Abstract
Menthae Haplocalycis herba, one kind of Chinese edible herbs, has been widely utilized for the clinical use in China for thousands of years. Over the last decades, studies on chemical constituents of Menthae Haplocalycis herba have been widely performed. However, less attention has been paid to non-volatile components which are also responsible for its medical efficacy than the volatile constituents. Therefore, a rapid and sensitive method was developed for the comprehensive identification of the non-volatile constituents in Menthae Haplocalycis herba using ultra-high performance liquid chromatography coupled with linear ion trap-Orbitrap mass spectrometry (UHPLC-LTQ-Orbitrap). Separation was performed with Acquity UPLC® BEH C18 column (2.1 mm × 100 mm, 1.7 μm) with 0.2% formic acid aqueous solution and acetonitrile as the mobile phase under gradient conditions. Based on the accurate mass measurement (<5 ppm), MS/MS fragmentation patterns and different chromatographic behaviors, a total of 64 compounds were unambiguously or tentatively characterized, including 30 flavonoids, 20 phenolic acids, 12 terpenoids and two phenylpropanoids. Finally, target isolation of three compounds named Acacetin, Rosmarinic acid and Clemastanin A (first isolated from Menthae Haplocalycis herba) were performed based on the obtained results, which further confirmed the deduction of fragmentation patterns and identified the compounds profile in Menthae Haplocalycis herba. Our research firstly systematically elucidated the non-volatile components of Menthae Haplocalycis herba, which laid the foundation for further pharmacological and metabolic studies. Meanwhile, our established method was useful and efficient to screen and identify targeted constituents from traditional Chinese medicine extracts.
Keywords: Menthae Haplocalycis herba, UHPLC-ESI-MS/MS, non-volatile constituents, target isolation
1. Introduction
As there is growing interest in the use of traditional Chinese medicines (TCMs), systematic screening and identification of chemical components is essential for revealing the material basis of their therapeutic effects and ensuring their safety [1]. However, TCM extracts composed of multi-components are difficult to be comprehensively analyzed. Ultra-high performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry (UHPLC-ESI-MS/MS) has been widely used as a powerful means for the analysis of multi-components in TCMs. Recently, with the development of various data acquisition methods, high-resolution mass spectrometry (HRMS), especially linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap MS), has exhibited excellent performances in constituents detection owing to its high-speed and detection sensitivity [2]. UHPLC provides effective chromatographic separation, while LTQ-Orbitrap provides multi-stage mass spectra using data-dependent analysis with a higher mass resolution and mass accuracy than many other mass spectrometers [2]. Therefore, UHPLC-ESI-MS/MS remarkably facilitates the identification of known and unknown components in TCM extracts with high sensitivity and accuracy [3,4,5].
Menthae Haplocalycis herba, one popular TCM, derived from the aerial part (stem and leaf) of Menthae haplocalyx Briq., is commonly used for the treatment of wind-heat cold, pharyngitis, measles, rubella in dispelling wind and heat. It has been officially documented in Chinese Pharmacopoeia (Version 2015) named “Bo He” and used as a Chinese edible herb [6]. In the past few decades, systematic chemical and pharmacological studies have been performed mainly on the volatile constituents [7,8]. However, studies in recent years have suggested that volatile constituents of Menthae Haplocalycis herba can lead to a series of toxic effects, such as liver injury and other toxic symptoms [9,10]. It is well known that Menthae Haplocalycis herba is clinically used as decoction, which indicates that non-volatile components should be responsible for its efficacy. For instance, flavonoids, phenolic acids and some terpenoids from Menthae Haplocalycis herba show various activities of anti-viral, anti-inflammatory and anti-oxidation, which is probably closely related to its traditional efficacy [11,12]. Limited knowledge about the non-volatile constituents hinders its application in clinical practice and retards its modernization process. Hence, it will be of great importance to characterize the non-volatile constituents in Menthae Haplocalycis herba.
Herein, a rapid and sensitive UHPLC-ESI-MS/MS method was established to systematically profile the non-volatile constituents in Menthae Haplocalycis herba, which may contribute to new drug development and effective substance basis clarification.
2. Results and Discussion
Positive ion mode was employed for the comprehensive analysis and the base peak chromatograms (BPC) are shown in Figure 1. Based on MS spectra and retention time (tR), a total of 64 compounds (Table 1, Figure 2) were unambiguously identified or tentatively characterized. Among them, 10 constituents were positively identified by comparing retention times and MS data with respective reference compounds. The representative mass spectra of chemical constituents identified in Menthae Haplocalycis herba in positive ion mode were displayed in Figure 3 as well.
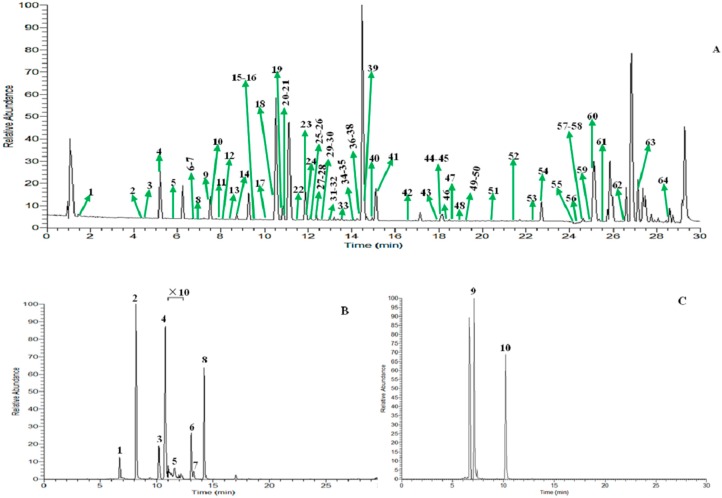
Figure 1.
(A) Base peak chromatogram of Menthae Haplocalycis herba in positive ion mode; (B) base peak chromatogram of mixed reference solution of Menthae Haplocalycis herba. (1. Bohecineole A, 2. Luteolin-7-O-glucoside, 3. Diosmin, 4. Hesperidin, 5. Rosmarinic acid, 6. Lithospermic Acid, 7. Salvianolic acid B, 8. Buddleoside); (C) base peak chromatogram of mixed reference solution of Menthae Haplocalycis herba. (9. (1R,2R,4S)-trans-1,8-cineole-2-O-β-d-glucopyranoside, 10. Naringin). “×10” magnified ten-fold.
Table 1.
Summary of chemical constituents identified in Menthae Haplocalycis herba by UHPLC-ESI-MS/MS.
