Abstract
The plasma membrane is a highly complex multicomponent system that is central to the functioning of cells. Cholesterol, a key lipid component of the plasma membrane, promotes the formation of nanodomains. These nanodomains are often correlated across the two membrane leaflets, but the underlying physical mechanism remains unclear. Using coarse-grained molecular dynamics simulations, we investigate the influence of cholesterol flip-flop on membrane properties, in particular, the interleaflet coupling of cholesterol-enriched domains. We show that the cholesterol density correlation between the leaflets of an average mammalian plasma membrane is significantly reduced by suppressing interleaflet cholesterol population. Our results suggest an amplifying role of cholesterol in signal transduction across the leaflets.
The plasma membrane (PM) plays a central role in biology by serving as the border of living cells, separating the cell interior from its environment. Typically, hundreds of different lipid types are present in the PM.1,2 The complexity of the PM gives rise to structural and compositional heterogeneity, as expressed by the raft concept.3,4
A major component of cellular membranes is cholesterol (CHOL). In various model membranes and over a substantial range of compositions, cholesterol induces phase separation into cholesterol-enriched liquid ordered (Lo) and cholesterol-depleted liquid disordered (Ld) domains.5,6 Such domains are also observed in membranes extracted from real cells, pointing to their potential biological relevance as a membrane organizing principle.7,8
Interestingly, in many cases, the domains in both leaflets appear to be correlated.9−11 Currently, multiple theories exist about the major driving force for this domain registration, including curvature coupling, interleaflet midplane tension, line tension of the domain boundary, chain interdigitation, electrostatic coupling, and cholesterol flip-flop.12−14 Molecular dynamics (MD) simulations are, in principle, suited to unravel these driving forces. In fact, a number of recent simulation studies address the issue of domain registration and point to a variety of factors that contribute to alignment or even anti-alignment of the domains.15−21 The extent of domain registration therefore involves multiple factors and appears highly system-dependent.
Whereas the molecular mechanism of CHOL flip-flop has been investigated in detail by means of MD simulations,22−24 a few studies have started to look at the CHOL influence on domain registration exploiting net CHOL translocation between the leaflets,25 the aliphatic chain length of CHOL,16 or increasing CHOL concentration.26 Chloroform, which, like CHOL, rapidly flip-flops between the leaflets, has been shown to drive phase registration in ternary model membranes.19 Here, using coarse-grain (CG) MD simulations, we investigate the contribution of cholesterol flip-flop to the domain registration without changing the chemical nature of CHOL or the bilayer composition. We use simple model systems consisting of three to four lipid types27 and our recently developed realistic mammalian PM model28,29 and compare simulations where the CHOL flip-flop was suppressed with ones allowing CHOL flip-flop. We applied two different flat-bottomed potentials to suppress CHOL flip-flopping (Figure S1): “wider” potentials (denoted fb3.5), allowing CHOL to populate the region between both leaflets,22 and “thinner” potentials (fb1.6), restricting CHOL to the individual leaflets. Besides the influence on domain registration, we analyze global membrane properties like area per lipid (APL), lipid tail order, and bilayer thickness.
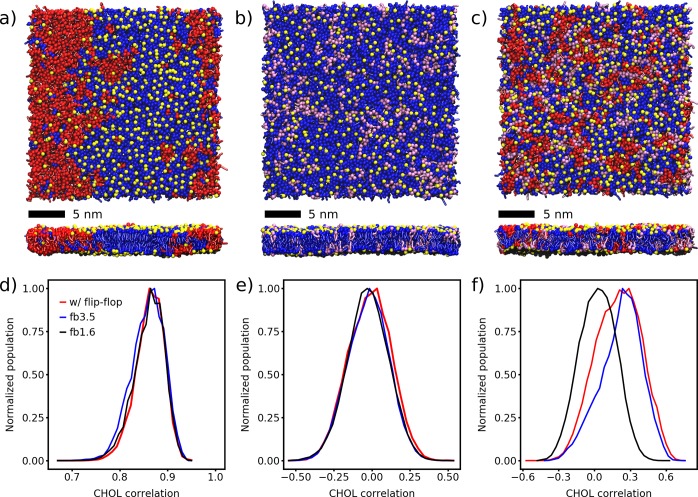
First, we will discuss the behavior of the ternary mixtures, followed by the quaternary mixture and the mammalian PM model. The ternary mixture consists of either dipalmitoylphosphatidylcholine (DPPC)/dilinoleoylphosphatidylcholine (DLiPC)/CHOL at a molar ratio of 0.42/0.28/0.30 or DPPC/dioleoylphosphatidylcholine (DOPC)/CHOL at a molar ratio of 0.64/0.16/0.20. The first mixture is strongly phase-separating due to the presence of the polyunsaturated DLiPC lipid, whereas the second mixture forms only small transient domains at the CG resolution of the employed Martini force field. Figure 1a shows the final snapshot of the DPPC/DLiPC/CHOL bilayer after 30 μs. The phase separation of DPPC (saturated tails) and DLiPC (double unsaturated tails) is clearly visible; CHOL preferentially resides in the DPPC-enriched Lo domains. The two domains are registered across the leaflets.
