Abstract
Fibrolamellar carcinomas are characterized by activation of protein kinase A, a kinase composed of catalytic and regulatory subunits. PRKACA encodes a catalytic subunit of protein kinase A and almost all fibrolamellar carcinomas have a heterozygous 400kb deletion that leads to the fusion of DNAJB1 and PRKACA. The resulting DNAJB1-PRKACA fusion transcript is believed to activate protein kinase A by dysregulation of the catalytic portion of the protein. In contrast, PRKAR1A encodes one of the regulatory subunits of protein kinase A. We hypothesized that loss of function of this regulatory unit could also lead to protein kinase A activation and thus to fibrolamellar carcinoma. Since PRKAR1A mutations underlie the Carney complex, we searched for liver tumors in individuals with the Carney complex.
We identified 3 individuals with fibrolamellar carcinomas and a personal history of the Carney complex. All 3 tumors displayed the typical morphology of fibrolamellar carcinoma and were positive for Arginase, CK7 and CD68. FISH was negative for PRKACA rearrangements. However, PRKAR1A sequencing identified pathogenic mutations in two of two cases with successful sequencing. In addition, all 3 cases were negative for PRKAR1A protein expression, consistent with inactivation of this key regulatory unit of Protein kinase A. We also identified one additional fibrolamellar carcinoma in an individual without a documented history of the Carney Complex that was negative for PRKACA rearrangements but had loss of PRKAR1A protein expression as well as PRKAR1A mutations.
In conclusion, fibrolamellar carcinoma can be part of the Carney complex. In this setting, fibrolamellar carcinomas have inactivating PRKAR1A mutations instead of the DNAJB1-PRKACA fusion gene found in sporadic fibrolamellar carcinomas, providing an alternate means for activation of protein kinase A.
Introduction
Fibrolamellar carcinoma is a unique type of primary liver carcinoma that is characterized by distinctive clinical features, including a predilection for young patients, a lack of underlying chronic liver disease, and normal serum alpha-fetoprotein levels (1). Fibrolamellar carcinoma is also characterized by distinctive histologic morphology, with tumor cells having abundant granular eosinophilic cytoplasm, prominent nucleoli, and abundant intratumoral fibrosis (2). At the immunohistochemical level, fibrolamellar carcinomas continue to show distinctive features, with co-expression of keratin 7 and CD68 (3).
Recently, the somatic fusion gene DNAJB1-PRKACA has been identified as the key molecular event in fibrolamellar carcinoma (4, 5). This fusion gene leads to overexpression of PRKACA, and thus activation of protein kinase A (6, 7). Protein kinase A is a serine threonine kinase tetramer composed of two catalytic and two regulatory subunits. The regulatory subunits can be composed of 4 distinct proteins coded by different genes, while the catalytic subunit can be composed of 3 distinct proteins coded by different genes. PRKACA, a component of the DNAJB1-PRKACA somatic gene fusion, encodes one of the catalytic subunits while PRKAR1A encodes one of the major regulatory subunits. Protein kinase A is activated when cyclic AMP molecules in the cytoplasm bind to the regulatory subunits. The catalytic subunits are then able to phosphorylate serine and threonine residues and, in so doing, activate downstream proteins (8, 9).
In fibrolamellar carcinomas, the fusion protein encoded by DNAJB1-PRKACA results in increased Protein Kinase A activity via over-expression/dysregulation of the catalytic unit, PRKACA (4–7). We hypothesized that loss of function of the regulatory subunit, PRKAR1A, could also lead to Protein kinase A activation and thus to fibrolamellar carcinoma. Since germline PRKAR1A mutations frequently underlie the Carney complex (10, 11), we searched for liver tumors that arose in individuals with the Carney complex.