| Peak | tR (min) | Compound Formula | Identification | Experimental Mass m/z | Theoretical Mass m/z | Mass Error (× 10−6) | MS2 Data(Measured) | |
|---|---|---|---|---|---|---|---|---|
| 1 | PA1 | 1.52 | C7H7O2 | Benzoic acid | 123.04371 | 123.04405 | −2.81 | 95(100),82(2),81(16),67(3),57(2) |
| 2 | T1 | 4.41 | C16H26O8Na b | Petroside | 369.15112 | 369.15198 | −2.35 | 352(7),351(47),328(27),307(12),285(9),207(100),203(34),185(22),149(6) |
| 3 | T2 | 4.63 | C10H17O3 | (1R*,2S*)-1,2-dihydroxy-ρ-menth-4(8)-en-3-one | 185.11687 | 185.11722 | −1.89 | 168(4),167(100),157(6),149(18),139(19),125(32),121(26) |
| 4 | T3 | 5.21 | C16H28O8Na b | (2R,3R,4S,5S,6R)-2-(((1S,2R,3S)-2,3-dihydroxy-3-methyl-6-(propan-2-ylidene)cyclohexyl)oxy)-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol | 371.16702 | 371.16763 | −1.66 | 355(2),353(0.2),203(100),201(6),193(4),191(2) |
| 5 | T4 | 5.82 | C16H30O8Na b | Bohenoside A | 373.18250 | 373.18328 | −2.11 | 357(11),356(10),355(100),346(4),211(4) |
| 6 | T5 | 6.71 | C10H17O2 | Bohecineole A a | 169.12190 | 169.12230 | −2.40 | 151(71),133(7),123(86),113(5),109(56),107(100),58(4) △ |
| 6.75 | C10H16O2Na b | 191.10388 | 191.10425 | −1.94 | ||||
| 7 | P1 | 6.71 | C25H32O11Na b | Clemastanin A | 531.18329 | 531.18368 | −0.74 | 514(10),513(100),502(4),501(46),369(9),351(3) |
| 8 | PA2 | 7.01 | C9H11O3 | Phloretic acid | 167.06995 | 167.07027 | −1.92 | 149(100),139(23),126(42),125(57),121(40),109(10),107(49),97(10) |
| 9 | T6 | 7.11 | C16H28O7Na b | (1R,2R,4S)-trans-1,8-cineole-2-O-β-d-glucopyranoside a | 355.17206 | 355.17272 | −1.87 | 337(14),285(100),268(53),193(6),185(9),135(2) △ |
| 7.30 | 355.17209 | −1.78 | 337(12),285(3),267(36),257(2),203(21),201(100),193(4),185(2) | |||||
| 10 | P2 | 7.62 | C26H32O12Na b | (+)-1-Hydroxypinoresinol-1-O-β-d-glucoside | 559.17682 | 559.17859 | −3.17 | 541(19),536(4),515(28),437(7),398(9),397(100),396(15),395(65),337(67),309(24) |
| 11 | F1 | 7.95 | C27H31O15 | Luteolin-7-O-rutinoside | 595.16522 | 595.16574 | −0.88 | 577(0.3),549(0.4),449(100),287(12) |
| 12 | F2 | 8.19 | C21H21O11 | Luteolin-7-O-glucoside a | 449.10611 | 449.10783 | −3.84 | 287(100) △ |
| 8.16 | 449.10635 | −3.31 | 431(3),416(1),287(100),269(2) | |||||
| 13 | F3 | 8.45 | C21H19O12 | 2-(2,5-Dihydroxyphenyl)-5-hydroxy-4-oxo-4H-chromen-7-yl β-d-glucopyranosiduronic acid | 463.08578 | 463.08710 | −2.85 | 445(0.3),427(0.2),375(0.2),287(100) |
| 14 | T7 | 8.75 | C19H34O7Na b | Linarionoside B | 397.21912 | 397.21967 | −1.39 | 379(100),326(28),217(26),203(88),187(30) |
| 15 | F4 | 9.48 | C27H31O14 | Apigenin-7-O-rutinoside | 579.16974 | 579.17083 | −1.88 | 561(1),433(100),417(1),271(9) |
| 16 | F5 | 10.23 | C27H33O14 | Naringin a | 581.18488 | 581.18648 | −2.75 | 563(59),545(50),527(30),509(11),435(99),419(100),417(92),401(15),383(9),315(27),273(30) △ |
| 9.58 | 581.18573 | −1.29 | 563(20),545(12),527(7),419(100),273(24) | |||||
| 17 | F6 | 10.19 | C16H13O6 | Chrysoeriol | 301.07007 | 301.07066 | −1.97 | 286(6),286(100),270(0.49),255(3),183(0.5),121(0.23) |
| 18 | F7 | 10.35 | C28H33O15 | Diosmin a | 609.17963 | 609.18139 | −2.90 | 591(1),573(0.2),463(100),447(1),301(11) △ |
| 10.50 | 609.17999 | −2.30 | 591(0.1),463(100),447(1),429(0.3),301(11),286(0.1),258 | |||||
| 19 | PA3 | 10.64 | C36H30O16Na | Sodium salvianolic acid E | 741.14203 | 741.14260 | −0.77 | 723(1),561(100),543(28),381(3),363(4),345(2),317(2) |
| 20 | F8 | 10.77 | C28H35O15 | Hesperidin a | 611.19617 | 611.19704 | −1.43 | 593(36),575(34),491(12),489(25),465(97),449(100),447(75),431(26),345(30),303(45) △ |
| 10.92 | 611.19635 | −1.14 | 593(19),575(33),557(14),491(10),489(21),465(62),449(100),447(59),431(22),345(17),303(42) | |||||
| 21 | F9 | 10.98 | C16H15O6 | Homoeriodictyol | 303.08563 | 303.08630 | −2.25 | 285(13),179(46),177(100),153(28),151(5),136(1) |
| 22 | PA4 | 11.53 | C9H9O4 | 2,4-Dihydroxycinnamic acid | 181.04900 | 181.04953 | −2.95 | 163(100),153(0.4),139(3),137(0.2),135(2),121(0.1),117(0.1) |
| 23 | PA5 | 11.56 | C18H17O8 | Rosmarinic acid a | 361.09134 | 361.09179 | −1.25 | 347(10),343(24),333(7),328(12),293(11),263(9),191(13),164(5),163(100),145(7) △ |
| 11.84 | 361.09122 | −1.58 | ||||||
| 24 | F10 | 11.94 | C21H19O12 | Luteolin-7-O-glucuronide | 463.08563 | 463.08710 | −3.17 | 445(0.5),427(0.4),405(0.3),288(1),287(100) |
| 25 | PA6 | 12.14 | C27H23O12 | Cis-salvianolic acid J | 539.11725 | 539.11840 | −2.13 | 539(7),521(7),495(6),493(6),393(5),341(100),323(3),297(8),295(9),179(5) |
| 26 | PA7 | 12.28 | C18H15O8 | Bis(3,4-dihydroxybenzylidene)succinic acid | 359.07587 | 359.07614 | −0.76 | 341(100),323(3),313(34),297(4),295(10),285(7),221(15),179(1),123(16) |
| 27 | PA8 | 12.28 | C36H30O16Na | Sodium lithospermate B | 741.14203 | 741.14260 | −0.77 | 723(0.3),579(100),561(25),533(19),517(19),381(4),355(3) |
| 28 | PA9 | 12.39 | C18H13O7 | 2,4,12-Trihydroxy-7-oxo-8,9-dihydro-7H-benzo[f]naphtho[1,8-bc]oxepine-8-carboxylic acid | 341.06497 | 341.06557 | −1.78 | 323(96),313(18),297(63),295(100),279(18),277(22),267(2),253(12),249(18) |
| 29 | PA10 | 12.61 | C36H30O16Na | Sodium lithospermate B | 741.14233 | 741.14260 | −0.37 | 579(100),561(27),533(25),517(20),399(5),355(3) |
| 30 | T8 | 12.65 | C16H28O7Na b | (1R,2S,5R)-(−)-methol β-d-Glucuronide | 355.17181 | 355.17272 | −2.57 | 340(7),339(3),337(11),325(5),323(13),309(4),295(6),285(13),267(100),257(16),205(6),183(3) |
