Abstract
The chloroplast genome (CPG) of Pinus massoniana belonging to the genus Pinus (Pinaceae), which is a primary source of turpentine, was sequenced and analyzed in terms of gene rearrangements, ndh genes loss, and the contraction and expansion of short inverted repeats (IRs). P. massoniana CPG has a typical quadripartite structure that includes large single copy (LSC) (65,563 bp), small single copy (SSC) (53,230 bp) and two IRs (IRa and IRb, 485 bp). The 108 unique genes were identified, including 73 protein-coding genes, 31 tRNAs, and 4 rRNAs. Most of the 81 simple sequence repeats (SSRs) identified in CPG were mononucleotides motifs of A/T types and located in non-coding regions. Comparisons with related species revealed an inversion (21,556 bp) in the LSC region; P. massoniana CPG lacks all 11 intact ndh genes (four ndh genes lost completely; the five remained truncated as pseudogenes; and the other two ndh genes remain as pseudogenes because of short insertions or deletions). A pair of short IRs was found instead of large IRs, and size variations among pine species were observed, which resulted from short insertions or deletions and non-synchronized variations between “IRa” and “IRb”. The results of phylogenetic analyses based on whole CPG sequences of 16 conifers indicated that the whole CPG sequences could be used as a powerful tool in phylogenetic analyses.
Keywords: conifer species, genome annotation, structural inversion, comparative genomics, phylogenetic analysis
1. Introduction
The chloroplast genome (CPG) has multiple copies in a chloroplastid, which consists of 110–210 kb of circular DNA [1,2,3]. It has a quadripartite structure containing large and small single copy (LSC and SSC) regions, and two inverted repeats (IRs). There are 110–130 genes in most land plant CPGs [3,4]. With the development of next-generation sequencing, more than 1,000 CPGs have been reported in NCBI (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=2759&opt=plastid).
Large IRs are typical structures in CPGs, and have sizes ranging from 15 kb to 30 kb [3,5]. Some tRNA and rRNA genes (e.g., trnaI-GAU, trnV-GAC and 16S rRNA) are located in the large IRs. However, there are some differences in the gene numbers among species because of the contraction or expansion of IRs [5]. In addition, large IRs play important roles in stabilizing the CPG structure [6,7] due to the low rate of nucleotide substitution and enhanced copy-correction activity [5,8]. Thus, the loss of large IRs could result in the shortening of intergenic spaces [6], gene loss, and structural variations in CPGs [7,9]. The loss of large IRs has been confirmed in species in Pinaceae, Taxodiaceae [10,11], Cephalotaxaceae [12] and Legumes [8].
The contraction or expansion of large IRs could cause gene loss in some species’ CPGs, as mentioned above. In addition, gene transfer among chloroplast, mitochondrial, and nuclear genomes could lead to the transfer of some CPG genes to the nuclear genome [13]. A copy of the accD gene, which has been lost in the CPGs of Sciadopitys verticillata, has been found in the nuclear genome [10]. The infA gene has also transferred from the CPGs to the nuclear genome [14]. Additionally, the CPGs of species in Orchidaceae [15], Geraniaceae [16], and Pinaceae [17] lack all of the 11 intact ndh genes, which can also be observed in many land plants [15], and is related to gene transfer or ndh gene functions [15,17,18]. Besides those mentioned previously, the coding regions of matK, rbcL, and rpoB genes and non-coding regions of atpF-atpH, trnH-psbA, and psbK-psbI have been widely used in phylogenetic analyses of plants [19,20]. Additionally next-generation sequencing allows the LSC, SSC, and IRs regions, and shared protein-coding genes (PCGs), to be used as powerful tools in phylogenetic analyses [5,21].
Pinaceae is the largest family in gymnosperms, which includes three subfamilies, 10 genera and more than 230 species. Most of the species in Pinaceae are forest and timber species distributed in the northern hemisphere. Pinus is the only genus in the subfamily Pinoideae, which consists of more than 80 species. The species in Pinus are also the main wood and turpentine producers worldwide. In southern China, Pinus massoniana is an outstanding wood resource due to its economic value and broad geographic distribution. Additionally, the chloroplast genome plays an important role in studies of phylogenetic analysis, parental analysis, genetic structure, and germplasm resources evaluation, because of its characteristics of uniparental inheritance and conserved sequence.
