Abstract
Pseudomonas aeruginosa is a common Gram-negative bacterium and opportunistic human pathogen. The distinctive structure of its outer membrane (OM) and outer membrane vesicles (OMVs) plays a fundamental role in bacterial virulence, colonization ability, and antibiotic resistance. To provide critical insights into OM and OMV functionality, we conducted an all-atom molecular dynamics study of asymmetric membranes that are biologically relevant to P. aeruginosa. We hybridized a GLYCAM06-based lipopolysaccharides force field with the Stockholm lipids force field (Slipids) to model bilayer membranes with Lipid A molecules in one leaflet and physiologically relevant phospholipid molecules in the other, including 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG), and 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG). In particular, a membrane with phospholipid composition representing the P. aeruginosa OM was constructed and modeled by mixing the physiologically dominant components. The detailed structure of membranes was characterized by area per lipid, transmembrane mass and charge densities, radial distribution function (RDF), deuterium order parameter (SCD) of acyl chains, and inclination angles of phosphates and disaccharide in Lipid A. The membrane fluidity in equilibrium and the hydration of functional groups were probed and characterized quantitatively. The consistent properties of the Lipid A leaflet in different membranes demonstrates its compatibility with various phospholipids present in the P. aeruginosa OM. The more ordered acyl chains of Lipid A compared to the cytoplasmic cell membrane contribute to the low permeability of bacterial outer membrane. The findings of this computational investigation of P. aeruginosa OM will further the understanding of microbial pathogenesis and enable future study of OMV biogenesis.
Introduction
The Gram-negative bacterium Pseudomonas aeruginosa is notorious for causing serious infections among patients with burn wounds, cystic fibrosis, acute leukemia, organ transplants, and intravenous-drug addiction.1 As an opportunistic human pathogen, P. aeruginosa ranked as the 6th most common cause of healthcare-associated infections occurring from 2011–2014. Even worse, its resistance to selected antimicrobial agents can reach 34.5% of isolates tested.2 The prevalence as a hospital-associated and multi-drug resistant pathogen has earned P. aeruginosa recognition among the ‘ESKAPE’ group of pathogens, which represents the greatest concern in the clinic.3 The virulence of this microorganism is not only attributed to its extracellular products, such as exotoxin A and deoxyribonuclease, but also involves the release of Outer Membrane Vesicles (OMVs).4
Bacterial OMVs are spherical structures containing an aqueous lumen that is enclosed by a bilayer membrane, with typical diameters ranging from 50 to 250 nm.5 OMVs perform a variety of pathogenic functions, including the delivery of toxic factors (e.g., phospholipases, proteases, hemolysins, etc.) and the shielding of parent bacteria against attacks from the immune system as well as antibiotics.5–8 Previous evidence has indicated that OMVs originate from the bulging and pinching of the bacterial outer membrane (OM), which makes the bilayer membrane of OMVs closely resemble the composition of OM.6,9–11 Although we do not fully understand the mechanisms behind OMV biogenesis, the properties and dynamics of lipopolysaccharides (LPS) found in the OM have been implicated to play an important role. In P. aeruginosa, a self-produced small molecule is known to interact with the OM through preferential association with LPS in order to induce OMV formation.7,9 In other organisms, OMVs have been observed to preferentially package modified LPS chemotypes, leading to a suggestion that the altered properties of the modified LPS facilitate OMV biogenesis.12,13 The versatile functions of OMVs extend the significance of P. aeruginosa OM in the pathogenesis of infectious diseases from cell surface to extracellular space.
The asymmetric bilayer OM of P. aeruginosa has an outer leaflet consisting of lipopolysaccharides and an inner leaflet composed of phospholipids. LPS is a complex macromolecule comprising an endotoxic Lipid A, covalently bonded core oligosaccharide, and an O-polysaccharide (also referred to as O-antigen) chain attached to the core domain.14 Notably, the detailed compositions and structures of LPS vary among different gram-negative bacteria. Lipid A presents a peculiar conformation and unique architecture not present in other biomolecules.15 It serves not only as a hydrophobic anchor of LPS molecule to form the bilayer structure but also an activator of innate immune system.14
Although experimental observations provide insightful details of the OM structure, the difficulties of acquiring a clear physical picture of three-dimensional dynamic membranes on the atomistic scale remain to be solved.16–19 With its ability of capturing atomistic information, molecular dynamics (MD) simulation emerges as a powerful tool to study biological membranes. The MD studies on gram-negative bacterial OMs have been conducted extensively in the past decades, at both all-atom and coarse-grained levels.18–25 Specifically, the computational models for the P. aeruginosa OMs have been continuously matured and improved to reproduce experimental data more accurately.19,26–29
Despite extensive literature on biomembrane modeling, the past research was heavily focused on the development of new force fields (FFs) for LPS macromolecules. The simulations were limited to model membranes with single-component phospholipid leaflets. The fundamental understanding of the structural properties of OMs having diverse outer membrane phospholipids is not complete. More importantly, all-atom models of biologically relevant outer membranes are still lacking. Herein, we report an all-atom MD study of asymmetric Lipid A–phospholipid membranes with phospholipid species and compositions closely matching the experimental data for P. aeruginosa. We implement two well-established FFs for LPS and phospholipids to model five different asymmetric membranes. Four physiologically relevant outer membrane phospholipids, namely 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG), 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG) are considered (see Fig. 1). In addition, we also simulated a membrane with a mixture of DPPE, DOPE, and DPPG to reproduce head group and fatty acid compositions similar to those in physiological systems. The structural properties of these membranes were analyzed with atomistic details and compared with the existing experimental and computational data.
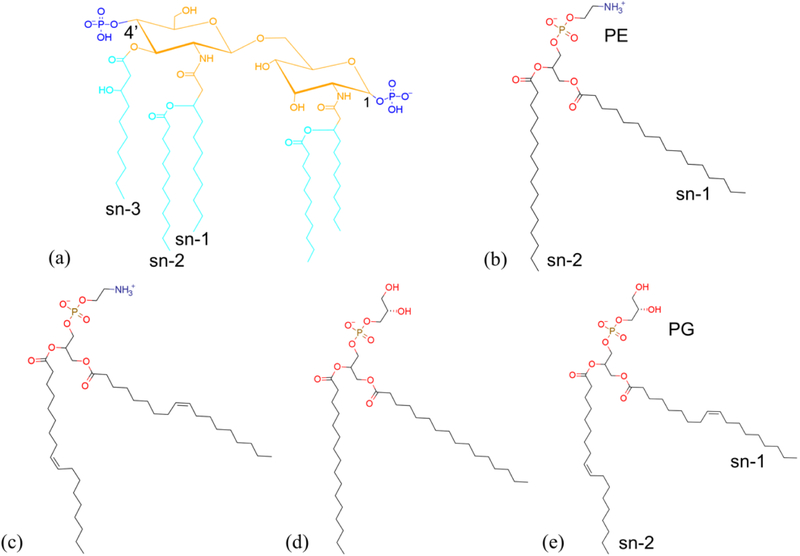
Fig. 1.
Chemical structure of (a) the P. aeruginosa Lipid A, (b) DPPE, (c) DOPE, (d) DOPE, and (e) DOPG modeled in the simulations. Functional groups are represented by different colors: blue for phosphate, orange for 3-(acetyl amino)-3-deoxy-D-glucose, cyan for dodecanoic acid (12:0) and 3-hydroxydecanoic acid 10:0 (3-OH). The annotation of sn-1, sn-2 and sn-3 chains of Lipid A are labeled. The DPPE and DOPG are selected to be the representative phospholipids for the head group and acyl chains (sn-1 and sn-2) annotation.
Computational Methods
To simulate asymmetric membranes with Lipid A and phospholipids, we hybridize a GLYCAM06-based LPS FF28 and Stockholm lipids (Slipids) FF.30–32 The GLYCAM FF was developed for simulating carbohydrates based on the AMBER FF family.33 The latest parameter set GLYCAM06 was further expanded to incorporate new parameters for unusual monosaccharides in the inner core region of LPS.28 This FF has been applied to model asymmetric P. Aeruginosa outer membranes with different LPS chemotypes, including Lipid A.18,24,28,29 On the other hand, the Slipids FF was particularly developed for a variety of fully saturated and unsaturated phospholipids.30–32 The empirical parameters of all non-bonded and bonded interactions in the FFs were tuned for a large set of training and test molecules to reproduce experimental and quantum mechanical data. The fidelity of the FFs has been further validated by simulating different lipid membranes and comparing simulation results with experimental data, including Nuclear Magnetic Resonance (NMR) measured structural profiles and membrane properties, Fourier-Transform Infrared spectroscopy (FT-IR) quantified molecular orientations, and so on.18,31,34 Both full-atom FFs apply the restricted electrostatic potential approach (RESP)35,36 with quantum mechanics calculations at the same level of theory to derive partial atomic charges, and thus are compatible with most members of the AMBER FF family for amino acids and small organic molecules.18,30,33 The water model is the three-site TIP3P model, while ion parameters are adapted from the standard AMBER FF. Considering that the asymmetric membranes with DPPE leaflets have been extensively studied, a Lipid A–DPPE membrane is modeled first as a benchmark simulation.