Methods and Materials
Cases and Controls
After obtaining IRB approval, we reviewed histologic slides of liver tumors that arose in patients with a personal or family history of the Carney complex at the authors’ institutions. Slides of other tumors from the patients were also retrieved and reviewed where possible. Each case was reviewed for diagnostic classification (RPG and MST liver tumors, JJM cardiac tumor) using standard histological criteria. Fibrolamellar carcinomas were characterized by large tumor cells with granular eosinophilic cytoplasm, prominent nucleoli, and intratumoral fibrosis. A representative formalin-fixed paraffin-embedded tissue block from the liver tumor was selected for ancillary immunohistochemistry, fluorescence in situ hybridization (FISH) and reverse transcription polymerase chain reaction (RT-PCR). A control group for immunohistochemistry and FISH was created from five randomly selected sporadic fibrolamellar carcinomas, all of which were known to harbor the DNAJB1-PRKACA fusion gene from prior studies(5).
Immunohistochemistry
Immunostains were performed on all cases and controls using steam antigen retrieval and standard techniques. Immunostains were performed for Keratin 7 (1:100; Clone OV-TL 12/30; Dako; CA), CD68 (1: 1500; Clone KP1; Dako, CA), Arginase (predilute; Clone SP156; Cell Marque, CA) and PRKAR1A (1:2000; Clone OTi6C7; Origene, MD).
FISH
Break apart FISH for rearrangement of the PRKACA locus was performed as previously described in all cases and controls (5). Normal and malignant hepatocytes can show polyploidy and thus in many cases will show >2 probe signals. To determine if there was PRKACA locus or chromosomal 19 copy number abnormalities, an enumerator FISH probe strategy was performed on three cases with sufficient tissue.
PRKAR1A sequencing
Formalin-fixed paraffin-embedded tumor tissue was macrodissected for tumor DNA extraction using the QiaAmp kit. After quantitation, the DNA was used for library preparation. Next generation sequencing of the coding regions and splice junctions of PRKAR1A was performed on tumor DNA from all cases using a targeted custom assay. In order to be considered valid, each variant needed to be supported by a minimum of 30 reads with a total read depth of more than 1000, an allele frequency of >10% and meet American College of Medical Genetics criteria for very strong evidence of pathogenicity or be predicted pathogenic by each of 3 prediction algorithms (PolyPhen, SIFT and Mutation Taster).
Results
Three individuals were identified who had both a history of the Carney complex and a liver tumor (Table 1). The patients were 7, 14 and 53 years of age at the diagnosis of the liver tumor and included 1 man and 2 women. The patients had other tumors including a hepatic adenoma, cardiac myxoma, pituitary adenoma and adrenocortical disease. These additional tumors were identified prior to (case 1) or at the same time as the diagnosis of the fibrolamellar carcinomas (cases 2 and 3).
Table 1:
Clinical characteristics of patients with the Carney complex and liver tumors
| Case | Age | Sex | Tx size (cm) |
Carney complex |
Other tumors |
|---|---|---|---|---|---|
| 1 | 7 | M | N/A | Yes | Pituitary adenoma and primary pigmented adrenocortical disease |
| 2 | 14 | F | 7.4 | Yes | Hepatocellular adenoma |
| 3 | 53 | F | 9.0 | Yes | Cardiac myxoma |
Histologically, each of the fibrolamellar carcinomas showed typical features, being composed of large eosinophilic neoplastic cells that had nuclei with open chromatin and prominent nucleoli (Figure 1). Each also showed abundant intratumoral fibrosis. In addition, each of the cases showed the characteristic immunophenotype of fibrolamellar carcinoma - expression of Arginase, Keratin 7 (Figure 1) and CD68. In each of the three Carney complex-associated cases, FISH was negative for the PRKACA rearrangement that is found in sporadic fibrolamellar carcinomas, instead showing intact PRKACA loci. PRKACA locus amplification is rare in fibrolamellar carcinomas, being found in less than 1% of cases,(12) but to investigate this possibility, additional FISH studies were performed. In case 1, 36% of tumor nuclei showed gains in the PRKACA locus as well as chromosome 19, consistent with aneusomy, while cases 2 and 4 showed normal PRKACA copy numbers. Case number 3 had insufficient tissue for testing.