In the present study, we obtained the complete CPG sequence of Pinus massoniana, and describe its gene content and microsatellite distribution. Comparisons with related species for gene content, gene rearrangements, ndh gene loss, and the contraction and expansion of short IRs were also performed. A phylogenetic analysis was performed on the basis of the whole CPGs of 16 conifers.
2. Results and Discussion
2.1. Genome Organization and Comparison with Other Species
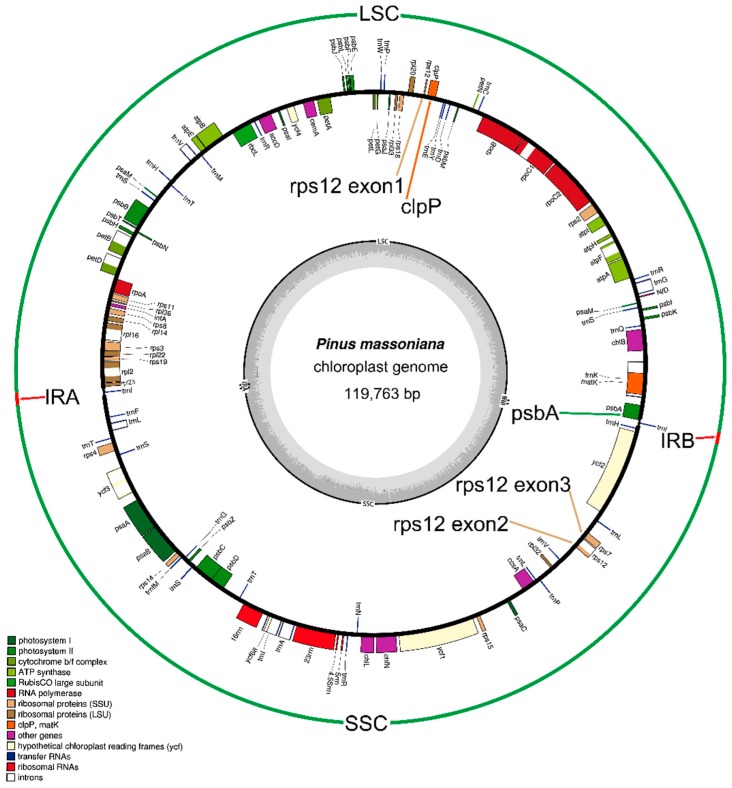
Using 35 primer pairs [22], we obtained the whole CPG sequence (119,763 bp) of P. massoniana (GenBank accession number: MF564195), which possesses a typical quadripartite structure including the LSC (65,563 bp) and SSC (53,230 bp) regions and the IRa and IRb pair (485 bp) (Figure 1). Compared with species in Magnoliaceae (159–161 kb), Salicaceae (Angiosperms) (156–171 kb) and Ginkgoaceae (Gymnosperms) (~157 kb), all of the species in Pinus have shorter CPGs (116–122 kb) (Supplementary Table S1) [3,23,24]. In addition, we observed differences in CPG sizes among species (Pinus) as follows: the CPGs of species in section Pinus (119–122 kb) > CPGs of species in section Parrya and section Cembra (116–118 kb) [25,26]; and the CPGs of species originating from the Americas (>120 kb) > CPGs of species originating from the Eurasian continent (119–120 kb) [22,26,27]. Compared with species in Magnoliaceae, Salicaceae, and Ginkgoaceae (SSC: 18–23 kb and IR: 17–28 kb), the CPGs of species in Pinus have longer SSCs (51–55 kb) and shorter IRs (400–500 bp). The previous studies revealed that the contraction of IRs could result in the expansion of SSC regions in Pinus [5,21].
Figure 1.
Chloroplast genome annotation map for Pinus massoniana. Genes lying outside the circle are transcribed in a clockwise direction, whereas genes inside are transcribed in a counterclockwise direction. Different colors represent different functional groups. The dashed darker and lighter gray in the inner circle denote GC and AT contents of chloroplast genome, respectively. LSC, SSC and IRs means long single copy, small sngle copy, and inverted repeat regions, respectively.
The CPG of P. massoniana has a similar GC content to those of other Pinus species. However, few differences in the GC contents were found among the CPGs. The GC content is highest in the SSC region (39.4%), moderate in the LSC region (37.9%), and lowest in the IR region (36.3%). The base contents are different from Oryza minuta CPGs, in which the GC content is unequally distributed in different regions, and the IR regions have the highest GC percentage [1].