Lipid A is the characteristic lipid component of bacterial outer membranes. The fluidity of Lipid A has significant impact on the bioactivity.37 Our work aims at studying the properties of Lipid A membranes in the liquid-crystalline phase. Experimentally, the phase transition temperatures of LPS from P. aeruginosa was found in the range between 303 K and 313 K.17,38 The transition temperature of the corresponding Lipid A has not been reported due to the lack of sharp transition in membrane properties and morphology while temperature varies.38 Nevertheless, the phase transition temperatures of free Lipid A have been reported to be approximately 10 K higher than the corresponding LPS for many other bacterial species (e.g., Escherichia coli and Salmonella Minnesota).37–39 Numerically, previous MD simulations have used a wide range of temperatures to investigate bacterial OMs in the liquid-crystalline phase, such as 310 K,21 313 K,40 323 K,40 328 K,18,23 and even 350 K.28 In order to maintain Lipid A fluidity and validate our force field by comparing our results with the references, we chose a moderate temperature of 323 K.
The GROMACS 5.1.2 package41–47 is used for carrying out all MD simulations. The initial geometries of five membranes are optimized by a steepest-descent energy minimization, followed by an isothermal−isochoric (NVT) equilibration for 100 ps at 323 K. The velocity-rescaling thermostat is applied in which lipids and solvent are separately coupled. A relative long isothermal−isobaric (NPT) equilibration is then performed for 1 ns at 1 bar with semi-isotropic pressure coupling using the Parrinello-Rahman algorithm48 while the temperature is maintained through the Nose-Hoover thermostat.49 All bonds are constrained by the linear constraint solver (LINCS) algorithm.50 A cutoff of 1.2 nm is used for short-range van de Waals and electrostatic interactions, as well as for building the neighbor list which is updated every five time steps. Long-range electrostatic interactions are solved by the particle-mesh Ewald (PME) method.51 In all simulations, the leap-frog integrator with a time step of 2 fs is used and the motion of bilayer relative to solvent is removed every time step. The production run for each configuration is performed for 1 μs in the NPT ensemble at 323 K and 1 bar.
Fig. 1(a) shows the Lipid A structure used in all simulations of this study.52 The penta-acyl Lipid A domain of the P. aeruginosa LPS contains a β (1–6)-linked D-glucosamine disaccharide decorated with two negatively charged phosphates at 1 and 4’ positions and five asymmetrically distributed fatty acids. The acylation and phosphorylation patterns of Lipid A dictate the endotoxic bioactivity of LPS.53 In contrast to Lipid A, phospholipids in the OM are more ubiquitous types in biological systems. We select the composition of glycerophospholipids based on the experimental data. Typical P. aeruginosa OM contains derivatives having 59.7% phosphatidylethanolamine, 27.1% phosphatidylglycerol, and 13.2% phosphatidylcholine with a variety of fatty acids, including 43.4% palmitic acid, 36.4% oleic acid, 13.5% palmitoleic acid, 6.5% stearic acid, as well as other minor species below the detection limit.54 To simplify the model development but still capture the physiological composition with high fidelity, we consider only the symmetric acyl chains and the possible combinations of two most populated head and tail groups, respectively. In particular, DPPE, DOPE, DPPG, and DOPG are selected for simulations with single-component phospholipid leaflets. In order to mimic physiological OM, we also develop a model with a multi-component phospholipid leaflet by mixing the above species. Using the experimentally measured ratios of PE versus PG and oleic acid versus palmitoleic acid, we construct a mixture of DPPE, DOPE, and DPPG that matches the ratios by ignoring the phospholipid with lowest possibility of presence, i.e. DOPG. The resulting phospholipid composition is 23% DPPE, 46% DOPE, and 31% DPPG.
The numbers of Lipid A and phospholipids used to construct the membranes must be chosen with caution to prevent the onset of computational artifacts.55 For Lipid A–DPPE membranes that have been widely studied, the common practice is matching the numbers of acyl chains in two leaflets.16,18,21,28 For example, for a penta-acyl Lipid A leaflet with 72 molecules, the DPPE leaflet will have 180 molecules. However, we observe deformation and bulging of membranes when this empirical molecule number (180) is applied to DOPE and DOPG, as shown in Fig. S1. Because of the double bonds in the DO lipids, this behavior can be attributed to considerable differences in effective volume and hydrophilic-lipophilic balance of the DO lipids. Notably, a 400 ns simulation of the Lipid A–DPPG membrane with 180 DPPG molecules does not show any unphysical deformations (Fig. S2). This indicates that the membranes with DO lipids are more sensitive to the area mismatch than the DP lipids. Without double bonds, the DP lipids have a range of “comfortable” number within which the two leaflets are compatible with each other. Considering the diversity of phospholipids in this study, we decide to use area per lipid as the guideline for determining the phospholipid number in asymmetric membranes. In particular, we build membranes by matching the total areas of the Lipid A and phospholipid leaflets in equilibrium, based on the experimental values of area per lipid.56–59 The detailed steps of membrane construction follows the same procedure described in Ref. 16. The periodic boundary conditions are applied to all three directions so that a planar membrane is modeled.
This approach produces stable asymmetric membranes with a Lipid A leaflet having 72 molecules and a single-component phospholipid leaflet having 150 DOPE molecules, 140 DPPG molecules, and 120 DOPG molecules, respectively. Similarly, the membrane with a mixture of phospholipids is constructed based on the area per lipid and the experimental composition of head groups and acyl chains. The total number of phospholipid molecules is 150 with 35 DPPE, 68 DOPE, and 47 DPPG molecules randomly distributed in the initial configuration. In addition, each Lipid A molecule is neutralized by one calcium ion and each PG head group is balanced by one sodium ion in order to model a system with pH value of 7.
Results and Discussion
Benchmark Simulation
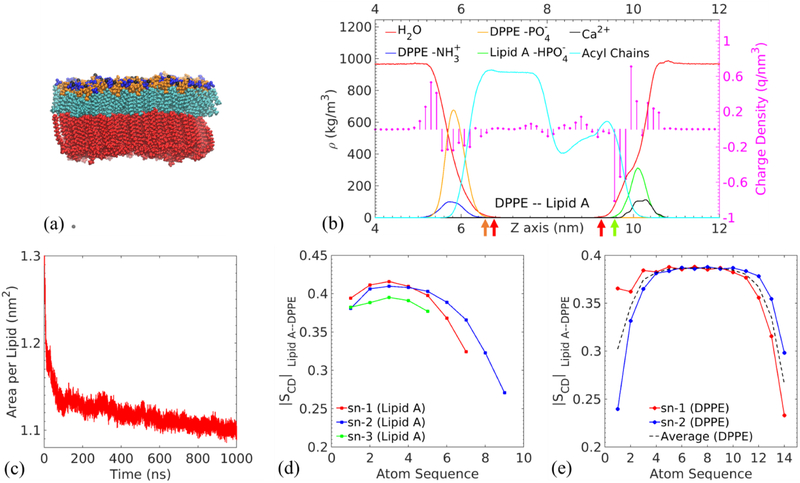
To validate the computational setup with the hybridized force fields for modeling asymmetric biomembranes, we simulate a Lipid A–DPPE membrane and benchmark its characteristic properties, including the area per lipid and deuterium order parameter (SCD) against existing computational and experimental data. Fig. 2(a) and (b) provide an overview of the membrane structure. The detailed position of the red DPPE layer shown in Fig. 2(a) is designated by the density profiles of zwitterionic PE head groups. Specifically, the positively charged amines are heavily exposed to water with mass and charge density peaks at z=5.71 nm and 5.30 nm, respectively. The negatively charged phosphates are embedded deeper in the membrane, and the corresponding density peaks are located at z=5.82 nm and 5.56 nm. Considering the mass density is calculated based on the averaged atom positions of the entire functional group while the partial charges of individual atoms contribute to the charge density, the deviations of the peak positions between the mass and charge densities are expected and consistent with previous simulations.18 Negatively charged phosphate groups of Lipid A colored in blue identify the upper leaflet with calcium ions present in their vicinity. From the density profiles, the mass density peaks of phosphates are selected to characterize the hydrophobic thickness (DHH) of the membrane.60 The hydrophobic inner region of thickness 4.279 nm is filled with acyl chains.