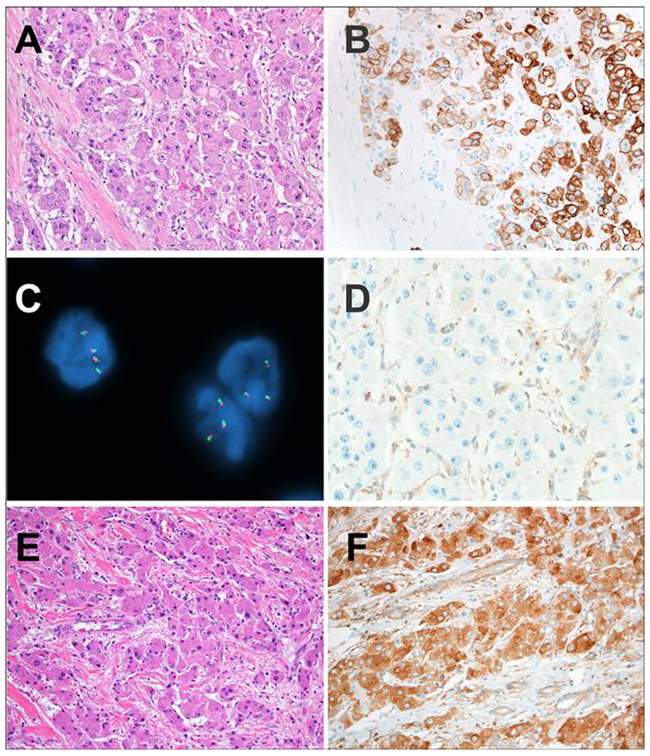
Figure 1 –
A: The liver tumors in the patients with the Carney complex were fibrolamellar carcinomas. The classic histologic features of fibrolamellar carcinoma are shown in this case including eosinophilic cells with abundant cytoplasm and prominent macronucleoli. Pale bodies were seen. Lamellar bands of fibrosis traversed the tumor.
B: The tumor cells of fibrolamellar carcinoma in patients with the Carney complex were positive for Keratin 7, as expected for fibrolamellar carcinoma
C: PRKACA break apart FISH revealed intact PRKACA loci with closely opposed red and green signals in Carney complex-associated fibrolamellar carcinomas. This finding indicates absence of PRKACA rearrangement in the tumor cells.
D: PRKAR1A expression was absent in the tumor cells of fibrolamellar carcinomas in patients with the Carney complex. The Kupffer cells served as an internal control and revealed retained expression.
E: Sporadic control fibrolamellar carcinomas also showed the classic morphology of fibrolamellar carcinoma.
F: PRKAR1A expression was retained in the neoplastic cells of all sporadic control fibrolamellar carcinomas.
No PRKAR1A protein expression was found in the tumors by immunostaining, with intact expression within Kupffer cells, which served as internal controls (Figure 1D). By contrast, the control group of sporadic fibrolamellar cases (Figure 1E) all showed positive expression of PRKAR1A protein (Figure 1F). The background non-neoplastic liver was available for review in two cases of fibrolamellar carcinoma in individuals with the Carney complex. In both cases, the background liver was histologically normal and showed intact PRKAR1A protein expression by immunohistochemistry (Figure 2C).
Figure 2 –
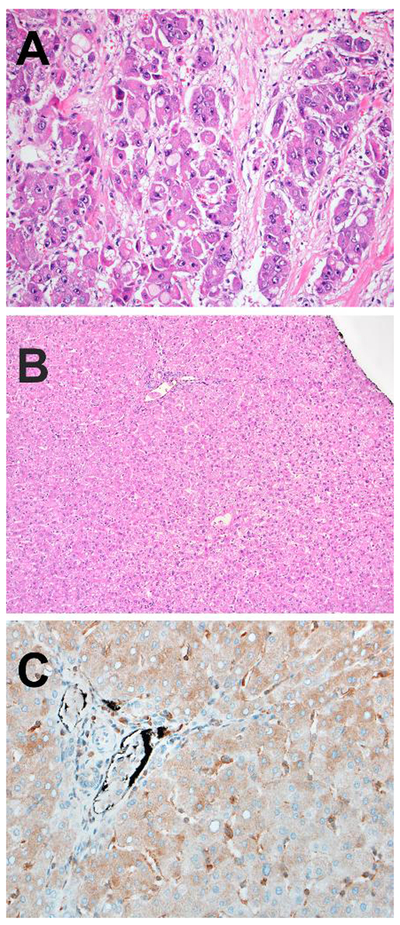
A: Another case of fibrolamellar carcinoma in a patient with the Carney complex showing numerous pale bodies and the characteristic features of fibrolamellar carcinoma.