Figure 1.
Lipid mixtures of DPPC/DLiPC/CHOL (a,d), DPPC/DOPC/CHOL (b,e), and DPPC/DOPC/DLiPC/CHOL (c,f). (a–c) Top and side views of the membrane organization after 30 μs of simulation without any restrictions on CHOL. DPPC, blue; DOPC, purple; DLiPC, red; CHOL, yellow. The phospholipid headgroups are omitted for clarity. (d–f) Distributions of the Pearson correlation of the CHOL densities of the two leaflets evaluated for the last 20 μs of the simulations. Snapshots were taken every 500 ps without averaging.
Table 1 lists the global membrane properties of the DPPC/DLiPC/CHOL bilayer. The average APL remains almost unchanged upon restricting CHOL flip-flop. A similar behavior is observed for the area compressibility, the tail order, as well as the bilayer thickness. Overall, the global membrane properties are virtually unaffected by the restriction of the CHOL flip-flop.
Table 1. Membrane Properties of the Ternary Mixtures DPPC/DLiPC/CHOL and DPPC/DOPC/CHOLa.
| DPPC/DLiPC/CHOL |
DPPC/DOPC/CHOL |
|||||
|---|---|---|---|---|---|---|
| w/o flip-flop |
w/o flip-flop |
|||||
| w/flip-flop | fb3.5 | fb1.6 | w/flip-flop | fb3.5 | fb1.6 | |
| average APL (nm2)b | 0.736 | 0.737 | 0.738 | 0.659 | 0.660 | 0.659 |
| average area compressibility (mN/m)b | 399 ± 5 | 407 ± 5 | 411 ± 7 | 389 ± 4 | 392 ± 6 | 391 ± 7 |
| average tail order DPPCc | 0.634 | 0.626 | 0.639 | 0.533 | 0.534 | 0.535 |
| average tail order DLiPC/DOPCc | 0.244 | 0.245 | 0.241 | 0.380 | 0.382 | 0.382 |
| average bilayer thickness (nm)c | 4.071 | 4.075 | 4.061 | 4.191 | 4.190 | 4.190 |
| CHOL flip-flop rate (106 s–1)b | 5.45 ± 0.08 | 0.0 | 0.0 | 1.77 ± 0.05 | 0.0 | 0.0 |
All errors are standard errors and were omitted if ≤0.002.
Averaged over the last 10 μs.
Averaged over the last 2 μs.
To analyze the extent by which cholesterol flip-flop affects the composition of the Lo and Ld domains, we evaluated the relative number of neighboring lipids for each lipid type (listed in Table S2). DPPC and CHOL prefer to be surrounded by each other or themselves, whereas they try to avoid DLiPC. On the contrary, DLiPC prefers itself in its surrounding, in line with the strong phase separation apparent from the snapshots (Figure 1a). This trend is unaffected by the CHOL flip-flop.
Finally, we analyzed the spatial correlation of the CHOL densities of the upper and lower leaflet, which provides a measure for the registration of the CHOL domains. In doing so, we extract the positions of the polar CHOL headgroups in each leaflet, convert it to a continuous density using a Gaussian kernel of σ = 15 Å, and calculate the Pearson correlation between the CHOL densities of the lower and upper leaflet (for details, see the SI). Figure 1d illustrates the distribution of the Pearson correlation for the DPPC/DLiPC/CHOL system in the last 20 μs of the simulations. The distributions exhibit a Gaussian shape and show no changes if CHOL flip-flop is restricted (maximum at 0.866 ± 0.001 with flip-flop, 0.863 ± 0.001 with fb3.5, 0.866 ± 0.001 with fb1.6). The maxima were determined by fitting a Gaussian function. The temporal evolution of the correlation of all three systems is depicted in Figure S6, showing a convergence of these data on a time scale of ∼4 μs. Apparently, restricting the CHOL flip-flop has virtually no effect on the interleaflet registration. To verify the CHOL density as a suitable measure of the domain registration, we also calculated the interleaflet correlation of the saturated (DPPC) and unsaturated (DLiPC) lipid densities, which are depicted in Figure S8.