| 31 | PA11 | 13.04 | C36H30O16Na | Sodium lithospermate B | 741.14197 | 741.14260 | −0.85 | 561(100),543(59),517(23),363(11),362(19),319(3) |
| 32 | PA12 | 13.06 | C27H23O12 | Lithospermic acid a | 539.11725 | 539.11840 | −2.13 | 539(31),538(28),521(100),493(19),481(15),452(18),393(23),231(21),199(21) △ |
| 13.07 | C27H23O12 | 539.11768 | −1.34 | 539(13),538(42),521(66),516(38),495(32),494(30),493(22),377(33),341(11),297(5),265(43),199(86),177(52) | ||||
| 33 | PA13 | 13.16 | C36H30O16Na b | Salvianolic acid B a | 741.14142 | 741.14260 | −1.60 | 561(100),543(51),517(19),363(11),362(17),319(2) △ |
| 13.18 | 741.14099 | −2.18 | 579(2),561(100),543(56),517(19),363(12),362(17),361(5) | |||||
| 34 | F11 | 14.21 | C28H33O14 | Buddleoside a | 593.18536 | 593.18648 | −1.89 | 575(0.2),447(100),431(1),413(0.3),285(12),257,242 △ |
| 14.42 | 593.18439 | −3.52 | 575(0.2),447(100),431(1),395(0.3),285(10),270,242 | |||||
| 35 | F12 | 14.23 | C16H13O5 | 6,7-Dihydroxy-4’-methoxyisoflavone | 285.07495 | 285.07575 | −2.80 | 285(35),271(8),270(100),242(14),239(3),158(2),152(6),132(3) |
| 36 | PA14 | 14.42 | C18H15O8 | Prolithospermic acid | 359.07535 | 359.07614 | −2.21 | 341(100),315(1),313(1.4),249(3),187(1),181(21),179(25),163(11) |
| 37 | PA15 | 14.43 | C9H9O4 | Caffeic acid | 181.04904 | 181.04953 | −2.73 | 163(100),153(0.4),139(1),135(0.6),117(0.1) |
| 38 | F13 | 14.45 | C16H13O6 | Hispidulin | 301.06982 | 301.07066 | −2.80 | 301(19),286(100),269(1),241(1),183(1) |
| 39 | PA16 | 14.63 | C18H13O7 | Salvianolic acid G | 341.06479 | 341.06557 | −2.31 | 323(100),305(3),297(21),295(8),267(1),279(3),231(5),195(11),163(11) |
| 40 | F14 | 14.98 | C28H35O14 | Didymin | 595.20074 | 595.20213 | −2.33 | 577(16),559(25),541(12),449(39),433(100), 287(34) |
| 41 | F15 | 15.11 | C17H15O7 | Jaceosidin | 331.08057 | 331.08122 | −1.99 | 316(100),303(1),288(0.3),285(1),183(0.2) |
| 42 | F16 | 16.64 | C17H15O7 | 5,6,4’-Trihydroxyl-7,8-dimethoxy flavone | 331.08026 | 331.08122 | −2.92 | 316(75),301(34),298(100),213(2),121(0.4) |
| 43 | F17 | 17.92 | C18H17O8 | Sideritiflavone | 361.09082 | 361.09179 | −2.69 | 347(8),346(84),331(39),328(100),300(1),213(3) |
| 44 | PA17 | 18.14 | C27H23O12 | trans-salvianolic acid J | 539.11700 | 539.11840 | −2.60 | 521(100),493(5),479(4),411(5),360(5),341(22),181(9),163(5) |
| 45 | PA18 | 18.14 | C36H30O16Na | Sodium lithospermate B | 741.14111 | 741.14260 | −2.01 | 561(100),543(52),515(2),383(19),363(3),319(2) |
| 46 | F18 | 18.35 | C16H13O6 | Diosmetin | 301.07025 | 301.07066 | −1.37 | 287(3),286(100),258(1) |
| 47 | T9 | 18.69 | C10H15O2 | (S)-(−)-Perillic acid | 167.10649 | 167.10665 | −0.99 | 149(67),139(72),125(16),121(100),95(18),93(13) |
| 48 | F19 | 18.98 | C16H15O6 | Hesperetin | 303.08572 | 303.08630 | −1.96 | 285(13),179(35),177(100),153(20),151(3),137(1),117(1) |
| 49 | PA19 | 19.22 | C36H30O16Na | Sodium lithospermate B | 741.14081 | 741.14260 | −2.42 | 579(37),561(14),533(100),517(7),399(3),353(4) |
| 50 | PA20 | 19.23 | C36H31O16 | Salvianolic acid E | 719.15924 | 719.16066 | −1.97 | 701(47),700(56),673(34),655(30),621(14),609(74),539(100),493(26),297(28) |
| 51 | F20 | 20.44 | C18H17O7 | Xanthomicrol | 345.09613 | 345.09687 | −2.17 | 345(15),330(100),329(85),315(1),301(14),300(3) |
| 52 | F21 | 21.51 | C18H17O8 | Thymonin | 361.09085 | 361.09179 | −2.61 | 346(100),331(75),328(53),313(33),300(13),299(4),227(1) |
| 53 | F22 | 22.49 | C19H19O8 | 5,6-Dihydroxy-7,8,3’,4’-tetramethoxyflavone | 375.10632 | 375.10744 | −2.99 | 360(70),359(8),345(47),343(12),342(100),314(1),270(2),213(3),165(1) |
| 54 | T10 | 22.78 | C10H15O2 | (4S*)-4-hydroxy-ρ-mentha-1,8-dien-3-one | 167.10641 | 167.10665 | −1.47 | 149(100),139(100),131(13),126(35),125(47),121(84),95(26),93(14) |
| 55 | T11 | 24.27 | C42H73O15 | Floralquinquenoside C | 817.49176 | 817.49439 | −3.22 | 817(53),816(93),801(16),799(33),771(100),728(28),656(28),582(20),563(28),256(15) |
| 56 | F23 | 24.43 | C18H17O7 | Nevadensin | 345.09686 | 345.09687 | −0.05 | 330(100),315(69),312(50),301(1),297(28),284(12) |
| 57 | F24 | 24.72 | C19H19O8 | 5,7-Dihydroxy-6,8,3’,4’-tetramethoxyflavone | 375.10712 | 375.10744 | −0.86 | 360(100),345(78),342(46),331(2),213 |
| 58 | F25 | 24.78 | C16H13O5 | Acacetin | 285.07047 | 285.07575 | −0.98 | 285(35),271(9),270(100),243(3),242(15),152(5) |
| 59 | F26 | 24.99 | C18H17O7 | 5,6-Dihydroxy-7,3’,4’-trimethoxy flavone | 345.09662 | 345.09687 | −0.75 | 330(64),315(40),312(100),284(1),240(1),213(3) |
| 60 | F27 | 25.08 | C16H13O5 | Genkwanin | 285.07538 | 285.07575 | −1.29 | 285(100),270(97),243(6),242(34),167(24),145(4) |
| 61 | F28 | 25.58 | C19H19O7 | 5-Dydroxy-6,7,3’,4’-tetramethoxy flavones | 359.11194 | 359.11252 | −1.64 | 345(9),344(82),327(11),326(100),315(2),298(5),165(0.15) |
| 62 | F29 | 26.43 | C20H21O8 | 5-Hydroxy-6,7,8,3’,4’-pentamethoxyflavone | 389.12198 | 389.12309 | −2.86 | 374(100),360(14),359(99),356(45),341(42),328(16),327(4),227(1.39),165(0.34) |
| 63 | F30 | 27.21 | C19H19O7 | Gardenin B | 359.11182 | 359.11252 | −1.97 | 344(100),329(92),326(53),311(37),298(15),297(5),227(1),135(1) |
| 64 | T12 | 28.55 | C30H49O3 | Ursolic acid | 457.36615 | 457.36762 | −3.21 | 439(67),411(100),393(4),356(3),227(3),191(6) |
Flavonoids (F); Phenolic acids (PA); Terpenoids (T); Phenylpropanoids (P); a Identified by comparison with standards; △ ESI-MS2 spectra of standards; b [M + Na]+ ions.
Figure 2.
The structures of chemical constituents identified in Menthae Haplocalycis herba by UHPLC-ESI-MS/MS.