Fig. 2.
Lipid A–DPPE membrane as the benchmark: (a) representative snapshot of an equilibrated membrane. The color scheme of Lipid A functional groups is the same as in Fig. 1. Black atoms represent calcium ions. DPPE phospholipid molecules are colored in red. (b) Mass density ρ (continuous lines) and charge density (circles terminating each stem) profiles of the selected functional groups along the direction normal to the membrane (the z direction). The values are averaged over the last 100 ns of the MD simulations. The left part of the figure represents the DPPE leaflet and the right corresponds to the Lipid A leaflet. (c) Area per lipid of Lipid A molecules. Carbon-deuterium order parameters of (d) sn-1, sn-2 and sn-3 acyl chains of Lipid A and (e) sn-1 and sn-2 acyl chains of DPPE.
As indicated by the two red arrows in Fig. 2(b), the water density drops below 5 kg/m3 (this small value is picked to define the water-free region) at z=6.74 nm in the DPPE leaflet and z=9.20 nm in the Lipid A leaflet. Correspondingly, the cutoff positions for the phosphate groups in the two leaflets are 6.60 nm (DPPE) as indicated by the orange arrow in Fig. 2(b) and 9.53 nm (Lipid A) as shown by the green arrow. An extension of 0.33 nm in the water density profile with respect to the phosphate cutoff position indicates that water penetrates much deeper into the Lipid A leaflet to solvate its phosphate groups. In contrast, the water penetration ceases within 0.14 nm from the DPPE phosphates. This behavior is consistently observed for other asymmetric membranes detailed below.
The area per lipid of Lipid A molecule is calculated by measuring the instantaneous area of simulation box in the plane parallel to the membrane. Fig. 2(c) demonstrates the variation of area per lipid in time as the membrane reaches equilibrium. The averaged value over the last 100 ns is given as 1.101±0.004 nm2. This value agrees well with existing computational work,18 which confirms that the hybrid force field produces a physical membrane. It is noteworthy that the simulation value is slightly lower than the experimental value (1.29 nm2) for the hexa-acyl Lipid A of Salmonella Minnesota at 40 degrees Celsius,56 which could be attributed to smaller tail volume of the penta-acyl Lipid A of P. aeruginosa.
In addition to the area per lipid, we also calculate the deuterium order parameter SCD per atom in the carbon tails to characterize the detailed structural properties of membrane on the single molecule level. SCD = (1/2)(3 cos2 θ – 1) measures the average angle θ between the bilayer normal and each carbon-deuterium bond in experiment or carbon-hydrogen bond in simulation.61–63 The angular bracket represents the ensemble average. The value of SCD varies between −0.5 to 1 depending on the average orientation of bond. SCD = −0.5 corresponds to bonds perpendicular to the normal, indicating a fully ordered acyl chain in all-trans conformation. When the bonds are disordered with random orientations, SCD takes the value of 0.
The absolute values of SCD collected in our simulation are plotted in Fig. 2(d) and (e). The flexibility of the chain terminal accredits to the general descending trend of the ∣SCD∣ profile. The averaged ∣SCD∣ values over all sn chains are 0.382 for Lipid A, obtained from the last 100 ns of the production run. The previous MD simulations reported values of 0.32 for Lipid A in a Lipid A–DPPE membrane at 328 K.16,18,64 The small discrepancy between our result and the value in Refs. 16 and 18 obtained at similar conditions may be induced by the use of full-length 16:0 DPPE molecules in this study instead of the truncated DPPE (9:0/6:0) model in the references.
The ordering of Lipid A from P. aeruginosa has been characterized experimentally by Fourier-transform infrared (FT-IR) spectroscopy, which reported an order parameter of 0.28 at 310.15 K.38 However, we note that the FT-IR determined order parameter in those experiments is different from the carbon-deuterium order parameter SCD probed in this study. The experimental order parameter is based on fluorescence anisotropy measurement using fluorescent probes, and ranges from 0 (isotropically aligned) to 1 (perfectly aligned).39,65–69 To the extent of our knowledge, no experimental SCD for the P. Aeruginosa Lipid A is available.
While the order parameter of symmetric DPPE bilayers has been reported by nuclear magnetic resonance (NMR) spectroscopy and MD simulations,57,64 the ordering of the DPPE leaflet in a Lipid A–DPPE asymmetric membrane has not been probed before. The average ∣SCD∣ of the DPPE leaflet is 0.362 in our membrane system. We note the low value of first ∣SCD∣ in the sn-2 chain of DPPE, which may relate to the fact that the sn-2 chain is closer to the head group and water than the sn-1 chain. The relatively higher average ∣SCD∣ indicates a more ordered DPPE leaflet in the asymmetric membrane, compared with experimental value of 0.202 at 342.15 K57 and simulation results of 0.329 at 300 K and 0.216 at 340 K for pure DPPE bilayers.16,70 We list the data in Table 1. We speculate that the highly ordered acyl chains of Lipid A modulate the tail structure of DPPE, and the interaction between the hydrocarbon chains of the two leaflets influences the overall ordering. Based on the values of area per lipid and ∣SCD∣ reported in previous studies,16,18,57,64,70 the benchmark models a Lipid A–DPPE membrane in the liquid-crystalline phase.
Table 1.
The order parameters of DPPE reported in literature and this study.
Area per lipid
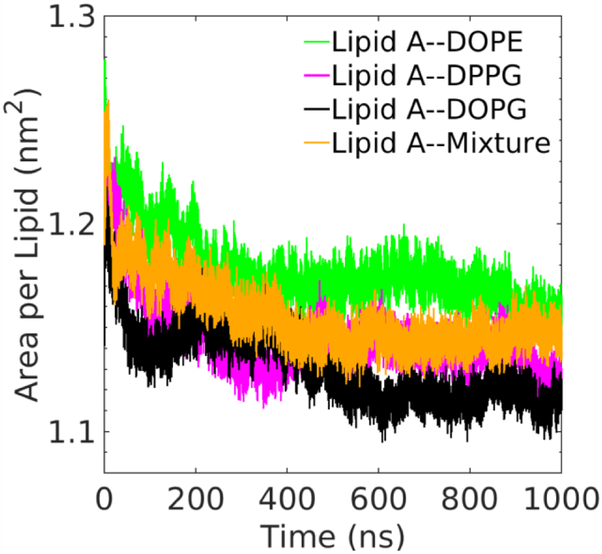
Using the hybrid force field, we simulate four Lipid A–phospholipid asymmetric membranes with biologically relevant species and compositions of phospholipid and probe their structural properties in atomistic detail. Fig. 3 demonstrates three membranes with single-component phospholipids and the one with multi-component phospholipids for reproducing the physiological composition in P. aeruginosa. Fig. S3 shows the bottom views of the Lipid A–Mixture membrane at different times during the simulation. Different species of phospholipids are homogeneously distributed in the lower leaflet of the membrane with no aggregation observed. The uniform distribution of phospholipids confirms their compatibility in the asymmetric Lipid A membrane, which is expected for these physiologically relevant lipid molecules. To determine if the acquired membranes are equilibrated, the area per lipid of Lipid A in asymmetric membranes is monitored during the simulation (Fig. 4). The averaged values over last 100 ns are respectively 1.157 nm2, 1.129 nm2, 1.115 nm2, 1.148 nm2 for the membranes with DOPE, DPPG, DOPG, and mixed phospholipids as the lower leaflet. For all membranes, the standard deviations of area per lipid are below 0.007 nm2. The well-equilibrated membrane set is exemplified by the small standard deviations.
Fig. 3.
Representative snapshots of four Lipid A–phospholipid asymmetric bilayer membranes at equilibrium, including: (a) Lipid A–DOPE, (b) Lipid A–DPPG, (c) Lipid A–DOPG, and (d) Lipid A–Mixture membranes. The color scheme of Lipid A functional groups is the same as in Fig. 1. Black and yellow atoms respectively represent calcium and sodium ions. DPPE, DOPE, DPPG, DOPG molecules are colored in red, green, purple, and silver, respectively.
Fig. 4.
Time evolution of area per lipid of the Lipid A leaflet in the corresponding asymmetric membranes in 1 μs MD simulations.