An overall similar picture arises for the second ternary mixture investigated (DPPC/DOPC/CHOL). The final snapshot of the bilayer after 30 μs is depicted in Figure 1b. A non-ideal mixing of DPPC and DOPC is visible, but there is no clear domain formation. Because of the higher DPPC content, the CHOL flip-flop rate is smaller by a factor of ∼3. The average APL, area compressibility, tail order, and bilayer thickness are listed in Table 1. As in the previous example, these global membrane properties are unaffected by restricting the CHOL flip-flop.
The lower tendency of the DPPC/DOPC/CHOL bilayer to form defined lateral domains is also reflected in the relative number of neighboring lipids (Table S3). The unsaturated lipid DOPC still prefers itself over DPPC or CHOL as a neighbor, but the relative preference is reduced to 1.38 compared to 2.18 for DLiPC. Again, CHOL flip-flop has no significant effect on the relative numbers of neighboring lipids.
Because the bilayer does not show a clear tendency to phase-separate in our simulations, the Pearson correlation of the CHOL densities is slightly negative (−0.011 ± 0.001 for the simulations with flip-flop; Figure 1e). It has a broader distribution compared to the DPPC/DLiPC/CHOL system and does not change upon CHOL restriction (fb3.5: −0.020 ± 0.001, fb1.6: −0.025 ± 0.001). Together, our data on ternary mixtures show that the degree of domain registration is sensitive to system composition, in line with previous results,17 but that cholesterol flip-flop has no significant contribution.
To approach the complexity of the PM, we investigate the quaternary lipid mixture DPPC/DOPC/DLiPC/CHOL 0.37/0.14/0.19/0.30. The number of unsaturated, singly unsaturated, and double unsaturated tails mimics their fraction in our idealized mammalian PM mixture. The final snapshot of the bilayer after 30 μs is depicted in Figure 1c. Here the formation of nanodomains is visible. In contrast to DPPC/DLiPC/CHOL (see Figure 1a), where full macroscopic phase separation occurs, the lipid domains of the DPPC/DOPC/DLiPC/CHOL mixture are smaller and more dynamic (Figure S7 and Table S1).
The average APL, area compressibility, tail order, and bilayer thickness are listed in Table S6. These global membrane properties show an overall similar picture compared to the ternary mixtures and are unaffected by restricting the CHOL flip-flop. In the unrestraint simulation, the CHOL flip-flop rate is 4.82 × 106 s–1, which is slightly reduced compared to DPPC/DLiPC/CHOL (5.48 × 106 s–1).
The tendency to form lateral domains is also reflected in the relative number of neighboring lipids (Table S4). The unsaturated lipids DLiPC and DOPC prefer themselves as neighbors over DPPC and CHOL. In addition, they prefer the same unsaturated lipid more than the other type. DPPC prefers itself and CHOL as neighbors; CHOL prefers DPPC and clearly disfavors DLiPC. Again, CHOL flip-flop has no significant effect on the relative numbers of neighboring lipids.
Because the bilayer does not show full macroscopic phase separation, the Pearson correlation of the CHOL densities ρCHOL is smaller than that in the DPPC/DLiPC/CHOL system. However, the tendency to form small lateral domains can be clearly recognized in the distributions of the Pearson correlation of the CHOL densities in Figure 1f. The simulations with flip-flop show a mean value of 0.208 ± 0.002 (fb3.5: 0.246 ± 0.003, fb1.6: 0.030 ± 0.001). Whereas by restricting CHOL with the wider flat-bottomed potential, the interleaflet correlation slightly increases, the narrower one results in a drastic decrease. A control simulation using the final snapshot of the unrestrained simulation as the starting configuration shows that after 2 μs the correlation decreases to −0.013 ± 0.005 (Figure S9). This assigns a key role to the CHOL interleaflet population in driving domain registration.