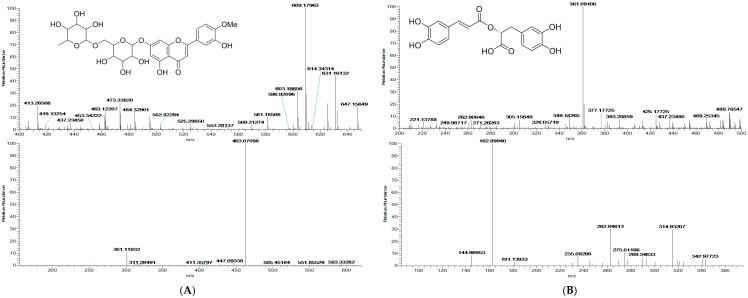
Figure 3.
The representative mass spectra of chemical constituents identified in Menthae Haplocalycis herba in positive ion mode of Diosmin (A); Rosmarinic acid (B); Bohenoside A (C) and Clemastanin A (D).
2.1. Characterization of Flavonoids
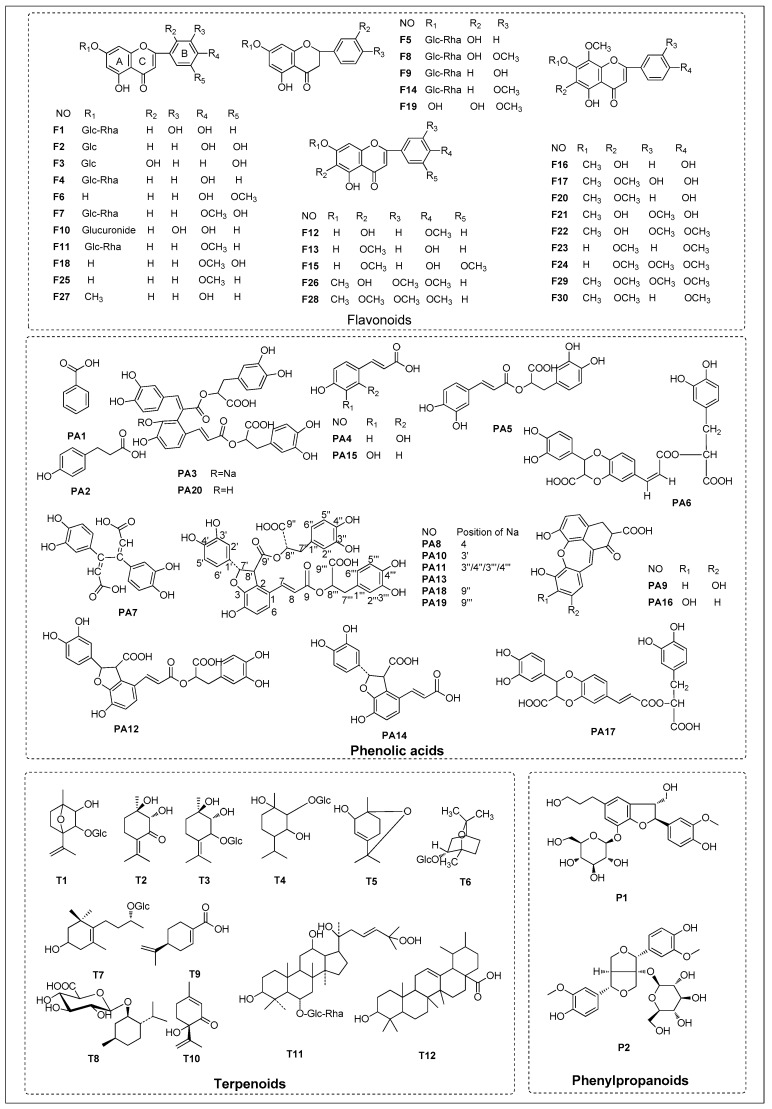
The developed UHPLC-ESI-MS/MS method is effective to obtain information on the carbohydrate sequence and aglycone moiety. Cleavage at the glycosidic O-linkages with a concomitant H-rearrangement leads to the elimination of monosaccharide residues, i.e. the loss of 146 u (deoxyhexose), 162 u (hexose) or 176 u (uronic acid), allowing the determination of carbohydrate sequence. Moreover, the fragment ions produced by Retro–Diels–Alder (RDA) reactions are useful in terms of flavonoid aglycone identification since they can provide information on the number and type of substituents on A-and B-rings [13]. Finally, 30 flavonoids were identified, including five flavanones (two flavanone aglycones and three flavanone glycosides) and 25 flavonoids (18 flavonoid aglycones and seven flavonoid glycosides).
2.1.1. Structural Characterization of Flavanones
Compounds F9 and F19 produced [M + H]+ ions at m/z 303.08572 (C16H15O6, error < 5 ppm). In the MS/MS spectra, they generated the similar fragment ions at m/z 117 corresponding to loss of B-ring from [M + H]+ ion, which indicated they might be flavonones. The fragment ions at m/z 153, m/z 151 and m/z 137 were yielded by RDA cleavage, indicating there were two -OH on A-ring and one -OH and one -OCH3 on B-ring. Combining the literature data and Clog P values, F9 and F19 were identified as Homoeriodictyol and Hesperetin, respectively [14,15].
Compound F8 showed [M + H]+ ion at m/z 611.19635 (C28H35O15, error < 5 ppm). The major fragment ions at m/z 465 and m/z 303 were yielded by successive neutral loss of Rha (146 Da) and Glc (162 Da) from [M + H]+ ions. By comparing retention time, fragmentation behaviors with reference substance, it was unambiguously identified as Hesperidin [15,16].
Compounds F5 and F14 yielded their respective [M + H]+ ions at m/z 581.18488 (C27H33O14) and m/z 595.20213 (C28H35O14) with mass errors within 5 ppm. Both of them generated [M + H − H2O]+, [M + H − Rha]+, [M + H − Glc]+ and [M + H − Rha − Glc]+ ions at m/z 563, m/z 577; m/z 435, m/z 449; m/z 419, m/z 433; m/z 273, m/z 287, respectively. Finally, F5 was unambiguously identified as Didymin by comparing with the authentic compound, while F14 was tentatively deduced as Naringin by analyzing the fragment ions produced by RDA cleavage [15,16,17].
2.1.2. Structural Characterization of Flavonoids
Compound F6, F13 and F18 showed [M + H]+ ions at m/z 301.07066 (C16H13O6, error < 5 ppm). The fragment ions yielded by RDA cleavage demonstrated that their substituent positions were remarkably different. By referring to the literature data, they were respectively identified as Chrysoeriol, Hispidulin and Diosmetin [16,18,19,20]. Likewise, compounds F12, F25, F27; F15, F16; F17, F21; F20, F23, F26; F22, F24 and F28, F30 were finally identified as 6,7-Dihydroxy-4’-methoxyisoflavone, Acacetin and Genkwanin [16,17]; Jaceosidin, 5,6,4’-Trihydroxyl-7,8-dimethoxy flavone [21]; Sideritiflavone, Thymonin [22]; Xanthomicrol, Nevadensin, 5,6-Dihydroxy-7,3’,4’-trimethoxy flavone [17,23]; 5,6-Dihydroxy-7,8,3’,4’-tetrame-thoxyflavone, 5,7-Dihydroxy-6,8,3’,4’-tetramethoxyflavone [16]; 5-Hydroxy-6,7,3’,4’-tetramethoxyflavones, Gardenin B [17,24], respectively.
Compound F29 generated [M + H]+ ion at m/z 389.12309 (C20H21O8, error < 5 ppm). Once reaching CID (collision-induced dissociation) mode, it further underwent a series of methyl units losing and RDA cleavage, which generated product ions at m/z 374, m/z 359, m/z 227 and m/z 165. Therefore, F29 was tentatively identified as 5-Hydroxy-6,7,8,3’,4’-pentamethoxyflavone.
Compound F1 gave [M + H]+ ion at m/z 595.16574 (C27H31O15, error < 5 ppm). The major ions at m/z 449 and m/z 287 in its ESI-MS2 spectrum indicated the presence of rutinoside. By analyzing fragment ions and comparing with literature data, F1 was plausibly defined as Luteolin-7-O-rutinoside [16].