Despite the diversity of lower leaflet composition, the membrane structure is stable and the packing of Lipid A is consistent among different membranes. This stable structural arrangement of the asymmetric membranes is confirmed by the direct observation of simulation trajectories and the equilibrium values of area per lipid regardless of the lower leaflet composition. From the consistent area per lipid, we infer that individual acyl chain in Lipid A occupies a smaller volume than most of the phospholipids at the same temperature, which could be associated with their closer packing. As mentioned in the Computational methods section, DPPE and DPPG are the two phospholipids that have stable membranes when matching the acyl chain numbers in the two leaflets. The gel-to-liquid crystalline phase transition temperature of phospholipids may provide an explanation. As mentioned before, the simulation temperature 323 K is below the DPPE phase transition temperature 337 K71 and close to DPPG phase transition temperature 314.15 K,72 but is well above the transition temperatures for DOPE and DOPG, which are around 265.65 K73 and 255.15 K74 respectively. Under this circumstance, the more fluidic DOPE and DOPG molecules possess larger occupation areas, which demand disparate numbers of acyl chains to match the area of Lipid A acyl chains. This fluidity is also verified by comparing the order parameters present in the following section.
Transmembrane mass and charge densities
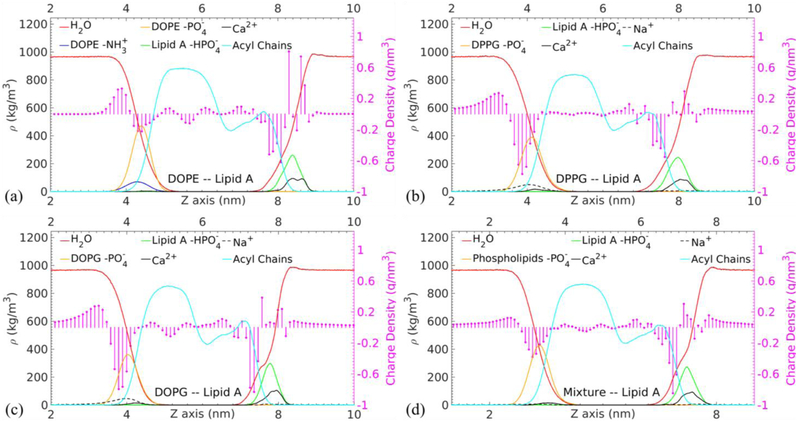
The mass and charge density distributions in the membrane normal direction are examined to provide transmembrane structural information of the equilibrated membranes. As shown in Fig. 5, the DHH values are 3.994 nm, 3.851 nm, and 3.747 nm for the DOPE, DPPG, and DOPG membranes, respectively. The membrane composed of unsaturated 18:1 oleic acids is thinner than the one formed by unsaturated 16:0 palmitic acids with the same head groups. This implies that the unsaturated acyl chains are more relaxed than the saturated acyl chains. Notably, the DHH of the membrane with mixed phospholipids in Fig. 5(d) is 3.88 nm, which is close to the value of Lipid A–DPPG membrane. This result suggests that the thickness of the mixture membrane is mainly determined by the constituent phospholipid that makes the thinnest membrane.
Fig. 5.
Mass density ρ profiles of the selected functional groups and total charge density ρc profiles in circles terminating each stem along the membrane normal direction (the z axis) for the selected functional groups in the (a) Lipid A–DOPE, (b) Lipid A–DPPG, (c) Lipid A–DOPG, and (d) Lipid A–Mixture membranes. The values are averaged over the last 100 ns of the MD simulations.
The density profiles of solvent, fatty chains, and phosphates in Fig. 5 provide compelling information about the overall hydration state of membrane. Our results show that the density distributions of these functional groups are consistent among the modeled membranes. The water molecules are able to penetrate deeper into the Lipid A leaflet than the phospholipid leaflet in all cases similar to the Lipid A–DPPE benchmark. This hydration behavior of Lipid A leaflet has also been observed experimentally, pertaining to the presence of divalent cations.75 The hydration of ion promotes the penetration of water molecules into the hydrophobic region of the membrane. Fig. 5 shows that all kinks in the water density lines, which indicate the enhanced water penetration, are located around z=8 nm in the vicinity of the peaks of both Lipid A phosphates and calcium ions.
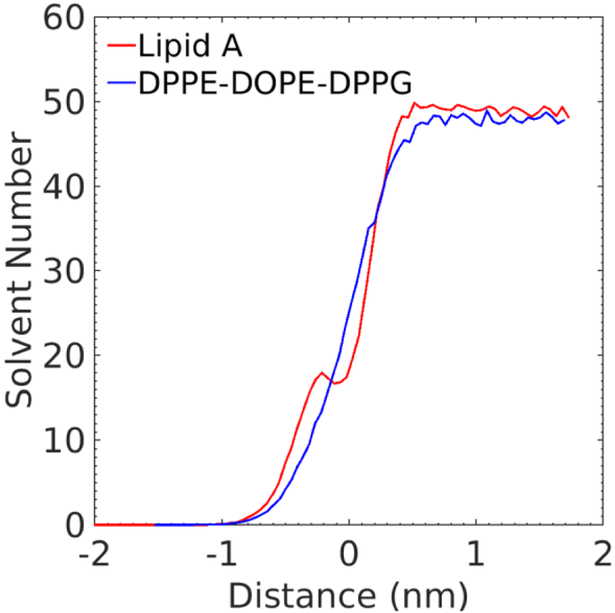
In order to confirm the significance of calcium ions on the membrane hydration, we apply the intrinsic surface approach76–81 to analyze the water penetration with respect to the rough membrane surfaces of Lipid A–Mixture membrane. We construct two membrane surfaces using Voronoi tessellation based on the positions of phosphorus atoms in Lipid A and phospholipids. The transverse solvent distribution as a function of the distance to the approximated rough membrane surface is acquired and shown in Fig. 6. The distributions clearly show more water molecules penetrate below phosphorous into the Lipid A leaflet than the phospholipid leaflet. A sudden drop of water number is also observed at the surface of Lipid A leaflet (corresponding to the zero distance), which is attributed to the close interaction between calcium ions and the Lipid A phosphate groups. This result provides evidence of the ion-assisted hydration of Lipid A leaflet.
Fig. 6.
Time-averaged water distribution of Lipid A–mixture membrane. The x axis is the distance between the roughed membrane surfaces to a selected transverse plane while the y axis represents the corresponding solvent number. The positive (negative) distance corresponds to the exterior (interior) of membrane. The red line plots the water distribution around the Lipid A leaflet and the blue line is for the lower leaflet with DPPE, DOPE, and DPPG. The selected time frames are from the trajectory files of the last 100 ns simulation of the Lipid A–mixture membrane.
The average charge density along the membrane normal corresponds to distinct charge groups in the phospholipid (left side) and Lipid A (right side) molecules. For the PE membranes, the first positive and negative peaks appearing in the leftmost dictate the amino and phosphate groups, respectively. In contrast, the negative peak for the PG molecules is more pronounced, which indicates higher negative charge carried by the phosphate of PG at pH=7. The smearing of counterions thus presents broader positive peaks in Fig. 5(b-d).
In the center, the slightly positive charge densities located between 6 nm and 7 nm correspond to the acyl chains in Lipid A. The saturated and unsaturated acyl chains of different phospholipids demonstrate distinguishable charge distributions. The charge variations in the fatty chain region of phospholipid are more pronounced for chains with unsaturated double bonds (i.e., DOPE and DOPG) than the ones with fully saturated single bonds (i.e., DPPE and DPPG), tallying with more reactive unsaturated compounds than the saturated ones.
The highest charge densities occur in the rightmost region of the profile and match the position of the phosphate groups in Lipid A and the associated calcium counterions. It is known that the divalent calcium ions could form salt bridges with the close proximity Lipid A phosphate groups.5 This binding is also tight enough that the charge density in the solvent can approach zero as shown in Fig. 5(a). In contrast, the presence of sodium ions leads to non-zero charge density in solvent (see Fig. 5(b-d)).
Radial distribution function
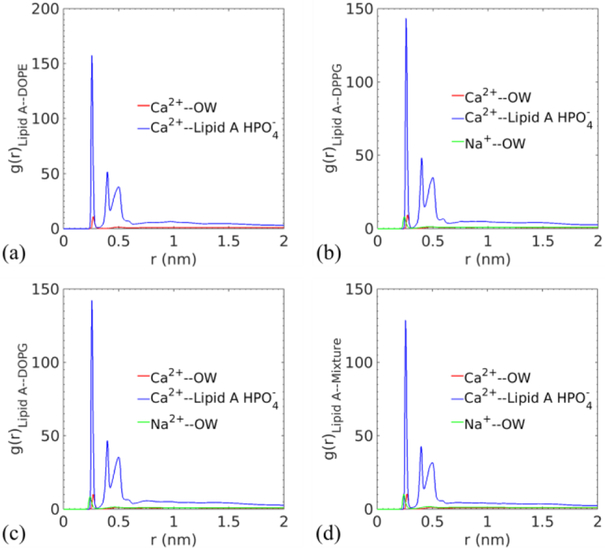
In addition to the density profiles, the selected radial distribution functions (RDFs) of ions are further analyzed to depict ion binding affinity to water and the Lipid A phosphate groups, which provides additional insight into the membrane structure. The ion binding affinity of Lipid A and LPS functional groups has been extensively studied by experiments and numerical simulations.11,17,18,26,82,83 Previous studies report that the divalent cations are able to increase the melting temperature of Lipid A, limit the water permeability of LPS-containing outer membranes, and improve lateral interactions between adjacent LPS molecules through binding with the phosphate groups of Lipid A or the phosphate groups in the inner core of LPS. Although MD models have insufficient quantitative accuracy to predict the cation binding affinity with phospholipid head groups when compared with noninvasive NMR experiments, which may also be true for the Lipid A phosphate groups, the simulation can capture qualitatively correct lipid-ion interactions.84
By measuring the relative density of atoms in concentric shells, the radial distribution function g(r) quantifies the probability of finding a specific group near a reference atom type. In Fig. 7, the hydration peaks of ions occur at 0.27 nm (Ca2+) and 0.24 nm (Na+, if present) for all cases, indicating consistent interactions between ions and water molecules. The larger distance between calcium ions and their first hydration shell could be explained by the dual role these ions play, namely being solvated by water while binding with phosphates.
