013;10:354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- 13.Bradley RW, Buck M, Wang B. Tools and Principles for Microbial Gene Circuit Engineering. J Mol Biol. 2016;428:862–888. doi: 10.1016/j.jmb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Wang B, Barahona M, Buck M. Amplification of small molecule-inducible gene expression via tuning of intracellular receptor densities. Nucleic Acids Res. 2015;43:1955–1964. doi: 10.1093/nar/gku1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brödel AK, Jaramillo A, Isalan M. Engineering orthogonal dual transcription factors for multi-input synthetic promoters. Nat Commun. 2016;7:13858. doi: 10.1038/ncomms13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Zong Y, Zhang HM, Lyu C, Ji X, Hou J, Guo X, Ouyang Q, Lou C. Insulated transcriptional elements enable precise design of genetic circuits. Nat Commun. 2017;8:52. doi: 10.1038/s41467-017-00063-z. This study provides a set of core promoters and insulated operators for 11 different transcriptions factors which function independently of their genetic context. The characteristics of combinatorial promoters constructed from these elements can be accurately predicted within 1.5-fold induction range without any free parameters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Didovyk A, Borek B, Tsimring L, Hasty J. Transcriptional regulation with CRISPR-Cas9: Principles, advances, and applications. Curr Opin Biotechnol. 2016;40:177–184. doi: 10.1016/j.copbio.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Didovyk A, Borek B, Hasty J, Tsimring L. Orthogonal Modular Gene Repression in Escherichia coli Using Engineered CRISPR/Cas9. ACS Synth Biol. 2016;5:81–88. doi: 10.1021/acssynbio.5b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machens F, Balazadeh S, Mueller-Roeber B, Messerschmidt K. Synthetic Promoters and Transcription Factors for Heterologous Protein Expression in Saccharomyces cerevisiae. Front Bioeng Biotechnol. 2017;5:63. doi: 10.3389/fbioe.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keung AJ, Joung JK, Khalil AS, Collins JJ. Chromatin regulation at the frontier of synthetic biology. Nat Rev Genet. 2015;16:159–171. doi: 10.1038/nrg3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier JAH, Möhrle R, Jeltsch A. Design of synthetic epigenetic circuits featuring memory effects and reversible switching based on DNA methylation. Nat Commun. 2017;8:15336. doi: 10.1038/ncomms15336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roquet N, Soleimany AP, Ferris AC, Aaronson S, Lu TK. Synthetic recombinase-based state machines in living cells. Science. 2016;353:aad8559–aad8559. doi: 10.1126/science.aad8559. [DOI] [PubMed] [Google Scholar]
- 24•.Brophy JA, Voigt CA. Antisense transcription as a tool to tune gene expression. Mol Syst Biol. 2016;12:854–854. doi: 10.15252/msb.20156540. This study proposes a new type of regulatory element based on a uni-directional terminator and an anti-sense promoter. The authors also provide sets of terminators and anti-sense promoters that yield up to 30-fold repression. This is an example of a regulatory element that is easily combined with existing regulatory motifs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kushwaha M, Rostain W, Prakash S, Duncan JN, Jaramillo A. Using RNA as Molecular Code for Programming Cellular Function. ACS Synth Biol. 2016;5:795–809. doi: 10.1021/acssynbio.5b00297. [DOI] [PubMed] [Google Scholar]
- 26.Chappell J, Westbrook A, Verosloff M, Lucks J. Computational design of Small Transcription Activating RNAs (STARs) for versatile and dynamic gene regulation. Nat Commun. 2017;8:1051. doi: 10.1038/s41467-017-01082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: De-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espah Borujeni A, Mishler DM, Wang J, Huso W, Salis HM. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 2016;44:1–13. doi: 10.1093/nar/gkv1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 30.Grangeasse C, Stülke J, Mijakovic I. Regulatory potential of post-translational modifications in bacteria. Front Microbiol. 2015;6:500. doi: 10.3389/fmicb.2015.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen J, Benenson Y. Synthetic biology of cell signaling. Nat Comput. 2016;15:5–13. [Google Scholar]