2.2. Gene Contents
We identified 113 genes in the CPG of Masson pine, of which 108 are unique genes; these consist of 73 PCGs, 31 tRNA genes, and 4 rRNA genes (Table 1, Figure 1). The gene contents are similar to those of Pinus taiwanensis, but there are two additional genes (ycf12 and ycf68, unknown functions) than in Pinus bungeana [25,28]. In addition to the two PCGs (rps12 and ycf3) that have two introns each, six PCGs (atpF, petB, petD, rpl2, rpl16, and rpoC1) and six tRNA genes (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) contain single introns. Like the clade known as the IR-lacking clade (IRLC) of legumes, clpP intron loss was also observed in P. massoniana CPGs, although the clpP gene has two introns in several angiosperms and G. biloba [3,29]. The clpP intron loss was also observed in the Passiflora edulis CPGs [4,29]. trnK-UUU has the longest intron (2501 bp), in which the matK gene is located according to a previous report on Gentiana [30].
Table 1.
Gene contents of P.massoniana chloroplast genome based on genome annotation.
| Category | Gene Contents |
|---|---|
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ, psaMa |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Small subunit of ribosome | rps2, rps3, rps4, rps7, rps8, rps11, rps12b, rps14, rps15, rps18, rps19 |
| Large subunit of ribosome | rpl2b, rpl14, rpl16b, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Subunits of cytochrome b/f complex | petA, petBb, petDb, petG, petL, petN |
| Subunits of ATP synthase | atpA, atpB, atpE, atpFb, atpH, atpI |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1b, rpoC2 |
| ChlorophyII biosynthesis | chlB, chlL, chlN |
| Protease | clpP |
| Maturase | matK |
| Envelope membrane protein | cemA |
| Translation initiation factor | infA |
| Cytochrome c biogenesis | ccsA |
| Subunit Acetyl-CoA-Carboxylate | accD |
| Subunit of rubisco | rbcL |
| Ribosomal RNAs | rrn4.5, rrn5, rrn16, rrn23 |
| Conserved open reading frames | ycf1, ycf2, ycf3b, ycf4, ycf12, ycf68 |
| Transfer RNA | trnA-UGCb, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC, trnG-GCCb, trnH-GUGa, trnI-GAUab, trnK-UUUb |
| trnL-CAA, trnL-UAAb, trnL-UAG, trnM-CAU, trnN-GUU, trnP-GGG, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-CCG, trnR-UCU | |
| trnS-GCUa, trnS-GGA, trnS-UGA, trnT-GGUa, trnT-UGU, trnV-GAC, trnV-UACb, trnW-CCA, trnY-GUA |
a Gene-copies in genome; b Intro-containing gene.
The LSC of the P. massoniana CPG contains 73 genes, including 17 tRNA genes and 56 PCGs. Additionally, 17 tRNA genes, 18 PCGs, and 4 rRNA genes are located in the SSC, while IRs only contain trnI-GAU, which is different from the species containing large IRs in their CPGs [31]. The boundary of the LSC-IRb is located in the psbA gene with 5′-psbA located in the LSC and 3′-psbA in the IR region. In addition, the PCGs trans-spliced across the boundary of the LSC and IR regions were in the CPGs in some other plants (e.g., rps12 of P. edulis [4] and Gentiana) [30], in which the exon1 of rps12 gene was observed in the LSC region, and the other two exons were in the IR regions. On the other hand, in the P. massoniana CPG, the 5′ exon of the rps12 gene was located in the LSC region, far from the other two remaining exons, which were observed in the SSC region because of the loss of large IRs in the P. massoniana CPG (Figure 1) [32].