B: The corresponding background non-neoplastic liver was histologically normal on routine histology in Carney complex-associated cases.
C: PRKAR1A expression was retained in the background liver from this patient with the Carney complex.
PRKAR1A sequencing of fibrolamellar carcinomas identified deleterious mutations in PRKAR1A in cases 1, 2 and 4. In each case, there was at least one truncating mutation that would lead to premature termination of the PRKAR1A protein. In accordance with HUGO Gene Nomenclature Committee guidelines, an asterisk identifies these cases (Table 2). The mutations are predicted to have deleterious effects on protein function by three different mutation effect prediction algorithms (PolyPhen, SIFT and Mutation Taster). In case 1, only a single mutation was identified, however, the possibility of a large insertion or deletion cannot be excluded using this next generation sequencing analysis. Sanger sequencing confirmed the presence of this mutation.
Table 2:
Sequencing Results for PRKAR1A-deficient Fibrolamellar Carcinoma with Corresponding Patient Demographics.
| Case | Age | Gender | PRKAR1A protein expression |
Tumor purity in sequenced tissue |
PRKAR1A Mutations | allele frequency |
|---|---|---|---|---|---|---|
| 1 | 7 | M | loss | 40-50% | c.286C>T; p.R96* | 58% |
| 2 | 14 | F | loss | 40-50% | c. 101-105del; p.S34Cfs*9 c.1055G>T; p.R352L |
62% 28% |
| 3 | 53 | F | loss | 30-40% | NA | NA |
| 4 | 68 | M | loss | 20-30% | Mutation 1: c.709-1G>A; splicing mutation Mutation 2:c.11delG; p.G4fs* |
17% 10% |
NA: A possible c.626C>T; p.T209I was found at a low allele frequency of 3%, but there was insufficient DNA to repeat sequencing.
The slides from the hepatocellular adenoma (case 2) and cardiac myxoma (case 3) were both reviewed. Aspects of both lesions were described in prior publications (13), (14), (15). The hepatocellular adenoma showed a myxoid pattern (13), a rare and recently described morphology for hepatic adenomas (16). The cardiac myxoma showed typical morphologic features and the expected immunolabeling with calretinin (Figure 3). PRKAR1A protein expression was lost in the neoplastic cells of the hepatocellular adenoma and the cardiac myxoma (Figure 3).
Figure 3–
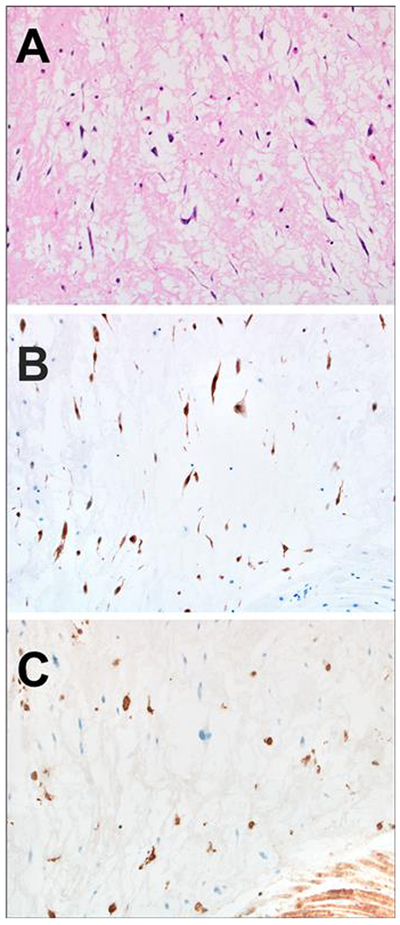
A: The cardiac myxoma was diagnosed at the same time as the fibrolamellar carcinoma in this case and shows neoplastic cells characterized by spindle cytoplasmic projections that imparted a spider-like appearance
B: Calretinin was positive in the spindle shaped neoplastic cells, as expected for the diagnosis of cardiac myxoma
C: PRKAR1A expression was absent in the neoplastic cells. Inflammatory cells and cardiac muscle served as positive internal controls.