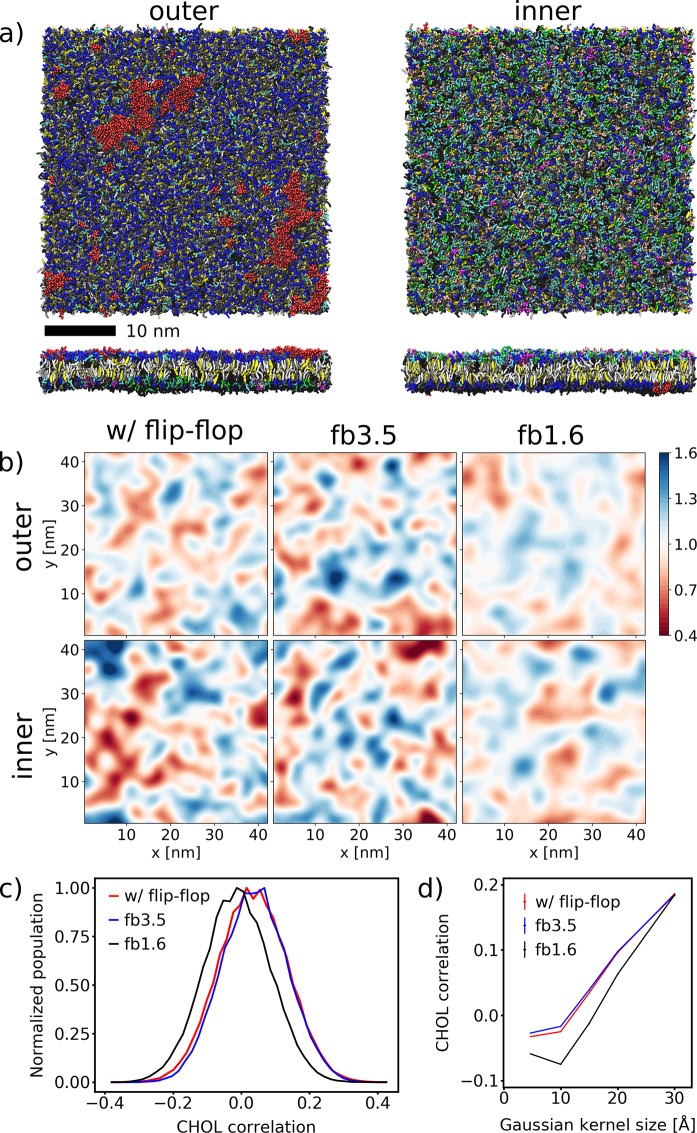
Let us now take a look at the impact of CHOL flip-flop on an idealized average PM mixture, consisting of >60 different lipid types asymmetrically distributed between the two leaflets.28 The final snapshot of the simulated PM patch after 100 μs with CHOL flip-flop is depicted in Figure 2a. The asymmetric composition can be easily recognized. Despite this, the clustering of the glycolipids (red) is clearly noticeable.28
Figure 2.
Final snapshot of the average PM simulated with CHOL flip-flop at 100 μs. (a) Top and side views of the membrane organization. The lipids are colored according to their headgroups: PC, blue; PE, cyan; SM, gray; PS, green; glycolipids, red; PI, pink; PA, white; PIPs, magenta; CER, ice blue; DAG, brown; CHOL, yellow. (b) Snapshot of the final CHOL density calculated with a Gaussian kernel of σ = 15 Å. (c) Distributions of the Pearson correlation of the CHOL densities of the two leaflets evaluated for the last 80 μs of the simulations. Snapshots of the CHOL densities were taken every 500 ps without averaging. (d) Mean Pearson correlation depending on the Gaussian kernel size σ used to calculate the CHOL density.
Table 2 summarizes the global membrane properties of the PM with and without CHOL flip-flop. Similar to the simpler mixtures, they are mostly unchanged. The APL is almost unaffected by suppressing CHOL flip-flop, with a maximum difference of 0.004 nm2 lower in the case of the wide flat-bottomed potential (fb3.5). Without CHOL flip-flop, the average tail order parameter in the outer leaflet slightly increases, while the trend in the inner leaflet is not so clear. The reason might be that the overall level of saturation is higher for the outer leaflet (Table S7). The average bilayer thickness measured at the phosphate groups decreases as well, especially for fb1.6. The area compressibility increases slightly when applying fb1.6. These observations could be traced back to the missing CHOL population in the bilayer middle, as fb1.6 does not allow the space between the leaflets to be populated. The resulting slightly higher CHOL concentration in the leaflets increases the lipid packing and thus the order parameter and the area compressibility. The CHOL flip-flop rate in the reference simulation is comparable to the one of the DPPC/DLiPC/CHOL mixture (Table 1).