- 32.Ganesh I, Ravikumar S, Lee SH, Park SJ, Hong SH. Engineered fumarate sensing Escherichia coli based on novel chimeric two-component system. J Biotechnol. 2013;168:560–566. doi: 10.1016/j.jbiotec.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Prindle A, Selimkhanov J, Li H, Razinkov I, Tsimring LS, Hasty J. Rapid and tunable post-translational coupling of genetic circuits. Nature. 2014;508:387–391. doi: 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron DE, Collins JJ. Tunable protein degradation in bacteria. Nat Biotechnol. 2014;32:1276–1281. doi: 10.1038/nbt.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Fernandez-Rodriguez J, Voigt CA. Post-translational control of genetic circuits using Potyvirus proteases. Nucleic Acids Res. 2016;44:6493–6502. doi: 10.1093/nar/gkw537. This study demonstrates how host-orthogonal proteases can be used to achieve multiple types of post-translational regulation by cleaving proteins in defined places to remove or expose a functional peptide, providing an interface with transcriptional and other types of regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F, Su RQ, Lai YC, Wang X. Engineering of a synthetic quadrastable gene network to approach Waddington landscape and cell fate determination. Elife. 2017;6:e23702. doi: 10.7554/eLife.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen AAK, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, Voigt CA. Genetic circuit design automation. Science. 2016;352:aac7341–aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 38.Rubens JR, Selvaggio G, Lu TK. Synthetic mixed-signal computation in living cells. Nat Commun. 2016;7:11658. doi: 10.1038/ncomms11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Green AA, Kim J, Ma D, Silver PA, Collins JJ, Yin P. Complex cellular logic computation using ribocomputing devices. Nature. 2017;548:117–121. doi: 10.1038/nature23271. This study demonstrates the power of post-transcriptional RNA regulation and describes how complex digital signal processing networks can be built from exclusively de-novo engineered parts. It is also an excellent example for successful forward-engineering and orthogonality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian Y, McBride C, Del Vecchio D. Programming Cells to Work for Us. Annu Rev Control Robot Auton Syst. 2018;1:4.1–4.30. [Google Scholar]
- 41.Fiore G, Perrino G, Di Bernardo M, Di Bernardo D. In Vivo Real-Time Control of Gene Expression: A Comparative Analysis of Feedback Control Strategies in Yeast. ACS Synth Biol. 2016;5:154–162. doi: 10.1021/acssynbio.5b00135. [DOI] [PubMed] [Google Scholar]
- 42.Menolascina F, Fiore G, Orabona E, De Stefano L, Ferry M, Hasty J, di Bernardo M, di Bernardo D. In-Vivo Real-Time Control of Protein Expression from Endogenous and Synthetic Gene Networks. PLoS Comput Biol. 2014;10:e1003625. doi: 10.1371/journal.pcbi.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milias-Argeitis A, Rullan M, Aoki SK, Buchmann P, Khammash M. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nat Commun. 2016;7:12546. doi: 10.1038/ncomms12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siu Y, Fenno J, Lindle JM, Dunlop MJ. Design and Selection of a Synthetic Feedback Loop for Optimizing Biofuel Tolerance. ACS Synth Biol. 2017 doi: 10.1021/acssynbio.7b00260. [DOI] [PubMed] [Google Scholar]
- 45.Kushwaha M, Salis HM. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat Commun. 2015;6:7832. doi: 10.1038/ncomms8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Vecchio D. Modularity, context-dependence, and insulation in engineered biological circuits. Trends Biotechnol. 2015;33:111–119. doi: 10.1016/j.tibtech.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Lee JW, Gyorgy A, Cameron DE, Pyenson N, Choi KR, Way JC, Silver PA, Del Vecchio D, Collins JJ. Creating Single-Copy Genetic Circuits. Mol Cell. 2016;63:329–336. doi: 10.1016/j.molcel.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ceroni F, Algar R, Stan G-B, Ellis T. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods. 2015;12:415–418. doi: 10.1038/nmeth.3339. [DOI] [PubMed] [Google Scholar]
- 49•.Venturelli OS, Tei M, Bauer S, Chan LJG, Petzold CJ, Arkin AP. Programming mRNA decay to modulate synthetic circuit resource allocation. Nat Commun. 2017;8:15128. doi: 10.1038/ncomms15128. This study demonstrates how cellular resources can be actively diverted towards a synthetic gene circuit, thereby providing a new perspective on resource limitation and the challenges it presents to modular circuit design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mishra D, Rivera PM, Lin A, Del Vecchio D, Weiss R. A load driver device for engineering modularity in biological networks. Nat Biotechnol. 2014;32:1268–1275. doi: 10.1038/nbt.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gyorgy A, Jiménez JI, Yazbek J, Huang HH, Chung H, Weiss R, Del Vecchio D. Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys J. 2015;109:639–646. doi: 10.1016/j.bpj.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carbonell-Ballestero M, Garcia-Ramallo E, Montañez R, Rodriguez-Caso C, Macía J. Dealing with the genetic load in bacterial synthetic biology circuits: Convergences with the Ohm’s law. Nucleic Acids Res. 2016;44:496–507. doi: 10.1093/nar/gkv1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian Y, Huang HH, Jiménez JI, Del Vecchio D. Resource Competition Shapes the Response of Genetic Circuits. ACS Synth Biol. 2017;6:1263–1272. doi: 10.1021/acssynbio.6b00361. [DOI] [PubMed] [Google Scholar]