In the course of this study, a 4th case was identified of a histologically classic fibrolamellar carcinoma, with CK7 and CD68 co-expression, but without PRKACA rearrangement and instead loss of PRKAR1A expression. This 4th case affected a 68-year-old man with no history of the Carney complex or other tumors. Unfortunately, the patient was lost to follow up and no additional clinical data were available for him. PRKAR1A sequencing revealed two mutations predicted to be pathogenic in this case.
Discussion
The data from this study have two important potential implications for Carney complex patients. First, fibrolamellar carcinoma appears to be part of the Carney complex. Of note, a prior study provides an additional independent observation of fibrolamellar carcinoma that developed in the context of the Carney complex (17). Danoff et al reported a Carney complex kindred containing one individual (Number 5) with fibrolamellar carcinoma. This patient was the only individual in the family who developed fibrolamellar carcinoma or a liver tumor. This observation suggests the expressivity of liver tumors in the Carney complex is variable and less common than endocrine tumors. An editorial by Dr. Carney accompanied this article and raised the possibility that fibrolamellar carcinomas could be part of the Carney complex (18). The current study supports that fibrolamellar carcinomas are indeed part of the Carney complex and identifies the underlying molecular lesion.
Second, these data suggest that patients with the Carney complex may benefit from screening for fibrolamellar carcinoma, to facilitate early detection. Radiologic screening is necessary, since fibrolamellar carcinoma is not associated with elevated serum alpha fetoprotein and no other diagnostic serum markers are validated for clinical use at this time (1).
Screening may additionally be helpful in detecting hepatic adenomas, which also have been reported in individuals with the Carney complex. The presence of hepatic adenomas in the Carney complex suggests the loss of PRKAR1A protein expression may play role in the biology of at least a subset of hepatic adenomas. Of note, the PRKAR1A deficient hepatic adenoma in this study had a distinctive and rare morphology, termed a myxoid hepatic adenoma. An additional hepatic adenoma has been reported, arising in a 26 year old woman with the Carney complex (19), but there is insufficient histological data to determine the morphological or molecular subtype. A further report of a hepatic adenoma in the setting of the Carney complex also lacked clinical and histological data (20). Thus, at this time it is unclear if PRKAR1A-deficient hepatic adenomas consistently have the distinctive morphology of a myxoid hepatic adenoma. Myxoid hepatic adenomas can also be sporadic (16), but it is unclear if sporadic myxoid adenomas are also driven by protein kinase A activation.
The presence of hepatic adenomas in individuals with the Carney complex suggests an additional basic biological observation: while protein kinase A activation appears to be the central biological driver of fibrolamellar carcinomas, other hepatic tumors might also be driven by protein kinase A over expression. This observation suggests that additional unique genetic/epigenetic changes are present in fibrolamellar carcinomas compared to hepatic adenomas.
The presence of fibrolamellar carcinoma as part of the Carney Complex reinforces the core understanding that fibrolamellar carcinomas are driven by protein kinase A activation. In the setting of the Carney complex, fibrolamellar carcinomas do not have the characteristic DNAJB1-PRKACA that is found in sporadic fibrolamellar carcinomas. Instead, protein kinase A is activated by an alternative method through the loss of the regulatory unit, PRKAR1A, as inactivating mutations lead to the loss of PRKAR1A protein expression. These PRKAR1A-deficient fibrolamellar carcinomas are histologically and immunophenotypically similar to fibrolamellar carcinomas with the DNAJB1-PRKACA fusion gene, except in regard to PRKAR1A expression. There is insufficient clinical follow-up at this point to know if the age at presentation and the prognosis is similar between these two groups of fibrolamellar carcinoma. Of the 360 cases of hepatocellular carcinoma evaluated in The Cancer Genome Atlas, there are no cases of PRKAR1A-deficient hepatocellular carcinomas (21). However, evidence of the tumorigenic potential of PRKAR1A loss is seen in a prkar1a-knockout mouse model, wherein 5 of 12 mice with tumors developed hepatocellular tumors (22).