Table 2. Membrane Properties of the Average PMa.
| w/o flip-flop |
|||
|---|---|---|---|
| w/flip-flop | fb3.5 | fb1.6 | |
| outer average APL (nm2)b | 0.503 | 0.499 | 0.502 |
| inner average APL (nm2)b | 0.542 | 0.538 | 0.541 |
| average area compressibility (mN/m)b | 367 ± 12 | 378 ± 12 | 396 ± 5 |
| outer average tail orderc | 0.429 | 0.430 | 0.436 |
| inner average tail orderc | 0.379 | 0.376 | 0.382 |
| average bilayer thickness (nm)c | 4.166 | 4.162 | 4.138 |
| CHOL flip-flop rate (106 s–1)b | 5.48 ± 0.02 | 0.0 | 0.0 |
All errors are standard errors and were omitted if ≤0.002.
Averaged over the last 10 μs.
Averaged over the last 40 μs.
To characterize the degree of lateral inhomogeneity in the PM patch, Table S5 lists the relative number of neighboring lipids in the average PM. The majority of the lipid neighbors do not change when the CHOL flip-flop is restricted. The most striking effect appears for the lipids with two unsaturated tails in the outer leaflet; they have a larger tendency to be surrounded by themselves, whereas lipids with one or two saturated tails are slightly more depleted in their environment. Surprisingly, the amount of CHOL in their surroundings is not influenced. For the inner leaflet, this effect is of only minor importance.
Finally, we take a look at the Pearson correlation of the CHOL densities in the outer and inner leaflet of the PM evaluated for the last 80 μs of the simulations. The CHOL densities were calculated using a Gaussian kernel of σ = 15 Å (cf. SI). Figure 2c shows their distributions evaluated for single snapshots. Their width is somewhat smaller than the ones of the DPPC/DOPC/CHOL bilayer. In the simulated patch with CHOL flip-flop, a correlation of 0.034 is observed. By applying the wider flat-bottomed potential the correlation remains almost unchanged (0.039), but it is reduced to −0.012 in the case of fb1.6 (all errors ≤0.0003). Corresponding to the quaternary mixture, the narrower flat-bottomed potential leads to a significant decrease in the CHOL density correlation. Figure 2d depicts the influence of the Gaussian kernel size σ on the interleaflet CHOL correlation. For σ < 30 Å, fb1.6 shows a reduced CHOL correlation. For larger σ, the difference becomes blurred.
We performed additional control simulations for the PM mixture where we suppressed CHOL flip-flop starting from the final snapshot after 100 μs of the PM patch simulated with flip-flop. These simulations were performed for 20 μs. Figure S10 depicts the distributions of the Pearson correlation of the CHOL densities of the two leaflets evaluated for the last 15 μs of these control simulations. The maximum of the distributions is at 0.015 ± 0.001 using fb3.5 and at −0.003 ± 0.001 using fb1.6, respectively, showing that suppressing the interleaflet CHOL population leads to a deregistration of the CHOL densities in both leaflets.
In summary, we investigated the effect of CHOL flip-flop by means of CG MD simulations of four different lipid bilayers: the ternary mixtures DPPC/DLiPC/CHOL and DPPC/DOPC/CHOL, the quaternary mixture DPPC/DOPC/DLiPC/CHOL, as well as a more complex idealized mammalian PM model. In all four cases, no striking changes of the global membrane properties or in terms of the lipid mixing were observed. For the ternary mixtures, the correlation of the CHOL densities in the upper and lower leaflet was also unaffected by CHOL flip-flop, but distinct changes were observed for the quaternary mixture and the PM model. While a restriction with the wider flat-bottomed potential fb3.5 inflicted no changes (or even an increase for the quaternary mixture), suppressing the interleaflet CHOL population (fb1.6) resulted in a significant decrease in the CHOL density correlation. This is remarkable because it shows that it is not the flip-flop process itself that increases correlation between the leaflets but that it results from the intermediate state where CHOL is sandwiched between the leaflets. This state is significantly populated, in particular, in the presence of (poly)unsaturated lipids in line with neutron scattering data.22
Taken together, our results demonstrate a remarkable impact of CHOL flip-flop on the domain registration, most pronounced in complex lipid bilayers. A possible explanation for the dependency of this effect on the system composition is obtained by considering the different nature of the domains in the four studied systems. The DPPC/DLiPC/CHOL mixture is strongly phase-separating and the interleaflet surface tension is likely the major driving force for the strong domain coupling.27 In the case of the DPPC/DOPC/CHOL mixture, only small transient DOPC clusters are formed, which might have a too small spatial extent and a too short lifetime (Figure S7 and Table S1) to be influenced by the CHOL flip-flop. Although the domains in the quaternary mixture and the PM are also transient, their larger extent together with their dynamic flexibility allows them to react to the presence of an interleaflet CHOL population (see Figure 3). This interleaflet CHOL prefers Ld domains in its surrounding because they offer more space and enable a better embedding of the CHOL molecules. Thus interleaflet CHOL leads to a weak repulsion of Lo domains, resulting in an increased interleaflet correlation (Figure 3, bottom). A recent simulation study showing that interleaflet CHOL prefers registered Ld domains over registered Lo domains and anti-registered domains in DPPC/DLiPC/CHOL mixtures26 supports this idea.