- 54.Liao C, Blanchard AE, Lu T. An integrative circuit–host modelling framework for predicting synthetic gene network behaviours. Nat Microbiol. 2017 doi: 10.1038/s41564-017-0022-5. [DOI] [PubMed] [Google Scholar]
- 55.Klumpp S, Hwa T. Bacterial growth: Global effects on gene expression, growth feedback and proteome partition. Curr Opin Biotechnol. 2014;28:96–102. doi: 10.1016/j.copbio.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, Hasty J, Bhatia SN. Programmable probiotics for detection of cancer in urine. Sci Transl Med. 2015;7:289ra84–289ra84. doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, Silver PA. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shipman SL, Nivala J, Macklis JD, Church GM. CRISPR–Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature. 2017 doi: 10.1038/nature23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaerli Y, Munteanu A, Gili M, Cotterell J, Sharpe J, Isalan M. A unified design space of synthetic stripe-forming networks. Nat Commun. 2014;5:4905. doi: 10.1038/ncomms5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kong W, Blanchard AE, Liao C, Lu T. Engineering robust and tunable spatial structures with synthetic gene circuits. Nucleic Acids Res. 2017;45:1005–1014. doi: 10.1093/nar/gkw1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hennig S, Rödel G, Ostermann K. Artificial cell-cell communication as an emerging tool in synthetic biology applications. J Biol Eng. 2015;9:13. doi: 10.1186/s13036-015-0011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y, Kim JK, Hirning AJ, Josi K, Bennett MR. Emergent genetic oscillations in a synthetic microbial consortium. Science. 2015;349:986–989. doi: 10.1126/science.aaa3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Din MO, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN, et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–85. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baumgart L, Mather W, Hasty J. Synchronized DNA cycling across a bacterial population. Nat Genet. 2017;49:1282–1285. doi: 10.1038/ng.3915. [DOI] [PubMed] [Google Scholar]
- 66.Scott SR, Hasty J. Quorum Sensing Communication Modules for Microbial Consortia. ACS Synth Biol. 2016;5:969–977. doi: 10.1021/acssynbio.5b00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant PK, Dalchau N, Brown JR, Federici F, Rudge TJ, Yordanov B, Patange O, Phillips A, Haseloff J. Orthogonal intercellular signaling for programmed spatial behavior. Mol Syst Biol. 2016;12:849–849. doi: 10.15252/msb.20156590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng W, Du P, Lou Q, Wu L, Zhang HM, Lou C, Wang H, Ouyang Q. Rational Design of an Ultrasensitive Quorum-Sensing Switch. ACS Synth Biol. 2017;6:1445–1452. doi: 10.1021/acssynbio.6b00367. [DOI] [PubMed] [Google Scholar]
- 69.Khakhar A, Bolten NJ, Nemhauser J, Klavins E. Cell–Cell Communication in Yeast Using Auxin Biosynthesis and Auxin Responsive CRISPR Transcription Factors. ACS Synth Biol. 2016;5:279–286. doi: 10.1021/acssynbio.5b00064. [DOI] [PubMed] [Google Scholar]
- 70.Hennig S, Clemens A, Rödel G, Ostermann K. A yeast pheromone-based inter-species communication system. Appl Microbiol Biotechnol. 2015;99:1299–1308. doi: 10.1007/s00253-014-6133-5. [DOI] [PubMed] [Google Scholar]
- 71.Urrios A, Macia J, Manzoni R, Conde N, Bonforti A, De Nadal E, Posas F, Solé R. A Synthetic Multicellular Memory Device. ACS Synth Biol. 2016;5:862–873. doi: 10.1021/acssynbio.5b00252. [DOI] [PubMed] [Google Scholar]
- 72.Cao Y, Ryser MD, Payne S, Li B, Rao CV, You L. Collective Space-Sensing Coordinates Pattern Scaling in Engineered Bacteria. Cell. 2016;165:620–630. doi: 10.1016/j.cell.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao Y, Feng Y, Ryser MD, Zhu K, Herschlag G, Cao C, Marusak K, Zauscher S, You L. Programmable assembly of pressure sensors using pattern-forming bacteria. Nat Biotechnol. 2017;35:1087–1093. doi: 10.1038/nbt.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johns NI, Blazejewski T, Gomes AL, Wang HH. Principles for designing synthetic microbial communities. Curr Opin Microbiol. 2016;31:146–153. doi: 10.1016/j.mib.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perry N, Nelson EM, Timp G. Wiring Together Synthetic Bacterial Consortia to Create a Biological Integrated Circuit. ACS Synth Biol. 2016;5:1421–1432. doi: 10.1021/acssynbio.6b00002. [DOI] [PubMed] [Google Scholar]