Fig. 7.
Radial distribution functions between two different cations and the oxygen atom of water, and between cations and the phosphate groups in Lipid A for (a) Lipid A–DOPE, (b) Lipid A–DPPG, (c) Lipid A–DOPG, and (d) Lipid A–Mixture membranes.
This insight can be drawn from the RDFs of calcium ions and Lipid A phosphates. The well-defined three peaks at 0.26 nm, 0.4 nm, and 0.5 nm for all membranes highlight the strong binding affinity between the divalent ions and Lipid A phosphates. The first peak represents the closest phosphates in the nearest-neighbor Lipid A molecules of a reference calcium ion, the second and third peaks come from the corresponding phosphates of the same group of Lipid A molecules but attached on the other glucosamines. The persistent RDFs of calcium ions not only illustrate their capabilities of hydrating and bridging but also confirm the stability of Lipid A leaflet when varying the lower leaflet composition.
Deuterium order parameters
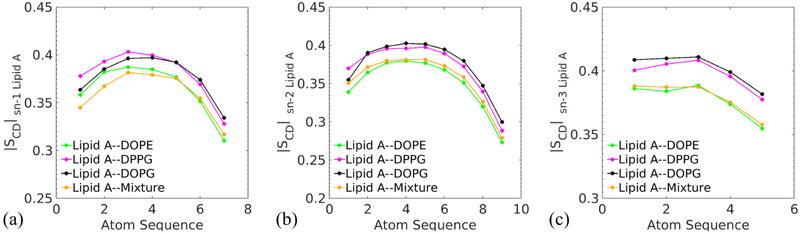
Through the evaluation of deuterium order parameters (SCD), the orientation and fluidity of hydrocarbon chains in the membranes can be revealed. Fig. 8 shows the ∣SCD∣ value of Lipid A as a function of carbon atom sequence for different acyl chains. The average values over all sn chains of the membranes with single-component phospholipids are 0.361 (Lipid A–DOPE), 0.380 (Lipid A–DPPG), and 0.382 (Lipid A–DOPG), which are inversely proportional to the areas per Lipid A of the corresponding membranes.16,85
Fig. 8.
Carbon−deuterium order parameter (SCD) for the acyl chains of Lipid A in four asymmetric membranes. The values of SCD are obtained by averaging over the last 100 ns of the MD simulations, and plotted as a function of the carbon sequence in the fatty acid, in which larger numbers represent positions closer to the tail of acyl chain.
The profiles also describe the detailed structure of each individual chain. For the sn-1 and sn-2 chains, they are overall disordered compared with the sn-3 chain, in particular near the disaccharide, which is reflected by small ∣SCD∣ values at low atom sequence numbers. As the distance from the membrane surface increases, the segment of chain first becomes more ordered. Then, the hydrocarbons near the tail of chain adopt more isotropic orientations. As for sn-3 chains, the acquired values are larger than those of the sn-1 and sn-2 chains. The ordering of sn-3 may be related to the formation of hydrogen bonding between water and hydroxyl groups, which in turn straightens the end of sn-3 chain near the membrane surface.28
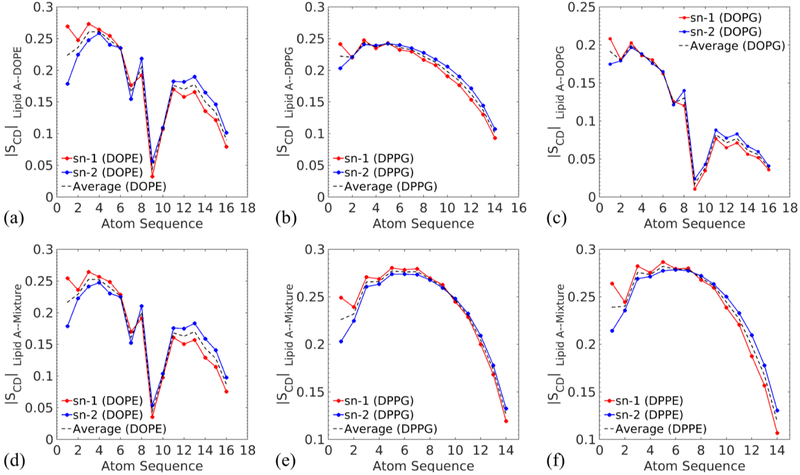
The averaged ∣SCD∣ values for the phospholipid acyl chains in the lower leaflets are 0.180 (DOPE), 0.204 (DPPG), and 0.112 (DOPG) as displayed in Fig. 9(a-c), indicating that the hydrocarbon chains of phospholipids are much disordered than the ones of Lipid A. The kinks in Fig. 9(a) and (c) are correlated with the unsaturated bonds in DOPE and DOPG. These double bonds keep the chains from close packing and lead to more fluidic and less ordered lower leaflets, which exhibit small ∣SCD∣.
Fig. 9.
∣SCD∣ for the phospholipid tails of (a) DOPE (b) DPPG and (c) DOPG in the corresponding single-component membranes, and ∣SCD∣ for each phospholipid component, (d) DOPE, (e) DPPG and (f) DPPE, in the Lipid A–Mixture membrane. The values are obtained as described previously.
For the membrane with mixed phospholipids, the average ∣SCD∣ of Lipid A has an intermediate value 0.363 as the result of interactions between Lipid A and a variety of fatty acids. In addition, the ∣SCD∣ of phospholipids in the Lipid A–mixture membrane are 0.174 (DOPE), 0.238 (DPPG), and 0.240 (DPPE), indicating convergence in the ordering of different phospholipids in the mixture leaflet. The combination of unsaturated long chains with saturated short chains as well as the mixture of zwitterionic PE with negatively charged PG reduce the overall ordering of acyl chains in the mixture leaflet. This could be attributed to the pronounced variation in the packing state of chains. Consequently, the profile of DPPE in Fig. 9(f) transforms into a DPPG-like shape, which is significantly different from the profile exhibited in Fig. 2(e). We observe all 16:0 DP molecules demonstrate a similar level of ordering in the mixture leaflet.
Inclination angle of Lipid A
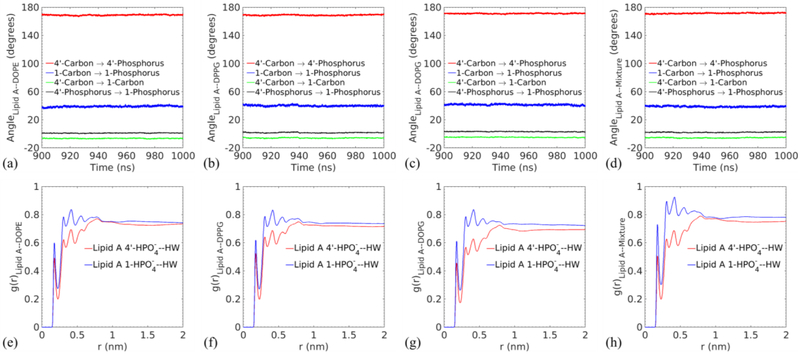
Besides the fluidity, the molecular conformation of Lipid A is also a critical indicator for its bioactivity.38,86 The backbone inclination angle of asymmetric penta-acyl Lipid A influences the position and orientation of the 1 and 4’ phosphates. As shown in Fig. 10(a-d), the averaged angles between the vector from 4’-phosphrous to 1-phosphrous and the membrane surface are 0.976° (DOPE), 1.565° degree (DPPG), 3.032° (DOPG) and 2.052° (Mixture). The positive angle implies that 1-phosphate can be more hydrated than 4’-phosphate, which is buried in the surrounding Lipid A molecules. The RDFs of the phosphates in Fig. 10(e-h) confirm this finding. In particular, the higher peaks in g(r) for 1-phosphate with respect to water hydrogen atoms is induced by stronger interactions between this phosphate and water. The observed inclination is consistent with experimental results.11,86 Notably, signaling molecules such as Pseudomonas Quinolone Signal (PQS) can bind strongly with 4’-phosphate and improve its hydration and mobility.11 Other angles are also provided to further the understanding of the Lipid A conformation. The angle between the membrane surface and the vectors from 4’-carbon to 4’-phosphorous, 1-carbon to 1-phosphorous, and 4’-carbon to 1 phosphorous are respectively around 170 °, 40 °, and −6 ° in Fig. 10(a-d). The 4’ phosphorous group and 1 phosphorous group lift upward from the membrane surface while the disaccharide carbon backbone sinks into the surface at a shallow angle, which corresponds to 4’ carbon at a higher position than 1 carbon.