Figure 3.
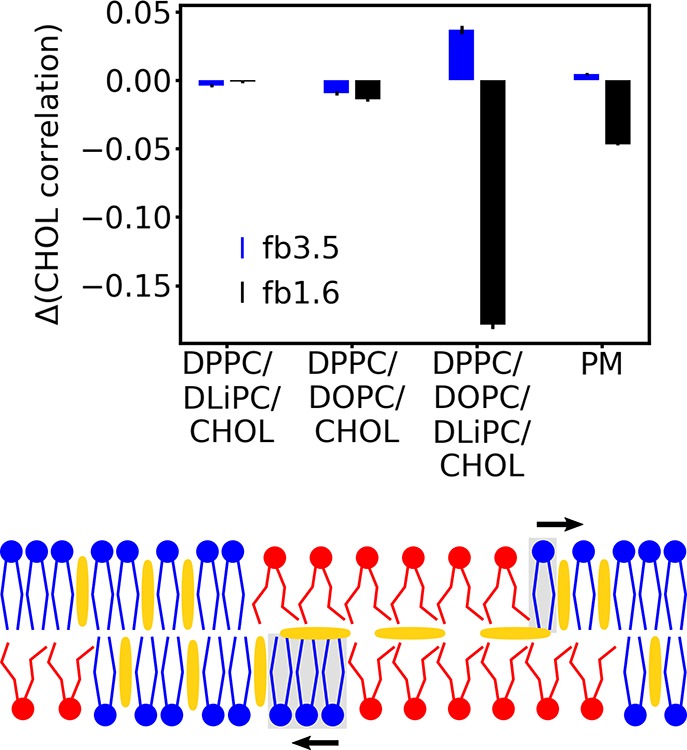
Changes of the interleaflet correlation of the CHOL densities by restricting CHOL flip-flop relative to the unrestricted bilayer (top). Schematic presentation of the effect of the interleaflet CHOL population to promote phase registration (bottom; CHOL, yellow): The interleaflet CHOL prefers to stay between registered Ld phases (red). It repels the lipids in the gray shaded part of the Lo phase (blue) and thus enhances registration.
On a more general note, our study shows that CHOL serves as an efficient signaling molecule transferring information between the leaflets by populating the interleaflet space. Through the alignment of (transient) domains, CHOL can quickly transfer local density gradients across the leaflets. Proteins being omnipresent in biological membranes might trigger such small local density gradients, for example, by their individual lipid fingerprint.33 Together with the help of CHOL, this could potentially steer a variety of cellular processes that depend on lateral membrane organization.
Computational Methods
All MD simulations were performed with the CG force field Martini (version 2.2)30,31 using the MD package GROMACS (versions 4.6.7 and 2016.1).32 We applied flat-bottomed potentials in the direction of the membrane normal to the CHOL molecules to suppress their flip-flopping between the leaflets (Figure S1). Two different widths were used: “wider” potentials with a flat region of 3.5 nm (denoted fb3.5), allowing CHOL to populate the region between both leaflets, and “thinner” potentials with a flat region of 1.6 nm (fb1.6), restricting the polar CHOL heads to the lipid linker region. As a reference, additional simulations without any restrictions to CHOL were performed for each bilayer. For further details of the bilayer compositions and the simulation setup, see the SI.