Compounds F3 and F10 produced [M + H]+ ions at m/z 463.01780 (C21H19O12, error < 5 ppm). After the CID cleavage, their further fragmentation all resulted in [M + H − H2O]+ ion at m/z 445, [M + H − 2H2O]+ ion at m/z 427 and [M + H − Glucuronic acid]+ ion at m/z 287. According to the fragmentation pathways and Clog P values, F3 and F10 were tentatively identified as 2-(2,5-Dihydroxyphenyl)-5-hydroxy-4-oxo-4H-chromen-7-yl-β-d-glucopyranosiduronic acid and Luteolin-7-O-glucuronide, respectively.
Compound F7 exhibited [M + H]+ ion at m/z 609.17999 (C28H33O15, error < 5 ppm). Its ESI-MS2 spectrum gave the fragment ions at m/z 463 and m/z 301, involving successive loss of rhamnosyl and glucosyl groups. The ion at m/z 301 further generated predominant fragment ions at m/z 286 and m/z 258, which were similar to Diosmetin [15,16,18,25]. The deduced result was further confirmed by comparing with an authentic compound.
Similarly, based on literature data or reference compounds, compounds F2, F4, and F11 were identified as Luteolin-7-O-glucoside, Apigenin-7-O-rutinoside and Buddleoside, respectively [15,16,17,18].
2.2. Characterization of Phenolic Compounds
In the preliminary study, we found that UV absorption spectra of some peaks in the fingerprints of Menthae Haplocalycis herba were similar to salvianolic acid compounds. The basic structure units of salvianolic acid are tanshinol and caffeic acid. Thus, in their ESI-MS/MS spectra, regular fragment ions including [M + H − C9H8O4]+ and [M + H − C9H10O5]+ are produced by the neutral loss of caffeic acid and tanshinol. Moreover, owing to the existence of carboxyl and carbonyl, it is common to observe the neutral loss of CO and CO2. According to these fragmentation patterns, 20 phenolic acids were positively or tentatively identified.
Compound PA2 produced [M + H]+ ion at m/z 167.07027 (C9H11O3, error < 5 ppm). It firstly produced the ESI-MS2 base peak ion at m/z 149 by losing H2O. Upon reaching CID mode, [M + H]+ ion further generated [M + H − CO]+, [M + H − H2O − CO]+ and C7H7O+ ions at m/z 139, m/z 121 and m/z 107, respectively. According to the above analysis, PA2 was tentatively defined as Phloretic acid. Similarly, based on the above analysis, PA1 was tentatively identified as Benzoic acid.
Compounds PA4 and PA15 exhibited the same [M + H]+ ions at m/z 181.04953 (C9H9O4, error < 5 ppm). Both of them firstly generated ESI-MS2 base peak ions at m/z 163 by loss of H2O. The major fragment ions in the ESI-MS2 spectra were m/z 163, m/z 153, m/z 135 and m/z 117, suggesting the presence of -COOH. Compared with literature data and Clog P values, PA4 and PA15 were plausibly characterized as 2,4-Dihydroxycinnamic acid and Caffeic acid, respectively [26]. Meanwhile, by comparing with authentic standards and literature, PA5 and PA13 were identified as Rosmarinic acid and Salvianolic acid B, respectively [12,27,28].
Compounds PA6, PA12 and PA17 showed [M + H]+ ions at m/z 539.11840 (C27H23O12, error < 5 ppm). After the CID cleavage, the further fragmentation of m/z 539 resulted in [M + H − C9H10O5]+ at m/z 341 and [M + H − C9H10O5 − CO2]+ at m/z 297, involving the presence of tanshinol. By referring to literature data, the properties of Cis-salvianolic acid J, Lithospermic acid and trans-salvianolic acid J were in accordance with the description. Therefore, PA12 was unambiguously defined as Didymin by comparing with reference compound, while PA6 and PA17 were finally deduced as Cis-salvianolic acid J and trans-salvianolic acid J according to the literature data and Clog p values [12,26,27,29].
Compounds PA7 and PA14 yielded [M + H]+ ions at m/z 359.07614 (C18H15O8, error < 5 ppm). The major fragment ions in their ESI-MS/MS spectra were m/z 341 [M + H − H2O]+, m/z 313 [M + H − H2O − CO]+ and m/z 179 [M + H − C9H8O4]+, indicating the presence of caffeic acid. By referring to the literature data and Clog p values, PA7 and PA14 were tentatively defined as Bis (3,4-dihydroxybenzylidene) succinic acid and Prolithospermic acid, respectively [26].
Compounds PA9 and PA16 displayed [M + H]+ ions at m/z 341.06557 (C18H13O7, error < 5 ppm). Both of their [M + H]+ ions generated a series of fragment ions at m/z 323 [M + H − H2O]+, m/z 297 [M + H − CO2]+, m/z 295 [M + H − H2O − CO]+, m/z 279 [M + H − CO2 − H2O]+ and m/z 267 [M + H − H2O − 2CO]+. According to the literature data and Clog P values, PA9 and PA16 were tentatively identified as Benzo[f] naphthol [1,8-bc] oxepine-8-carboxylic acid and Salvianolic acid G, respectively [29].
Compound PA20 showed [M + H]+ ion at m/z 719.16066 (C36H31O16, error < 5 ppm). The fragment ions observed at m/z 539 [M + H − C9H8O4]+, m/z 493 [M + H − C9H10O5 − CO]+ and m/z 297 [M + H − C9H8O4 − C9H10O5 − CO2]+ in the ESI-MS2 spectrum indicated the presence of caffeic acid and tanshinol. By comparison with the literature data, PA20 was finally identified as Salvianolic acid E [29]. Likewise, PA3, PA8, PA10, PA11, PA18 and PA19 all produced the identical [M + H]+ ions at m/z 741.14260 (C36H30NaO16, error < 5 ppm). They all have the similar fragmentation pathways and characteristic fragment ions. According to their Clog P values, they were tentatively identified and differentiated. As a result, the sodium was monitored at C-3, C-4, C-3’, C-3’’/4’’/3’’’/4’’’, C-9’’ and C-9’’’, respectively. By comparison with the bibliography and ESI-MS2 fragmentation data, PA3 was deduced as Sodium salvianolic acid E while PA8, PA10, PA11, PA18 and PA19 were tentatively assigned as and Sodium lithospermate B, respectively [29].
2.3. Characterization of Terpenoids
It is difficult to determine terpenoids by UHPLC-PDA analysis because of their weak UV absorption. UHPLC-MS/MS is a powerful technique to identify these kinds of constituents. In their ESI-MS/MS spectra, terpenoids usually lose a molecule of H2O or CH3 because it normally contains hydroxy and methyl groups. For terpenoids glycosides, [M + H − 162]+ was easily monitored as the characteristic ion by losing a dehydrated glucose. Moreover, fragment ions referred above often have high abundance. Based on these fragmentation pathways, four monoterpenoid aglycones, six monoterpenoids glycosides and two triterpenoids were finally identified.
2.3.1. Identification of Monoterpenoid Aglycones
Compound T2 gave [M + H]+ ion at m/z 185.11722 (C10H17O3, error < 5 ppm). Upon CID mode, it further generated [M + H − H2O]+, [M + H − CO]+, [M + H − 2H2O]+ and [M + H − H2O − CO]+ ions at m/z 167, m/z 157, m/z 149 and m/z 139, respectively. According to the above analysis, T2 was tentatively identified as (1R*,2S*)-1,2-dihydroxy-ρ-menth-4(8)-en-3-one. Similarly, based on the above analysis and authentic compound, T5 was unambiguously identified as Bohecineole A [30].