2.3. Structural and Gene Rearrangements
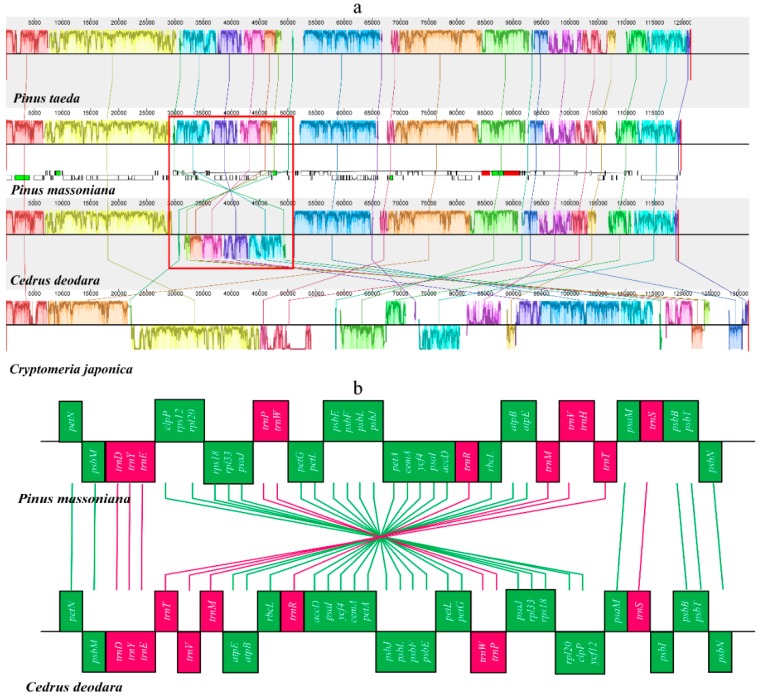
We compared P. massoniana organization with other conifers: P. taeda, Cedrus deodara, and Cryptomeria japonica. There were no structural variations between P. massoniana and P. taeda, while some inversions were observed among Pinus (P. massoniana and P. taeda), C. deodara, and C. japonica (Figure 2a). Figure 2a shows many structural variations between P. massoniana and C. japonica, but the former only showed one large inversion of 21,556 bp between the genes clpP and trnT-GGU in the LSC region based on a comparison with C. deodara [4]. The gene order in P. massoniana (clpP, rps12, rpl20, rps18, rpl33, psaJ, trnP, trnW, petG, petL, psbE, psbF, psbL, psbJ, petA, cemA, ycf4, psaI, accD, trnR, rbcL, atpB, atpE, trnM, trnV, trnH and trnT) changed to the inverted order in C. deodara (trnT, trnV, trnM, atpE, atpB, rbcL, trnR, accD, psaI, ycf4, cemA, petA, psbJ, psbL, psbF, psbE, petL, petG, trnW, trnP, psaJ, rpl33, rps18, rpl20 and clpP) (Figure 2b). Large IRs play important roles in stabilizing the CPGs against major structural variations, and the loss of large IRs could result in shorter intergenic spacers [6], more gene loss, and structural rearrangements [7,9]. Thus, the loss of the large IRs in the CPGs of P. massoniana and C. deodara may be the main cause of rearrangements in gene block order. The rearrangement of segments in CPGs are phylogenetically informative [4,6], and may be considered as useful tools in phylogenetic analyses.
Figure 2.
Synteny and rearrangements detected in chloroplast genome sequences of four Coniferous species using the Mauve multiple-genome alignment. (b) is a schematic illustration of the red frame part of the (a). (a) Color bars indicate syntenic blocks, and connecting lines indicate correspondence blocks; (b) Green boxes means protein-coding genes; red boxes means tRNAs. Boxes above and below the main line indicate the forward and reverse direction, respectively.
2.4. Microsatellite Polymorphisms
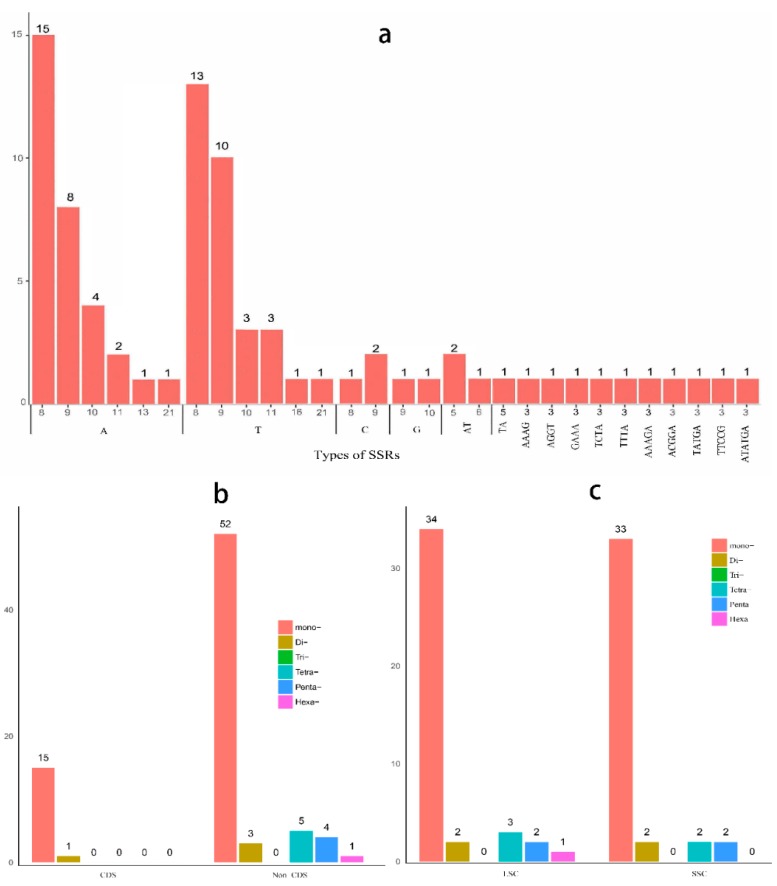
Microsatellites are SSRs with motifs of 1–6 bp that are distributed throughout the nuclear or plasmid genomes. In this study, 81 SSRs were detected in the P. massoniana CPG, with minimum SSRs of eight, five, four, three, three, and three for mono-, di-, tri-, tetra-, penta- and hexa-nucleotides, respectively (Figure 3). The majority of the SSRs are mono-; these account for 82.7% of the total SSRs (Figure 3a). Of these, 92.5% have A/T motifs, while only 7.5% have C/G motifs, which is in accordance with the trend of A/T-enrichment in CPGs [11,33]. Sixty-five of the 81 SSRs were found to be located in non-coding sequences (non-CDSs), while 19.75% of the total SSRs were located in the coding sequence (CDS) (Figure 3b). The similar distribution of numbers and types of SSRs were observed in the LSC and SSC regions of the P. massoniana CPG (Figure 3c). Among the 10 PCGs containing SSRs, two genes (ycf1 and ycf2) contained two or more SSRs motifs, while the other eight genes (chlL, petA, rpl32, rpl33, rpoC2, rps7, rps19 and ycf4) contained one mono-SSR of the A/T type. Although gene sequences are strongly conserved in CPGs, some SSR variations had already been detected in CDSs [34]. Some differences among numbers and types of SSRs were observed in CPG genes [4,35]. The clpP and ndhA genes contain tri- and tetra-SSRs, respectively, in the P. edulis CPG [4]. Additionally, five genes (psbC, accD, cemA, petA, ycf2 and ycf1) were found that contained two or more SSRs in the Ananas comosus CPG [35]. These all had different distributions from the SSRs in the P. massoniana CPG [35]. In addition, the SSR variations could result in frameshift mutations or loss of gene functions [34]. These are potential causes of gene loss or pseudogene formation.
Figure 3.
Distribution of each simple sequence repeats (SSR) category in chloroplast genome (CPG) of Pinus massoniana. (a) Distribution of each SSR category in whole chloroplast genome; (b) Distribution of each SSR category in the coding sequence (CDS) and non-CDS of CPG; (c) Distribution of each SSR category in LSC and SSC of CPG.
2.5. Loss of ndh Genes
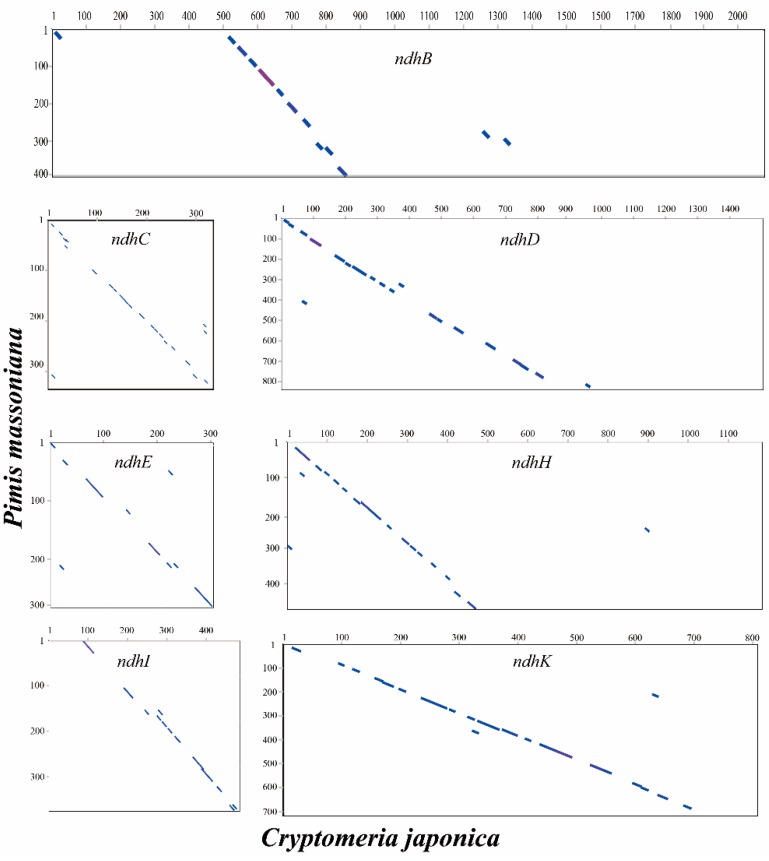
The ndh genes are located in nuclear, mitochondrial, and CPGs that can encode the NAD(P)H-dehydrogenase-like (NDH). With the exception of some species in Pinaceae [17], Orchidaceae [15], and Gnetales [36], 11 ndh genes, including ndhA–K, were found in most land plant CPGs. In this study, the comparisons with CPGs of C. japonica [9] and G. biloba [3] indicated that the masson pine CPG lacks all 11 intact ndh genes. Among these ndh genes in the masson pine CPG, the four genes ndhA, ndhF, ndhG and ndhJ have been lost completely, the ndhC and ndhE genes remain as pseudogenes because of short insertions or short deletions, and the other five ndh genes (ndhB, ndhH, ndhD, ndhI and ndhK) remain as truncated pseudogenes (Figure 4). As shown in Figure 4, the ndhI gene lost its 5′ end, while the other four truncated pseudogenes (ndhB, ndhH, ndhD and ndhK) lost their 3′ ends. ndhK lacks a short sequence from the 3′ end (79 bp), but it was not classified as a truncated pseudogene in a previous study of Pinus thunbergii [37].