The DNAJB1-PRKACA fusion gene characterizes essentially all cases of previously reported sporadic fibrolamellar carcinomas that have complete histological confirmation (4, 5, 7, 14, 23). The fusion gene is formed from a heterozygous ~400kb deletion and encodes a chimeric protein that retains the kinase domain of PRKACA (4). PRKACA encodes the catalytic subunit of Protein kinase A. Expression of the fusion gene results in activation of Protein kinase A and its downstream targets (6, 7). For example, the mRNA of several Protein kinase A downstream targets are activated in fibrolamellar carcinomas, including CREB3L, ETV1, and ERBB2 (6). However, the current study also identified a case of fibrolamellar carcinoma characterized by PRKAR1A loss in a 68-year-old man without a definite history of the Carney complex. This raises the possibility of very rare sporadic fibrolamellar carcinomas without the classic DNAJB1-PRKACA fusion gene, but with PRKAR1A loss (due to biallelic PRKAR1A inactivation). However, the clinical information available was insufficient to determine if this individual had the Carney complex. Nonetheless, the phenomenon of sporadic biallelic PRKAR1A inactivation has been described in cardiac myxomas (24) and remains a possibility in rare cases of sporadic fibrolamellar carcinomas.
The mutations identified in cases 1 and 2 have been previously identified as germline pathogenic alterations in the Carney complex. The remaining identified alterations have not been reported in the human gene mutation database (HGMD), but it is known that the Carney complex is frequently caused by a variety of germline small insertions or deletions. In the current study, missense mutations in PRKAR1A led to loss of protein expression. The precise mechanism is not evident from our data, but possibilities include instability of the mutated RNA transcript, loss of the epitope to which the antibody binds, or post-translational changes. Promoter methylation is also possible.
In conclusion, fibrolamellar carcinomas can be part of the Carney complex. In this setting, tumors lack the classic DNAJB1-PRKACA fusion gene and instead have protein kinase A activation by loss of PRKAR1A. Individuals with the Carney complex may benefit from screening for fibrolamellar carcinoma and other hepatic tumors.
Acknowledgements
The authors would like to thank the Division of Anatomic Pathology Research Committee for funding support and the Mayo Clinic Medical Genome Facility for excellent technical assistance.
Footnotes
Disclosure: The authors have no financial conflicts to disclose.
Disclosure – Some of the data presented herein were presented at the United States and Canadian Academy of Pathology Annual Meeting, San Antonio, TX in 2017
References:
- 1.Graham RP, Torbenson MS. Fibrolamellar carcinoma: A histologically unique tumor with unique molecular findings. Semin Diagn Pathol 2016. [DOI] [PubMed] [Google Scholar]
- 2.Craig JR, Peters RL, Edmondson HA, Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer 1980;46:372–379. [DOI] [PubMed] [Google Scholar]
- 3.Ross HM, Daniel HD, Vivekanandan P, Kannangai R, Yeh MM, Wu TT, Makhlouf HR, et al. Fibrolamellar carcinomas are positive for CD68. Mod Pathol 2011;24:390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim II, Gleason CE, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science 2014;343:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham RP, Jin L, Knutson DL, Kloft-Nelson SM, Greipp PT, Waldburger N, Roessler S, et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod Pathol 2015;28:822–829. [DOI] [PubMed] [Google Scholar]