Acknowledgments
This work was supported by the European Commission via an ERC grant to S.J.M. (COMP-MICR-CROW-MEM, grant agreement 669723) and a Marie Skłodowska-Curie Actions fellowship to S.T. (MicroMod-PSII, grant agreement 748895). Part of this work was performed under the auspices of the U.S. Department of Energy under contract number DE-AC52-07NA27344 (LLNL-JRNL-751356). We acknowledge computing time from the National Computing Facilities Foundation (NCF) of The Netherlands Organization for Scientific Research (NWO).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.8b01877.
Details of the bilayer setup, simulation details, evaluation of the Pearson correlation of the CHOL densities, additional snapshots of the membrane patches, density profiles along the membrane normal, the time-dependent behavior of the CHOL densities, Pearson correlation of the saturated and unsaturated lipid densities, control simulations of the quaternary bilayer and the PM, global membrane properties of the quaternary mixture, and the lipid mixing (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Yang Y.; Lee M.; Fairn G. D. Phospholipid Subcellular Localization and Dynamics. J. Biol. Chem. 2018, 293, 6230–6240. 10.1074/jbc.R117.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama T.; Riezman H. Understanding the Diversity of Membrane Lipid Composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- Lingwood D.; Simons K. Lipid Rafts as a Membrane-Organizing Principle. Science 2010, 327, 46–50. 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Sezgin E.; Levental I.; Mayor S.; Eggeling C. The Mystery of Membrane Organization: Composition, Regulation and Roles of Lipid Rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart T.; Hess S. T.; Webb W. W. Imaging Coexisting Fluid Domains in Biomembrane Models Coupling Curvature And Line Tension. Nature 2003, 425, 821–824. 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Wu J.; Heberle F. A.; Mills T. T.; Klawitter P.; Huang G.; Costanza G.; Feigenson G. W. Phase Studies of Model Biomembranes: Complex Behavior Of DSPC/DOPC/Cholesterol. Biochim. Biophys. Acta, Biomembr. 2007, 1768, 2764–2776. 10.1016/j.bbamem.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch S. L.; Cicuta P.; Sengupta P.; Honerkamp-Smith A.; Holowka D.; Baird B. Critical Fluctuations in Plasma Membrane Vesicles. ACS Chem. Biol. 2008, 3, 287–293. 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- Levental I.; Grzybek M.; Simons K. Raft Domains Of Variable Properties and Compositions in Plasma Membrane Vesicles. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 11411–11416. 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling V.; Wan C.; Tamm L. K. Domain Coupling in Asymmetric Lipid Bilayers. Biochim. Biophys. Acta, Biomembr. 2009, 1788, 64–71. 10.1016/j.bbamem.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belička M.; Weitzer A.; Pabst G. High-resolution Structure of Coexisting Nanoscopic And Microscopic Lipid Domains. Soft Matter 2017, 13, 1823–1833. 10.1039/C6SM02727J. [DOI] [PubMed] [Google Scholar]
- Eicher B.; Marquardt D.; Heberle F. A.; Letofsky-Papst I.; Rechberger G. N.; Appavou M.-S.; Katsaras J.; Pabst G. Intrinsic Curvature-Mediated Transbilayer Coupling in Asymmetric Lipid Vesicles. Biophys. J. 2018, 114, 146–157. 10.1016/j.bpj.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiantia S.; London E. Acyl Chain Length and Saturation Modulate Interleaflet Coupling in Asymmetric Bilayers: Effects on Dynamics and Structural Order. Biophys. J. 2012, 103, 2311–2319. 10.1016/j.bpj.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimzyanov T. R.; Kuzmin P. I.; Pohl P.; Akimov S. A. Undulations Drive Domain Registration from the Two Membrane Leaflets. Biophys. J. 2017, 112, 339–345. 10.1016/j.bpj.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.; Haataja M. Compositional interface dynamics within symmetric and asymmetric planar lipid bilayer membranes. Soft Matter 2013, 9, 2120–2124. 10.1039/C2SM27269E. [DOI] [Google Scholar]