Fig. 10.
Top panel: time variations in the angle between the selected vectors and the membrane surface for (a) Lipid A–DOPE, (b) Lipid A–DPPG, (c) Lipid A–DOPG, and (d) Lipid A–Mixture membranes. The legends indicate the vectors whose directions are defined by linking the position of the first atom to the second atom. Bottom panel: radial distribution functions between the 1 and 4’ position phosphate groups in Lipid A and the hydrogen atom of water in each membrane (e-h) corresponds to (a-d), respectively.
Conclusions
The antimicrobial-resistant bacterium P. aeruginosa causes severe healthcare-associated infections. This demanding situation drives researchers to probe virulence-related processes, in which the extraordinary outer membrane of Gram-negative bacteria is proven to play a key role. While the macroscopic properties can be captured in great detail through experimental approaches, all-atom molecular dynamics simulation provides an avenue to an atomistic understanding of bacterial outer membranes. Thus, in this work, we systematically investigated the biologically relevant outer membranes of P. aeruginosa using MD simulations. Based on the experimental data for cultured P. aeruginosa, we modeled five sets of membranes, Lipid A with DPPE, Lipid A with DOPE, Lipid A with DPPG, Lipid A with DOPG, and Lipid A with a mixture of DPPE, DOPE and DPPG, which reproduces the physical phospholipid composition of P. aeruginosa. The results reveal the consistency of the Lipid A leaflet over the heterogeneous phospholipid leaflets. The small area per lipid and large deuterium order parameter of Lipid A indicate a leaflet with low fluidity and highly organized molecules. Despite the model limitation, our findings also qualitatively support the view that calcium ions form salt bridges with phosphates in Lipid A and stabilize the moiety. Through further examination, we discovered that the overall hydrated Lipid A leaflet displays solvation differences due to the tilted Lipid A backbone. This inclination causes the 1-phosphate in Lipid A to project toward the outer aqueous environment and the 4’-phosphate to become buried between the acyl chains of Lipid A. As for the phospholipid leaflet, we observed an improvement in the packing and ordering of acyl chains in the asymmetric membranes compared to the pure phospholipid bilayers. While the area per lipid values for pure DOPE, DPPG and DOPG bilayers are 0.65 nm2 (at 266 K),73 0.67 nm2 (at 323 K),59 and 0.73 nm2 (at 323 K),59 respectively, the corresponding values for asymmetric Lipid A–phospholipid membrane are 0.56 nm2, 0.58 nm2, and 0.67 nm2. This could be attributed to the rigidity and ordering of Lipid A molecules, which restrict the fluctuation and movement of the phospholipid layer. We also observe a reduction in the overall ordering of acyl chains of the mixed phospholipids. In particular, the DPPE molecules are significantly less ordered in the mixture. This disruption of the chain ordering is caused by the combination of different phospholipids. In all, these findings of P. aeruginosa outer membranes from the atomistic simulation provide invaluable insight into the physical chemistry of bacteria OMs.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge financial support from the National Institute of Allergy and Infectious Diseases under grant R21AI121848. Generous allocation of computing time was provided by the Watson Data Center at Binghamton University. We also thank the Center for Computational Research at the University at Buffalo for allocating additional computing time for this project through the Virtual Infrastructure for Data Intensive Analysis (VIDIA). The authors also acknowledge Professor Paul R. Chiarot for helpful conversations.
Footnotes
Conflicts of interest There are no conflicts to declare.
References
- (1).Bodey GP; Bolivar R; Fainstein V; Jadeja L Infections Caused by Pseudomonas Aeruginosa. Clin. Infect. Dis 1983, 5 (2), 279–313. [DOI] [PubMed] [Google Scholar]
- (2).Weiner LM; Webb AK; Limbago B; Dudeck MA; Patel J; Kallen AJ; Edwards JR; Sievert DM Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol 2016, 37 (11), 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Rice LB Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis 2008, 197 (8), 1079–1081. [DOI] [PubMed] [Google Scholar]
- (4).Kadurugamuwa JL; Beveridge TJ Virulence Factors Are Released from Pseudomonas Aeruginosa in Association with Membrane Vesicles during Normal Growth and Exposure to Gentamicin: A Novel Mechanism of Enzyme Secretion. J. Bacteriol 1995, 177 (14), 3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Beveridge TJ Structures of Gram-Negative Cell Walls and Their Derived Membrane Vesicles. J. Bacteriol 1999, 181 (16), 4725–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Vella BD; Schertzer JW Understanding and Exploiting Bacterial Outer Membrane Vesicles In Pseudomonas; Ramos J-L, Goldberg JB, Filloux A, Eds.; Springer; Netherlands: Dordrecht, 2015; pp 217–250. [Google Scholar]
- (7).Mashburn LM; Whiteley M Membrane Vesicles Traffic Signals and Facilitate Group Activities in a Prokaryote. Nature 2005, 437 (7057), 422–425. [DOI] [PubMed] [Google Scholar]
- (8).Bomberger JM; MacEachran DP; Coutermarsh BA; Ye S; O’Toole GA; Stanton BA Long-Distance Delivery of Bacterial Virulence Factors by Pseudomonas Aeruginosa Outer Membrane Vesicles. PLoS Pathog. 2009, 5 (4), e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Schertzer J; Whiteley M A Bilayer-Couple Model of Bacterial Outer Membrane Vesicle. MBio 2012, 3 (2), e00297–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Schertzer JW; Boulette ML; Whiteley M More than a Signal: Non-Signaling Properties of Quorum Sensing Molecules. Trends Microbiol. 2009, 17 (5), 189–195. [DOI] [PubMed] [Google Scholar]
- (11).Mashburn-Warren L; Howe J; Garidel P; Richter W; Steiniger F; Roessle M; Brandenburg K; Whiteley M Interaction of Quorum Signals with Outer Membrane Lipids: Insights into Prokaryotic Membrane Vesicle Formation. Mol. Microbiol 2008, 69 (2), 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Elhenawy W; Bording-Jorgensen M; Valguarnera E; Haurat MF; Wine E; Feldman MF LPS Remodeling Triggers Formation of Outer Membrane Vesicles in Salmonella. MBio 2016, 7 (4), e00940–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rangarajan M; Aduse-Opoku J; Hashim A; McPhail G; Luklinska Z; Haurat MF; Feldman MF; Curtis MA LptO (PG0027) Is Required for Lipid A 1-Phosphatase Activity in Porphyromonas Gingivalis W50. J. Bacteriol 2017, 199 (11), e00751–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Raetz CRH; Whitfield C Lipopolysaccharide Endotoxins. Annu. Rev. Biochem 2002, 71 (1), 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zähringer U; Lindner B; Rietschel ET Molecular Structure of Lipid A, the Endotoxic Center of Bacterial Lipopolysaccharides. Adv. Carbohydr. Chem. Biochem 1994, 50, 211–276. [PubMed] [Google Scholar]
- (16).Soares TA; Straatsma TP Assessment of the Convergence of Molecular Dynamics Simulations of Lipopolysaccharide Membranes. Mol. Simul 2008, 34 (3), 295–307. [Google Scholar]
- (17).Abraham T; Schooling SR; Nieh M-P; Kucerka N; Beveridge TJ; Katsaras J Neutron Diffraction Study of Pseudomonas Aeruginosa Lipopolysaccharide Bilayers. J. Phys. Chem. B 2007, 111 (10), 2477–2483. [DOI] [PubMed] [Google Scholar]