Compounds T9 and T10 were a pair of isomers. Both of them gave the identical [M + H]+ ions at m/z 167.10665 (C10H15O2, error < 5 ppm) and similiar ESI-MS/MS fragment ions at m/z 149, m/z 139, m/z 125, m/z 121, m/z 95 and m/z 93. Nevertheless, their ion abundances were remarkably different. The ion at m/z 149 was yielded by neutral loss of H2O from [M + H]+ ion and further produced fragment ion at m/z 121 by losing carbonyl. According to these fragmentation pathways, T9 and T10 were tentatively identified as (S)-(−)-Perillic acid and (4S*)-4-hydroxy-ρ-mentha-1,8-dien-3-one, respectively.
2.3.2. Identification of Monoterpenoid Glycosides
With respect to compound T1, [M + Na]+ adduct ion at m/z 369.15198 (C16H26O8Na, error < 5 ppm). After the CID cleavage, m/z 351, m/z 328, m/z 207 and m/z 185 were produced by successively neutral loss of one molecular of water and dehydrated glucose. According to the above analysis, T1 was plausibly described as petroside [31]. Likewise, by comparing with the authentic compound, T6 was unambiguously identified as (1R,2R,4S)-trans-1,8-cineole-2-O-β-d-glucopyranoside.
Similarly, based on the above analysis, T3, T4, T7 and T8 were tentatively deduced as (2R,3R,4S,5S,6R)-2-(((1S,2R,3S)-2,3-dihydroxy-3-methyl-6-(propan-2-ylidene)cyclohexyl)oxy)-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol, Bohenoside A, Linarionoside B and (1R,2S,5R)-(−)-methol β-d-Glucuronide, respectively.
2.3.3. Identification of Triterpenoids
Compounds T11 and T12 produced their respective [M + H]+ ions at m/z 817.49439 (C42H73O15) and 457.36762 (C30H49O3) with mass errors within 5 ppm. The [M + H]+ ion of T11 generated [M + H − H2O]+ ion at m/z 799, [M + H − H2O − CO]+ ion at m/z 771 and [M + H − Glc]+ ion at m/z 655, etc. By comparing with the literature data, T11 was tentatively identified as Floralquinquenoside C [32]. Upon CID mode, T12 further generated a series of fragment ions at m/z 439, m/z 411, m/z 393 and m/z 191. The ion at m/z 439 was yielded by neutral loss of H2O from [M + H]+ ion and then produced fragment ions at m/z 191 by RDA cleavage. By comparing with the literature data, T12 was finally identified as Ursolic acid [33].
Our data demonstrated that some monoterpenoids are the glycosides of volatile constiuents in Menthae Haplocalycis herba. Taking T8 for example, it is the glucuronide of menthol, as we all know that menthol is the main effective component attributed to volatile constituents in Menthae Haplocalycis herba. Meanwhile, some monoterpenoid glycosides may be metabolized by intestinal flora after hydrolysis of aglycones (probably volatile constituents) and then absorbed into blood to display pharmacological effects. In this sense, it will be of great significance to carry out the study of non-volatile constituents in Menthae Haplocalycis herba.
2.4. Characterization of Phenylpropanoids
In the ESI-MS/MS spectra, phenylpropanoids always lose a molecule of H2O because they contain hydroxy groups. For phenylpropanoids glycosides, [M + H − 162]+ was easily monitored as their characteristic fragment ion. According to the fragmentation pathways, two phenylpropanoids glycosides were tentatively identified.
Compounds P1 and P2 generated their respective [M + Na]+ adduct ions at m/z 531.18368 (C25H32O11Na) and m/z 559.17859 (C26H32O12Na) with mass errors within 5 ppm. Both of them yielded [M + Na − H2O]+ and [M + Na − Glc]+ ions at m/z 513, m/z 369 and m/z 541, m/z 397, respectively. Thus, P1 and P2 were tentatively determined as Clemastanin A and (+)-1-Hydroxypinoresinol-1-O-β-d-glucoside, respectively.
2.5. Target Isolation and Verification
Acacetin, Rosmarinic acid and Clemastanin A were obtained from the effluent fraction of H2O–MeOH (50:50⟶75:25, v/v) by multiple isolation means. Their structures were verified combined with the 1H-NMR, 13C-NMR, which consistent with bibliographies [34,35,36,37]. The obtained MS data [35,36,37] of them were in accordance with the deduction of the MS/MS results, which further demonstrated the reliability of the deduced fragmentation patterns and identified the profile of non-volatile constituents in Menthae Haplocalycis herba.
3. Materials and Methods
3.1. Materials and Reagents
HPLC grade acetonitrile and formic acid were supplied by Fisher Scientific (Fisher, Fair Lawn, NJ, USA). Ultrapure water was purchased from Hangzhou Wahaha Group Co., Ltd. (Hangzhou, Zhejiang, China). All of the other reagents and chemicals were of analytical grade and commercially available.
Reference compounds including Luteolin-7-O-glucoside, Lithospermic acid, Bohecineole A, (1R,2R,4S)-trans-1,8-cineole-2-O-β-d-glucopyranoside and Salvianolic acid B were prepared from Menthae Haplocalycis herba by authors. Their structures were fully characterized by chemical and spectroscopic methods (UV, IR, NMR and MS) [38,39]. Hesperidin and Rosmarinic acid were purchased from National Institutes for Food and Drug Control (Beijing, China). Buddleoside, Diosmin and Naringin were purchased from Chengdu Must Bio-Technology Co., Ltd. (Chengdu, Sichuan, China). All of these reference compounds showed purities of above 98% by HPLC analysis.
Dried Herbal medicine samples of Menthae Haplocalycis herba were purchased from Anguo Linshi Medicinal Materials Co., Ltd. in Hebei, China and were authenticated as the aerial part of Menthae. haplocalyx Briq, which was harvested in Jiangsu at autumn by Professor Chunsheng Liu at the Beijing University of Chinese Medicine (BUCM, Beijing, China). All Menthae Haplocalycis herba samples were stored in Chinese medicine institutes of BUCM.
3.2. Sample Preparation
3.2.1. Standard Solutions
Stock solutions were prepared by dissolving appropriate amounts of 10 reference compounds in methanol. Proper amounts of each stock solution were then transferred to a 25 mL volumetric flask, and then methanol was added to make up the volume to obtain a final mixed reference solution. All the solutions were stored at 4 °C and brought to room temperature before use.
3.2.2. Sample Solutions
Sample (0.2 g) milled by 65 meshes beforehand were extracted with 10 mL methanol in an ultrasonic bath for 30 min. After being cooled to room temperature, it was weighed and adjusted to the original weight by adding methanol, and then filtered through a 0.22 μm nylon filter for analysis.
3.3. UHPLC-ESI-MS/MS System
UHPLC-ESI-MS/MS analysis was performed on a DIONEX Ultimate 3000 UHPLC system (Thermo Fisher Scientific, Waltham, MA, USA) with a binary pump and an autosampler. A series of preliminary experiments were performed to optimize mobile phase composition and elution conditions. Finally, analysis was carried out at 35 °C on an Acquity UPLC® BEH C18 column (2.1 mm × 100 mm, 1.7 μm, Waters Corporation, Milford, MA, USA). The mobile phase consisted of 0.2% formic acid aqueous solution (A) and acetonitrile (B). A gradient program was adopted as follows: 0–5 min, 5%–19.5% B; 5–8.5 min, 19.5% B; 8.5–11 min, 19.5–27% B; 11–15 min, 27% B; 15–22 min, 27–40% B; 22–24 min, 40–55% B; 24–26 min, 55–75% B; 26–28.5 min, 75% B; 28.5–30% min, 75–100% B. The flow rate was kept at 0.30 mL/min and the sample volume injected was 2 μL.