- Perlmutter J. D.; Sachs J. N. Interleaflet Interaction and Asymmetry in Phase Separated Lipid Bilayers: Molecular Dynamics Simulations. J. Am. Chem. Soc. 2011, 133, 6563–6577. 10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- Lin X.; Zhang S.; Ding H.; Levental I.; Gorfe A. A. The Aliphatic Chain of Cholesterol Modulates Bilayer Interleaflet Coupling and Domain Registration. FEBS Lett. 2016, 590, 3368–3374. 10.1002/1873-3468.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler P. W.; Williamson J. J.; Sansom M. S. P.; Olmsted P. D. Roles of Interleaflet Coupling and Hydrophobic Mismatch in Lipid Membrane Phase-Separation Kinetics. J. Am. Chem. Soc. 2016, 138, 11633–11642. 10.1021/jacs.6b04880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna M.; Javanainen M.; Monne H. M-S.; Gabius H.-J.; Rog T.; Vattulainen I. Long-Chain GM1 Gangliosides Alter Transmembrane Domain Registration Through Interdigitation. Biochim. Biophys. Acta, Biomembr. 2017, 1859, 870–878. 10.1016/j.bbamem.2017.01.033. [DOI] [PubMed] [Google Scholar]
- Reigada R. Alteration of Interleaflet Coupling Due to Compounds Displaying Rapid Translocation in Lipid Membranes. Sci. Rep. 2016, 6, 32934. 10.1038/srep32934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano D. A.; Moore P. B.; Klein M. L.; Discher D. E. Raft Registration Across Bilayers in a Molecularly Detailed Model. Soft Matter 2011, 7, 8182–8191. 10.1039/c1sm05490b. [DOI] [Google Scholar]
- Sornbundit K.; Modchang C.; Triampo W.; Triampo D.; Nuttavut N.; Sunil Kumar P. B.; Laradji M. Kinetics of Domains Registration in Multicomponent Lipid Bilayer Membranes. Soft Matter 2014, 10, 7306–7315. 10.1039/C4SM01059K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink S. J.; de Vries A. H.; Harroun T. A.; Katsaras J.; Wassall S. R. Cholesterol Shows Preference for the Interior of Polyunsaturated Lipid Membranes. J. Am. Chem. Soc. 2008, 130, 10–11. 10.1021/ja076641c. [DOI] [PubMed] [Google Scholar]
- Bennett W. F. D.; MacCallum J. L.; Hinner M. J.; Marrink S. J.; Tieleman D. P. Molecular View of Cholesterol Flip-Flop and Chemical Potential in Different Membrane Environments. J. Am. Chem. Soc. 2009, 131, 12714–12720. 10.1021/ja903529f. [DOI] [PubMed] [Google Scholar]
- Jo S.; Rui H.; Lim J. B.; Klauda J. B.; Im W. Cholesterol Flip-Flop: Insights from Free Energy Simulation Studies. J. Phys. Chem. B 2010, 114, 13342–13348. 10.1021/jp108166k. [DOI] [PubMed] [Google Scholar]
- Collins M. D. Interleaflet Coupling Mechanisms in Bilayers of Lipids and Cholesterol. Biophys. J. 2008, 94, L32–L34. 10.1529/biophysj.107.124362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner M. D.; Feigenson G. W. Presence and Role of Midplane Cholesterol in Lipid Bilayers Containing Registered or Antiregistered Phase Domains. J. Phys. Chem. B 2018, 122, 8193–8200. 10.1021/acs.jpcb.8b03949. [DOI] [PubMed] [Google Scholar]
- Risselada H. J.; Marrink S. J. The Molecular Face of Lipid Rafts in Model Membranes. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 17367–17372. 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Melo M. N.; van Eerden F. J.; Arnarez C.; Lopez C. A.; Wassenaar T. A.; Periole X.; de Vries A. H.; Tieleman D. P.; Marrink S. J. Lipid Organization of the Plasma Membrane. J. Am. Chem. Soc. 2014, 136, 14554–14559. 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Carpenter T. S.; Bhatia H.; Bremer P.-T.; Marrink S. J.; Lightstone F. C. Computational Lipidomics of the Neuronal Plasma Membrane. Biophys. J. 2017, 113, 2271–2280. 10.1016/j.bpj.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; de Vries A. H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; Tieleman D. P. Perspective on the Martini Model. Chem. Soc. Rev. 2013, 42, 6801–6822. 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High Performance Molecular Simulations Through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Corradi V.; Mendez-Villuendas E.; Ingólfsson H. I.; Gu R.-X.; Siuda I.; Melo M. N.; Moussatova A.; DeGagné L. J.; Sejdiu B. I.; Singh G.; Wassenaar T. A.; Delgado Magnero K.; Marrink S. J.; Tieleman D. P. Lipid–Protein Interactions Are Unique Fingerprints for Membrane Proteins. ACS Cent. Sci. 2018, 4, 709–717. 10.1021/acscentsci.8b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.