- (18).Dias RP; da Hora GCA; Ramstedt M; Soares TA Outer Membrane Remodeling: The Structural Dynamics and Electrostatics of Rough Lipopolysaccharide Chemotypes. J. Chem. Theory Comput 2014, 10 (6), 2488–2497. [DOI] [PubMed] [Google Scholar]
- (19).Wu EL; Engström O; Jo S; Stuhlsatz D; Yeom MS; Klauda JB; Widmalm G; Im W Molecular Dynamics and NMR Spectroscopy Studies of E. Coli Lipopolysaccharide Structure and Dynamics. Biophys. J 2013, 105 (6), 1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ma H; Irudayanathan FJ; Jiang W; Nangia S Simulating Gram-Negative Bacterial Outer Membrane: A Coarse Grain Model. J. Phys. Chem. B 2015, 119 (46), 14668–14682. [DOI] [PubMed] [Google Scholar]
- (21).Ma H; Cummins DD; Edelstein NB; Gomez J; Khan A; Llewellyn MD; Picudella T; Willsey SR; Nangia S Modeling Diversity in Structures of Bacterial Outer Membrane Lipids. J. Chem. Theory Comput 2017, 13 (2), 811–824. [DOI] [PubMed] [Google Scholar]
- (22).Kang Y; Barbirz S; Lipowsky R; Santer M Conformational Diversity of O-Antigen Polysaccharides of the Gram-Negative Bacterium Shigella Flexneri Serotype Y. J. Phys. Chem. B 2014, 118 (9), 2523–2534. [DOI] [PubMed] [Google Scholar]
- (23).Pontes FJS; Rusu VH; Soares TA; Lins RD The Effect of Temperature, Cations, and Number of Acyl Chains on the Lamellar to Non-Lamellar Transition in Lipid-A Membranes: A Microscopic View. J. Chem. Theory Comput 2012, 8 (10), 3830–3838. [DOI] [PubMed] [Google Scholar]
- (24).Nascimento A; Pontes FJS; Lins RD; Soares TA Hydration, Ionic Valence and Cross-Linking Propensities of Cations Determine the Stability of Lipopolysaccharide (LPS) Membranes. Chem. Commun 2014, 50 (2), 231–233. [DOI] [PubMed] [Google Scholar]
- (25).Dias RP; Li L; Soares TA; Alexov E Modeling the Electrostatic Potential of Asymmetric Lipopolysaccharide Membranes: The MEMPOT Algorithm Implemented in DelPhi. J. Comput. Chem 2014, 35 (19), 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lins RD; Straatsma TP Computer Simulation of the Rough Lipopolysaccharide Membrane of Pseudomonas Aeruginosa. Biophys. J 2001, 81 (2), 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lins RD; Vorpagel ER; Guglielmi M; Straatsma TP Computer Simulation of Uranyl Uptake by the Rough Lipopolysaccharide Membrane of Pseudomonas Aeruginosa. Biomacromolecules 2008, 9 (1), 29–35. [DOI] [PubMed] [Google Scholar]
- (28).Kirschner KN; Lins RD; Maass A; Soares TA A Glycam-Based Force Field for Simulations of Lipopolysaccharide Membranes: Parametrization and Validation. J. Chem. Theory Comput 2012, 8 (11), 4719–4731. [DOI] [PubMed] [Google Scholar]
- (29).Ravi HK; Stach M; Soares TA; Darbre T; Reymond J-L; Cascella M Electrostatics and Flexibility Drive Membrane Recognition and Early Penetration by the Antimicrobial Peptide Dendrimer BH1. Chem. Commun 2013, 49 (78), 8821. [DOI] [PubMed] [Google Scholar]
- (30).Jämbeck JPM; Lyubartsev AP Derivation and Systematic Validation of a Refined All-Atom Force Field for Phosphatidylcholine Lipids. J. Phys. Chem. B 2012, 116 (10), 3164–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Jämbeck JPM; Lyubartsev AP An Extension and Further Validation of an All-Atomistic Force Field for Biological Membranes. J. Chem. Theory Comput 2012, 8 (8), 2938–2948. [DOI] [PubMed] [Google Scholar]
- (32).Jämbeck JPM; Lyubartsev AP Another Piece of the Membrane Puzzle: Extending Slipids Further. J. Chem. Theory Comput 2013, 9 (1), 774–784. [DOI] [PubMed] [Google Scholar]
- (33).Kirschner KN; Yongye AB; Tschampel SM; González-Outeiriño J; Daniels CR; Foley BL; Woods RJ GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem 2008, 29 (4), 622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tessier MB; Demarco ML; Yongye AB; Woods RJ Extension of the GLYCAM06 Biomolecular Force Field to Lipids, Lipid Bilayers and Glycolipids. Mol. Simul 2008, 34 (4), 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Cornell WD; Cieplak P; Bayly CI; Gould IR; Merz KM; Ferguson DM; Spellmeyer DC; Fox T; Caldwell JW; Kollman PA A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc 1995, 117 (19), 5179–5197. [Google Scholar]
- (36).Bayly CI; Cieplak P; Cornell WD; Kollman PA A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model. J. Phys. Chem 1993, 97 (40), 10269–10280. [Google Scholar]
- (37).BRANDENBURG K; SEYDEL U Investigation into the Fluidity of Lipopolysaccharide and Free Lipid A Membrane Systems by Fourier-Transform Infrared Spectroscopy and Differential Scanning Calorimetry. Eur. J. Biochem 1990, 191 (1), 229–236. [DOI] [PubMed] [Google Scholar]
- (38).Brandenburg K; Andrä J; Müller M; Koch MHJ; Garidel P Physicochemical Properties of Bacterial Glycopolymers in Relation to Bioactivity. Carbohydr. Res 2003, 338 (23), 2477–2489. [DOI] [PubMed] [Google Scholar]
- (39).Brandenburg K; Seydel U Physical Aspects of Structure and Function of Membranes Made from Lipopolysaccharides and Free Lipid A. Biochim. Biophys. Acta - Biomembr 1984, 775 (2), 225–238. [Google Scholar]
- (40).Berglund NA; Piggot TJ; Jefferies D; Sessions RB; Bond PJ; Khalid S Interaction of the Antimicrobial Peptide Polymyxin B1 with Both Membranes of E. Coli: A Molecular Dynamics Study. PLOS Comput. Biol 2015, 11 (4), e1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Berendsen HJC; van der Spoel D; van Drunen R GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun 1995, 91 (1–3), 43–56. [Google Scholar]
- (42).Lindahl E; Hess B; van der Spoel D GROMACS 3.0: A Package for Molecular Simulation and Trajectory Analysis. J. Mol. Model. 2001, 7 (8), 306–317. [Google Scholar]
- (43).Van Der Spoel D; Lindahl E; Hess B; Groenhof G; Mark AE; Berendsen HJC GROMACS: Fast, Flexible, and Free. J. Comput. Chem 2005, 26 (16), 1701–1718. [DOI] [PubMed] [Google Scholar]
- (44).Hess B; Kutzner C; van der Spoel D; Lindahl E GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput 2008, 4 (3), 435–447. [DOI] [PubMed] [Google Scholar]
- (45).Pronk S; Páll S; Schulz R; Larsson P; Bjelkmar P; Apostolov R; Shirts MR; Smith JC; Kasson PM; van der Spoel D; et al. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29 (7), 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Páll S; Abraham MJ; Kutzner C; Hess B; Lindahl E Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); 2015; Vol. 8759, pp 3–27. [Google Scholar]
- (47).Abraham MJ; Murtola T; Schulz R; Páll S; Smith JC; Hess B; Lindahl E GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar]
- (48).Parrinello M; Rahman A Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys 1981, 52 (12), 7182–7190. [Google Scholar]
- (49).Berendsen HJC; Postma JPM; van Gunsteren WF; DiNola A; Haak JR Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys 1984, 81 (8), 3684–3690. [Google Scholar]
- (50).Hess B; Bekker H; Berendsen HJC; Fraaije JGEM LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem 1997, 18 (12), 1463–1472. [Google Scholar]
- (51).Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG A Smooth Particle Mesh Ewald Method. J. Chem. Phys 1995, 103 (19), 8577–8593. [Google Scholar]
- (52).Erridge C; Bennett-Guerrero E; Poxton IR Structure and Function of Lipopolysaccharides. Microbes Infect. 2002, 4 (8), 837–851. [DOI] [PubMed] [Google Scholar]
- (53).Rietschel ET; Kirikae T; Schade FU; Mamat U; Schmidt G; Loppnow H; Ulmer AJ; Zähringer U; Seydel U; Di Padova F Bacterial Endotoxin: Molecular Relationships of Structure to Activity and Function. FASEB J. 1994, 8 (2), 217–225. [DOI] [PubMed] [Google Scholar]