The optimized operating parameters in positive ion mode were listed as follows: capillary temperature of 350 °C; sheath gas flow rate of 40.0 arb; auxiliary gas flow rate of 20.0 arb; source voltage of 4 kV; capillary voltage of 25 V, and tube lense of 110 V. HRMS analysis was operated with a mass range of m/z 100–1000 at a resolving power of 30,000.
3.4. Peak Selections and Data Processing
Thermo Xcalibur 2.1 workstation (Thermo Fisher Scientific, San Jose, CA, USA) was used for data acquisition and processing. In order to obtain as many fragment ions of non-volatile compounds of Menthae Haplocalycis herba as possible, the peaks detected with intensity over 30,000 were selected for identification. The chemical formulas for all parent ions of the selected peaks were calculated from the accurate mass using a formula predictor by setting the parameters as follows: C (0–50), H (0–100), O (0–30), Cl (0–2), N (0–2), Na (0–2), K (0–1) and ring double bond (RDB) equivalent value (0–20). Other elements such as Br and P were not considered because they are rarely present in Menthae Haplocalycis herba.
3.5. Extraction and Isolation of Target Compounds
The air dried Menthae Haplocalycis herba samples (20.0 Kg) were extracted three times with tenfold excess of 70% EtOH under reflux for 1.5 h each at 80 °C. The combined extract was evaporated under reduced pressure to obtain a crude residue. This residue was further dispersed in H2O, and then successively passed through a Dianion HP (Mitsubishi Chemical Co., Kyoto, Japan) 2MGL macroporous resin column and then washed with extracted with H2O–MeOH (5:95⟶MeOH, v/v). The H2O–MeOH (50:50⟶75:25, v/v) extract was further purified by multiple isolation methods, such as silica gel column chromatography, C18 antiphase silica gel column chromatography, Sephadex LH-20 gel chromatography, HPD-400 macroporous resin column, etc. Acacetin, Rosmarinic acid and Clemastanin A were obtained finally.
4. Conclusions
Our study took advantage of the UHPLC-LTQ-Orbitrap HRMS system and firstly reported the identification of 64 non-volatile compounds with various structure types, including 30 flavonoids, 20 phenolic acids, 12 terpenoids and two phenylpropanoids in Menthae Haplocalycis herba. Finally, target isolation of three compounds named Acacetin, Rosmarinic acid and Clemastanin A were performed based on the obtained results, which further confirmed the deduced fragmentation patterns and identified the profile of non-volatile constituents in Menthae Haplocalycis herba. The results also clearly elucidated that there may exist some inevitable relations between volatile and non-volatile constituents. Meanwhile, our developed method has been shown to be an excellent tool for the systematic characterization of non-volatile constituents in Menthae Haplocalycis herba, which also benefits its further pharmacological research and clinical applications. Moreover, this study sets a good example for the rapid identification of chemical constituents in TCMs.
Acknowledgments
This work was supported by National Natural Science Foundation (81173520) and Beijing Nova Program (Z171100001117029).
Author Contributions
Bin Liu and Jia-Yu Zhang designed the experiments; Kun-Rui Zhong and Le Zhang contributed to the isolation and identification of Acacetin, Rosmarinic acid and Clemastanin A; Jing-Jing Xu, Lu-Lu Xu, Fei Wang and Zhan-Peng Shang contributed to the data collection and analysis; Jia-Yu Zhang and Bin Liu contributed reagents/materials/analysis tools; Lu-Lu Xu, Jia-Yu Zhang and Ru-Feng Wang wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Ling Y., Liu K.Y., Zhang Q., Liao L., Lu Y.H. High performance liquid chromatography coupled to electrosprayionization and quadrupole time-of-flight–mass spectrometry as a powerful analytical strategy for systematic analysis and improved characterization of the major bioactive constituents from radix dipsaci. J. Pharm. Biomed. 2014;98:120–129. doi: 10.1016/j.jpba.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J.Y., Cai W., Zhou Y., Liu Y., Wu X.D., Li Y., Lu J.Q., Qiao Y.J. Profiling and identification of the metabolites of baicalin and study on their tissue distribution in rats by ultra-high-performance liquid chromatography with linear ion trap-Orbitrap mass spectrometer. J. Chromatogr. B. 2015;985:91–102. doi: 10.1016/j.jchromb.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Prokudina E.A., Havlicek L., Al-Maharik N., Lapcik O., Strnad M., Gruz J. Rapid UPLC–ESI–MS/MS method for the analysis of isoflavonoids and other phenylpropanoids. J. Food Compos. Anal. 2012;26:36–42. doi: 10.1016/j.jfca.2011.12.001. [DOI] [Google Scholar]
- 4.Schummer C., Sassel J., Bonenberger P., Moris G. Low-Level Detections of Sudan I, II, III and IV in Spices and ChiliContaining Foodstuffs Using UPLC-ESI-MS/MS. J. Agric. Food Chem. 2013;61:2284–2289. doi: 10.1021/jf400602a. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J.Y., Wang Z.J., Li Y., Liu Y., Cai W., Li C., Lu J.Q., Qiao Y.J. A strategy for comprehensive identification of sequential constituents using ultra–High–performance liquid chromatography coupled with linear ion trap–Orbitrap mass spectrometer, application study on chlorogenic acids in Flos Lonicerae Japonicae. Talanta. 2016;147:16–39. doi: 10.1016/j.talanta.2015.09.039. [DOI] [PubMed] [Google Scholar]
- 6.Xu J.J., Zhu Y.C., Li L. Analysis of chemical constituents of traditional chinese medicinal formula Huang-Qi-Chi-Feng-Tang by high performance liquid chromatography-electrospray ionization-mass spectrometry. Asian J. Chem. 2013;25:4367–4371. [Google Scholar]
- 7.Christophe M., Christine S. Identification of monomenthyl succinate in natural mint extracts by LC-ESI-MS-MS and GC-MS. J. Agric. Food Chem. 2006;54:4814–4819. doi: 10.1021/jf060729c. [DOI] [PubMed] [Google Scholar]
- 8.Cao G., Shan Q.Y., Li X.M., Cong X.D., Zhang Y., Cai H., Cai B.C. Analysis of fresh mentha haplocalyx volatile components by comprehensive two-dimensional gas chromatography and high-resolution time-of-flight Mass Spectrometry. Analyst. 2011;136:4653–4661. doi: 10.1039/c1an15616k. [DOI] [PubMed] [Google Scholar]
- 9.Chen G.L., Yao D.Y., Wang Y.J., She Y.B., Fan F. Studies on the Pharmacologic Effects and Acute Toxicity of Peppermint Oil. Pharmacol. Clin. Chin. Mater. Med. 2001;17:10–12. [Google Scholar]
- 10.Zhang R.F., Yang Z.F., Jiang S.F. Research Progress on Pharmacological and Toxicological Effects of Peppermint Oil. J. China Pharm. 2012;21:1–3. [Google Scholar]
- 11.Shen M.F., Li X.M., Shan Q.Y. Research Progress of Chemical Compositions and Pharmacological Actions of Mentha Canadensis. Chin. Arch. Tradit. Chin. Med. 2012;30:1484–1487. [Google Scholar]