- 6.Simon EP, Freije CA, Farber BA, Lalazar G, Darcy DG, Honeyman JN, Chiaroni-Clarke R, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc Natl Acad Sci U S A 2015;112:E5916–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinh TA, Vitucci EC, Wauthier E, Graham RP, Pitman WA, Oikawa T, Chen M, et al. Comprehensive analysis of The Cancer Genome Atlas reveals a unique gene and non-coding RNA signature of fibrolamellar carcinoma. Sci Rep 2017;7:44653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnham RE, Scott JD. Protein kinase A catalytic subunit isoform PRKACA; History, function and physiology. Gene 2016;577:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beavo JA, Bechtel PJ, Krebs EG. Activation of protein kinase by physiological concentrations of cyclic AMP. Proc Natl Acad Sci U S A 1974;71:3580–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirschner LS, Carney JA, Pack SD, Taymans SE, Giatzakis C, Cho YS, Cho-Chung YS, et al. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nat Genet 2000;26:89–92. [DOI] [PubMed] [Google Scholar]
- 11.Carney JA, Gordon H, Carpenter PC, Shenoy BV, Go VL. The complex of myxomas, spotty pigmentation, and endocrine overactivity. Medicine (Baltimore) 1985;64:270–283. [DOI] [PubMed] [Google Scholar]
- 12.Graham RP, Yeh MM, Lam-Himlin D, Roberts LR, Terracciano L, Cruise MW, Greipp PT, et al. Molecular testing for the clinical diagnosis of fibrolamellar carcinoma. Mod Pathol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terracciano LM, Tornillo L, Avoledo P, Von Schweinitz D, Kuhne T, Bruder E. Fibrolamellar hepatocellular carcinoma occurring 5 years after hepatocellular adenoma in a 14-year-old girl: a case report with comparative genomic hybridization analysis. Arch Pathol Lab Med 2004;128:222–226. [DOI] [PubMed] [Google Scholar]
- 14.Graham RP, Terracciano LM, Meves A, Vanderboom PM, Dasari S, Yeh MM, Torbenson MS, et al. Hepatic adenomas with synchronous or metachronous fibrolamellar carcinomas: both are characterized by LFABP loss. Mod Pathol 2016;29:607–615. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez-Cantu YM, Rodriguez-Padilla C, Tena-Suck ML, Garcia de la Fuente A, Mejia-Banuelos RM, Diaz Mendoza R, Quintanilla-Garza S, et al. Synchronous Fibrolamellar Hepatocellular Carcinoma and Auricular Myxoma. Case Rep Pathol 2015;2015:241708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salaria SN, Graham RP, Aishima S, Mounajjed T, Yeh MM, Torbenson MS. Primary hepatic tumors with myxoid change: morphologically unique hepatic adenomas and hepatocellular carcinomas. Am J Surg Pathol 2015;39:318–324. [DOI] [PubMed] [Google Scholar]
- 17.Danoff A, Jormark S, Lorber D, Fleischer N. Adrenocortical micronodular dysplasia, cardiac myxomas, lentigines, and spindle cell tumors. Report of a kindred. Arch Intern Med 1987;147:443–448. [PubMed] [Google Scholar]
- 18.Carney JA. The complex of myxomas, spotty pigmentation, and endocrine overactivity. Arch Intern Med 1987;147:418–419. [PubMed] [Google Scholar]
- 19.Gennari M, Stratakis CA, Hovarth A, Pirazzoli P, Cicognani A. A novel PRKAR1A mutation associated with hepatocellular carcinoma in a young patient and a variable Carney complex phenotype in affected subjects in older generations. Clin Endocrinol (Oxf) 2008;69:751–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stratakis CA, Kirschner LS, Carney JA. Clinical and molecular features of the Carney complex: diagnostic criteria and recommendations for patient evaluation. J Clin Endocrinol Metab 2001;86:4041–4046. [DOI] [PubMed] [Google Scholar]
- 21.Atlas TCG. In: http://cancergenome.nih.gov/.
- 22.Veugelers M, Wilkes D, Burton K, McDermott DA, Song Y, Goldstein MM, La Perle K, et al. Comparative PRKAR1A genotype-phenotype analyses in humans with Carney complex and prkar1a haploinsufficient mice. Proc Natl Acad Sci U S A 2004;101:14222–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham RP, Garcia JJ, Greipp PT, Barr Fritcher EG, Kipp BR, Torbenson MS. FGFR1 and FGFR2 in fibrolamellar carcinoma. Histopathology 2016;68:686–692. [DOI] [PubMed] [Google Scholar]
- 24.Maleszewski JJ, Larsen BT, Kip NS, Castonguay MC, Edwards WD, Carney JA, Kipp BR. PRKAR1A in the development of cardiac myxoma: a study of 110 cases including isolated and syndromic tumors. Am J Surg Pathol 2014;38:1079–1087. [DOI] [PubMed] [Google Scholar]