- (54).TASHIRO Y; INAGAKI A; SHIMIZU M; ICHIKAWA S; TAKAYA N; NAKAJIMA-KAMBE T; UCHIYAMA H; NOMURA N Characterization of Phospholipids in Membrane Vesicles Derived from Pseudomonas Aeruginosa. Biosci. Biotechnol. Biochem 2011, 75 (3), 605–607. [DOI] [PubMed] [Google Scholar]
- (55).López Cascales JJ; Otero TF; Smith BD; González C; Márquez M Model of an Asymmetric DPPC/DPPS Membrane: Effect of Asymmetry on the Lipid Properties. A Molecular Dynamics Simulation Study. J. Phys. Chem. B 2006, 110 (5), 2358–2363. [DOI] [PubMed] [Google Scholar]
- (56).Brandenburg K; Funari SS; Koch MHJ; Seydel U Investigation into the Acyl Chain Packing of Endotoxins and Phospholipids under Near Physiological Conditions by WAXS and FTIR Spectroscopy. J. Struct. Biol 1999, 128 (2), 175–186. [DOI] [PubMed] [Google Scholar]
- (57).Thurmond RL; Dodd SW; Brown MF Molecular Areas of Phospholipids as Determined by 2H NMR Spectroscopy. Comparison of Phosphatidylethanolamines and Phosphatidylcholines. Biophys. J 1991, 59 (1), 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Rand RP; Parsegian VA Hydration Forces between Phospholipid Bilayers. Biochim. Biophys. Acta - Rev. Biomembr 1989, 988 (3), 351–376. [Google Scholar]
- (59).Pan J; Heberle FA; Tristram-Nagle S; Szymanski M; Koepfinger M; Katsaras J; Kučerka N Molecular Structures of Fluid Phase Phosphatidylglycerol Bilayers as Determined by Small Angle Neutron and X-Ray Scattering. Biochim. Biophys. Acta - Biomembr 2012, 1818 (9), 2135–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Kučerka N; Gallová J; Uhríková D; Balgavý P; Bulacu M; Marrink S-J; Katsaras J Areas of Monounsaturated Diacylphosphatidylcholines. Biophys. J 2009, 97 (7), 1926–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Heller H; Schaefer M; Schulten K Molecular Dynamics Simulation of a Bilayer of 200 Lipids in the Gel and in the Liquid Crystal Phase. J. Phys. Chem 1993, 97 (31), 8343–8360. [Google Scholar]
- (62).Seelig J; Niederberger W Deuterium-Labeled Lipids as Structural Probes in Liquid Crystalline Bilayers. Deuterium Magnetic Resonance Study. J. Am. Chem. Soc 1974, 96 (7), 2069–2072. [Google Scholar]
- (63).Seelig J; Seelig A Deuterium Magnetic Resonance Studies of Phospholipid Bilayers. Biochem. Biophys. Res. Commun 1974, 57 (2), 406–411. [DOI] [PubMed] [Google Scholar]
- (64).Leekumjorn S; Sum AK Molecular Studies of the Gel to Liquid-Crystalline Phase Transition for Fully Hydrated DPPC and DPPE Bilayers. Biochim. Biophys. Acta - Biomembr 2007, 1768 (2), 354–365. [DOI] [PubMed] [Google Scholar]
- (65).Brandenburg K; Seydel U Orientation Measurements on Membrane Systems Made from Lipopolysaccharides and Free Lipid A by FT-IR Spectroscopy. Eur. Biophys. J 1988, 16 (2), 83–94. [DOI] [PubMed] [Google Scholar]
- (66).Pottel H; van der Meer W; Herreman W Correlation between the Order Parameter and the Steady-State Fluorescence Anisotropy of 1,6-Diphenyl-1,3,5-Hexatriene and an Evaluation of Membrane Fluidity. BBA - Biomembr. 1983, 730 (2), 181–186. [Google Scholar]
- (67).Heyn MP Determination of Lipid Order Parameters and Rotational Correlation Times from Fluorescence Depolarization Experiments. FEBS Lett. 1979, 108 (2), 359–364. [DOI] [PubMed] [Google Scholar]
- (68).Jahnig F Structural Order of Lipids and Proteins in Membranes: Evaluation of Fluorescence Anisotropy Data. Proc. Natl. Acad. Sci 1979, 76 (12), 6361–6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Kinosita K; Kawato S; Ikegami A A Theory of Fluorescence Polarization Decay in Membranes. Biophys. J 1977, 20 (3), 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Leekumjorn S; Sum AK Molecular Simulation Study of Structural and Dynamic Properties of Mixed DPPC/DPPE Bilayers. Biophys. J 2006, 90 (11), 3951–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Petrov AG; Gawrisch K; Brezesinski G; Klose G; Möps A Optical Detection of Phase Transitions in Simple and Mixed Lipid-Water Phases. Biochim. Biophys. Acta 1982, 690 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- (72).Jacobson K; Papahadjopoulos D Phase Transitions and Phase Separations in Phospholipid Membranes Induced by Changes in Temperature, PH, and Concentration of Bivalent Cations. Biochemistry 1975, 14 (1), 152–161. [DOI] [PubMed] [Google Scholar]
- (73).Gruner SM; Tate MW; Kirk GL; So PTC; Turner DC; Keane DT; Tilcock CPS; Cullis PR X-Ray Diffraction Study of the Polymorphic Behavior of N-Methylated Dioleoylphosphatidylethanolamine. Biochemistry 1988, 27 (8), 2853–2866. [DOI] [PubMed] [Google Scholar]
- (74).Findlay EJ; Barton PG Phase Behavior of Synthetic Phosphatidylglycerols and Binary Mixtures with Phosphatidylcholines in the Presence and Absence of Calcium Ions. Biochemistry 1978, 17 (12), 2400–2405. [DOI] [PubMed] [Google Scholar]
- (75).Kučerka N; Papp-Szabo E; Nieh M-P; Harroun TA; Schooling SR; Pencer J; Nicholson EA; Beveridge TJ; Katsaras J Effect of Cations on the Structure of Bilayers Formed by Lipopolysaccharides Isolated from Pseudomonas Aeruginosa PAO1. J. Phys. Chem. B 2008, 112 (27), 8057–8062. [DOI] [PubMed] [Google Scholar]
- (76).Martinez H; Chacon E; Tarazona P; Bresme F The Intrinsic Interfacial Structure of Ionic Surfactant Monolayers at Water-Oil and Water-Vapour Interfaces. Proc. R. Soc. A Math. Phys. Eng. Sci 2011, 467 (2131), 1939–1958. [Google Scholar]
- (77).Willard AP; Chandler D Instantaneous Liquid Interfaces. J. Phys. Chem. B 2010, 114 (5), 1954–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Allen DT; Saaka Y; Pardo LC; Lawrence MJ; Lorenz CD Specific Effects of Monovalent Counterions on the Structural and Interfacial Properties of Dodecyl Sulfate Monolayers. Phys. Chem. Chem. Phys 2016, 18 (44), 30394–30406. [DOI] [PubMed] [Google Scholar]
- (79).Dominguez H; Berkowitz ML Computer Simulations of Sodium Dodecyl Sulfate at Liquid/Liquid and Liquid/Vapor Interfaces. J. Phys. Chem. B 2000, 104 (22), 5302–5308. [Google Scholar]
- (80).Pandit SA; Bostick D; Berkowitz ML An Algorithm to Describe Molecular Scale Rugged Surfaces and Its Application to the Study of a Water/Lipid Bilayer Interface. J. Chem. Phys 2003, 119 (4), 2199–2205. [Google Scholar]
- (81).Bresme F; Chacón E; Tarazona P Molecular Dynamics Investigation of the Intrinsic Structure of Water–fluid Interfaces via the Intrinsic Sampling Method. Phys. Chem. Chem. Phys 2008, 10 (32), 4704. [DOI] [PubMed] [Google Scholar]
- (82).Ferris FG; Beveridge TJ Physicochemical Roles of Soluble Metal Cations in the Outer Membrane of Escherichia Coli K-12. Can. J. Microbiol 1986, 32 (7), 594–601. [DOI] [PubMed] [Google Scholar]
- (83).Nikaido H Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev 2003, 67 (4), 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Catte A; Girych M; Javanainen M; Loison C; Melcr J; Miettinen MS; Monticelli L; Määttä J; Oganesyan VS; Ollila OHS; et al. Molecular Electrometer and Binding of Cations to Phospholipid Bilayers. Phys. Chem. Chem. Phys 2016, 18 (47), 32560–32569. [DOI] [PubMed] [Google Scholar]
- (85).Petrache HI; Dodd SW; Brown MF Area per Lipid and Acyl Length Distributions in Fluid Phosphatidylcholines Determined by 2H NMR Spectroscopy. Biophys. J 2000, 79 (6), 3172–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Seydel U; Oikawa M; Fukase K; Kusumoto S; Brandenburg K Intrinsic Conformation of Lipid A Is Responsible for Agonistic and Antagonistic Activity. Eur. J. Biochem 2000, 267 (10), 3032–3039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.