- 12.She G.M., Xu C., Liu B., Shi R.B. Polyphenolic Acids from Mint (the Aerial of Mentha haplocalyx Briq.) with DPPH Radical Scavenging Activity. J. Food Sci. 2010;75:359–362. doi: 10.1111/j.1750-3841.2010.01603.x. [DOI] [PubMed] [Google Scholar]
- 13.Lv S., Tian C.R., Wang H., Zhang X. Comparision on polyphenol, total flavonoids and antioxidant activity of different mint. Sci. Technol. Food Ind. 2011;32:160–163. [Google Scholar]
- 14.Filip C., Magda C. Mass spectrometry in the structural analysis of flavonoids. J. Mass Spectrom. 2004;39:1–15. doi: 10.1002/jms.585. [DOI] [PubMed] [Google Scholar]
- 15.Ye X.L., Cao D., Zhao X., Song F.Y., Huang Q.H., Fan G.R., Wu F.H. Chemical fingerprint and metabolic profile analysis of Citrus reticulate ‘Chachi’ decoction by HPLC-PDA-IT-MSn and HPLC-Quadrupole-Orbitrap-MS method. J. Chromatogr. B. 2014;970:108–120. doi: 10.1016/j.jchromb.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Chen X.Y., Zhang L., Wu Y., Wang W.H., Liu B. Rapid analysis of chemical compositions in the flavonoid fraction of Mentha haplocalyx Briq. with LCMS-IT-TOF. J. Beijing Univ. TCM. 2015;38:546–550. [Google Scholar]
- 17.Renu P., Brijesh K. HPLC-QTOF-MS/MS Based Rapid Screening of Phenolics and Triterpenic Acids in Leaf Extracts of Ocimum Species and their Interspecies Variation. J. Liq. Chromatogr. R. T. 2016;39:225–238. [Google Scholar]
- 18.Lin T., Duan J.A., Qian D.W., Wang D.W., Qian S.H. Identification of flavonoids from herb of Mentha haplocalyx by HPLC-MS/MS. Chin. J. Nat. Med. 2006;4:111–115. [Google Scholar]
- 19.Pascual-T J.D., Gonzalez M.S., Vicente S., Bellido I.S. Flavonoids from Chenopodium botrys. Planta Med. 1981;41:389–391. doi: 10.1055/s-2007-971732. [DOI] [PubMed] [Google Scholar]
- 20.Federico F., Branca M.S., Paula B.A., Rosa M.S., Margarida A.F. Approach to the Study of C-Glycosyl Flavones by Ion Trap HPLC-PAD-ESI/MS/MS: Application to Seeds of Quince (Cydonia oblonga) Phytochem. Anal. 2003;14:352–359. doi: 10.1002/pca.727. [DOI] [PubMed] [Google Scholar]
- 21.Song W.Y., Kim N.J., Kim S.Y., Lee H.S. Liquid chromatography-tandem mass spectrometry for the determination of jaceosidin in rat plasma. J. Pharm. Biomed. 2009;49:381–386. doi: 10.1016/j.jpba.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Chen X.Y., Zhang S.J., Xuan Z.N., Ge D.Y., Chen X.M., Zhang J.J., Wang Q., Wu Y., Liu B. The Phenolic Fraction of Mentha haplocalyx and Its Constituent Linarin Ameliorate Inflammatory Response through Inactivation of NF-κB and MAPKs in Lipopolysaccharide-Induced RAW264.7 Cells. Molecules. 2017;22:811. doi: 10.3390/molecules22050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fereshteh J., Ebrahimi S.A., Nahid R.R., Massoud M. Xanthomicrol is the main cytotoxic component of Dracocephalum kotschyii and a potential anti-cancer agent. Phytochemistry. 2005;66:1581–1592. doi: 10.1016/j.phytochem.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J.Y., Li N., Che Y.Y., Zhang Y., Liang S.X., Zhao M.B., Jiang Y., Tu P.F. Characterization of seventy polymethoxylated flavonoids (PMFs) in the leaves of Murraya paniculata by on-line high-performance liquid chromatography coupled to photodiode array detection and electrospray tandem mass spectrometry. J. Pharm. Biomed. 2011;56:950–961. doi: 10.1016/j.jpba.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Zheng J., Gao H.Y., Chen G.T., Yang X.K., Wu B., Wu L.J. Chemical constituents of the active parts of Mentha spicata L. (II) J. Shenyang Pharm. Univ. 2006;23:212–216. [PubMed] [Google Scholar]
- 26.Zhu Z.Y., Zhang H., Zhao L., Dong X., Li X., Chai Y.F., Zhang G.Q. Rapid separation and identification of phenolic and diterpenoid constituents from Radix Salvia miltiorrhizae by high-performance liquid chromatography diode-array detection, electrospray ionization time-of-flight mass spectrometry and electrospray ionization quadrupole ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:1855–1865. doi: 10.1002/rcm.3023. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C.X., Luo H.W., Danyu Z.W. Studies on isolation and identification of water-soluble constituents of Salvia miltiorrhiza. J. China Pharm. Univ. 1999;30:411–416. [Google Scholar]
- 28.Luo S.X., Liang Z.Y., Zhang D.L., Chen Z.X. Research on extraction and analysis of rosmarinnic acid in Mentha haplocalyx Briq. Food Sci. 2005;11:192–193. [Google Scholar]
- 29.Dong X., Xu L., Lou Z.Y. Determination of chemical components from Salvia miltiorrhiza Bunge by electrospray ion-trap mass spectrometry. Chin. Pharmacol. J. 2010;45:1048–1054. [Google Scholar]
- 30.She G.M., Xu C., Liu B. New monocyclic monoterpenoid glycoside from Mentha haplocalyx Briq. Chem. Cent. J. 2012;6:37. doi: 10.1186/1752-153X-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masayuki Y., Toshiaki U., Hiroshi S., Akinobu K., Yuzo K., Hisashi M. Medicinal Foodstuffs. XVIII.1) Phytoestrogens from the Aerial Part of Petroselinum crispum MILL. (PARSLEY) and Structures of 6’’-Acetylapiin and a New Monoterpene Glycoside, Petroside. Chem. Pharm. Bull. 2000;48:1039–1044. doi: 10.1248/cpb.48.1039. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y.J., Zhao Z.Z., Chen H.B., Yi T., Qin M.J., Liang Z.T. Chemical Differentiation and Quality Evaluation of Commercial Asian and American Ginsengs based on a UHPLC–QTOF/MS/MS Metabolomics Approach. Phytochem. Anal. 2015;26:145–160. doi: 10.1002/pca.2546. [DOI] [PubMed] [Google Scholar]
- 33.Wang H.L., Qi W., Wu X.D., He Z.M. Fragmentation mechanisms of phenolic acids from Danshen in an ion trap by electrospray ionization multi-stage tandem mass spectrometry. J. Chin. Mass Spectrom. Soc. 2008;29:129–136. [Google Scholar]
- 34.Chaves O.C., Gomes R.A., Tomaz A.C.A. Secondary Metabolites from Sida rhombifolia L. (Malvaceae) and the Vasorelaxant Activity of Cryptolepinone. Molecules. 2013;18:2769–2777. doi: 10.3390/molecules18032769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo E.R., Piao M.S. Antioxidative Constituents from Lycopus lucidus. Arch. Pharm. Res. 2004;27:173–176. doi: 10.1007/BF02980102. [DOI] [PubMed] [Google Scholar]
- 36.Gu X.J., Li Y.B., Li P., Qian S.H., Duan J.A. Studies on chemical constituents of Prunella vulgaris. China J. Chin. Mater. Med. 2007;32:923–925. [PubMed] [Google Scholar]
- 37.Kizu H., Shimana H., Tomimori T. Studies on the Constituent of Clematis Species.VI.1) The Constituents of Clematis stans SIEB. et Zucc.2) Chem. Pharm. Bull. 1995;43:2187–2194. doi: 10.1248/cpb.43.2187. [DOI] [PubMed] [Google Scholar]
- 38.Xu C. Master’s Thesis. Beijing University of Chinese Medicine; Beijing, China: 2009. Chemical Studies of Mentha Haplocalyx Briq. [Google Scholar]
- 39.Zhong K.R. Master’s Thesis. Beijing University of Chinese Medicine; Beijing, China: 2016. Studies on the Chemical Constituents and the Differences between Leaves and Stems of Mentha Haplocalyx Briq. [Google Scholar]