- 76•.Hoynes-O’Connor A, Moon TS. Programmable genetic circuits for pathway engineering. Curr Opin Biotechnol. 2015;36:115–121. doi: 10.1016/j.copbio.2015.08.007. This review describes examples and opportunities of synthetic circuit design in metabolic engineering. [DOI] [PubMed] [Google Scholar]
- 77.Zhou K, Qiao K, Edgar S, Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33:377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang H, Pereira B, Li Z, Stephanopoulos G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci. 2015;112:8266–8271. doi: 10.1073/pnas.1506781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79•.Gupta A, Reizman IMB, Reisch CR, Prather KLJ. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol. 2017;35:273–279. doi: 10.1038/nbt.3796. This study combines metabolic engineering with a synthetic quorum sensing system to create an artificial “valve” which autonomously downregulates glycolytic flux at a critical time point and optimizes product yield. It is a prominent example of a circuit which uses population-level coordination to achieve environmental adaptation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci. 2014;111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81•.Scott SR, Din MO, Bittihn P, Xiong L, Tsimring LS, Hasty J. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat Microbiol. 2017;2:17083. doi: 10.1038/nmicrobiol.2017.83. This study represents the idea that control of growth itself can shape multicellular behavior. It demonstrates that synthetic control of the population size via quorum-sensing induced lysis leads to self-limitation, increased ecological stability and tunable population dynamics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Izard J, Gomez Balderas CD, Ropers D, Lacour S, Song X, Yang Y, Lindner AB, Geiselmann J, de Jong H. A synthetic growth switch based on controlled expression of RNA polymerase. Mol Syst Biol. 2015;11:840–840. doi: 10.15252/msb.20156382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang S, Lee AJ, Tsoi R, Wu F, Zhang Y, Leong KW, You L. Coupling spatial segregation with synthetic circuits to control bacterial survival. Mol Syst Biol. 2016;12:1–13. doi: 10.15252/msb.20156567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Renda BA, Hammerling MJ, Barrick JE. Engineering reduced evolutionary potential for synthetic biology. Mol BioSyst. 2014;10:1668–1678. doi: 10.1039/c3mb70606k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bittihn P, Hasty J, Tsimring LS. Suppression of Beneficial Mutations in Dynamic Microbial Populations. Phys Rev Lett. 2017;118:28102. doi: 10.1103/PhysRevLett.118.028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Chan CTY, Lee JW, Cameron DE, Bashor CJ, Collins JJ. “Deadman” and “Passcode” microbial kill switches for bacterial containment. Nat Chem Biol. 2015;12:82–86. doi: 10.1038/nchembio.1979. This study demonstrates how containment of genetically modified microbes can be achieved through two artificial gene networks that couple sensing of specific environmental signals to cell survival. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gallagher RR, Patel JR, Interiano AL, Rovner AJ, Isaacs FJ. Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 2015;43:1945–1954. doi: 10.1093/nar/gku1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cai Y, Agmon N, Choi WJ, Ubide A, Stracquadanio G, Caravelli K, Hao H, Bader JS, Boeke JD. Intrinsic biocontainment: Multiplex genome safeguards combine transcriptional and recombinational control of essential yeast genes. Proc Natl Acad Sci. 2015;112:1803–1808. doi: 10.1073/pnas.1424704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, Church GM. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wright O, Delmans M, Stan GB, Ellis T. GeneGuard: A modular plasmid system designed for biosafety. ACS Synth Biol. 2015;4:307–316. doi: 10.1021/sb500234s. [DOI] [PubMed] [Google Scholar]
- 91.Torres L, Krüger A, Csibra E, Gianni E, Pinheiro VB. Synthetic biology approaches to biological containment: pre-emptively tackling potential risks. Essays Biochem. 2016;60:393–410. doi: 10.1042/EBC20160013. [DOI] [PMC free article] [PubMed] [Google Scholar]