Figure 4.
Dotpot analysis of seven ndh genes between P. massoniana and Cryptomeria japonica.
In previous studies, two phenomena of ndh gene loss have been observed. Some ndh genes are lost completely or transferred to nuclear genomes. For example, some non-functional plastid ndh gene fragments are found in the nuclear genome of Picea abies [17]. Other ndh genes are retained as pseudogenes in CPG which are absent in the nuclear genome. For example, most of the ndh genes lost from CPGs are not located in the nuclear genomes of particular orchids [15]. In addition, the chloroplast NDH complex encoded by ndh genes is not necessary in some photoautotrophic plants [15,16,17,18]. Thus, we hypothesize that the ndh genes (ndhA, ndhF, ndhG and ndhJ) that were absent completely in the CPG of P. massoniana have been transferred to the nuclear genome. However, this still needs further verification.
2.6. Contraction and Expansion of Short IRs in Pinus
Large IRs play vital roles in maintaining the stability of the CPG [6,7], and the loss of a large IR could result in some variations in the gene content and genome structures in CPGs [6,11,12]. IR expansion and contraction exists in many land plants [5]. However, there are no large IR regions in the CPGs of the conifers (e.g., Pinaceae and Taxodiaceae). They have been replaced by short IRs (400–500 bp) [21,25,38], but these do not exist in some conifers [7,9].
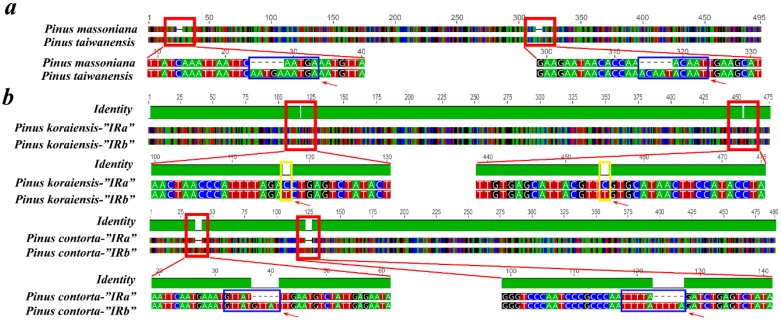
In the present study, we observed a pair of short IRs (485 bp) and an absence of large IRs. However, some variations in size and sequence were also found in short IRs among species in Pinus. The alignment results of “IRa” and “IRb” (screened using the reference sequences from P. taiwanensis) showed that the main causes of the variations are as follows: (1) Insertion or deletion of short sequences (variations in repeat numbers of microsatellite motifs). The IRs of P. massoniana are 10 bp shorter than those of P. taiwanensis because of the loss of a 5-bp microsatellite motif (AATGA and ACAAT) in two loci of IRs of P. massoniana (Figure 5a); (2) Non-synchronized variations between “IRa” and “IRb” (e.g., single nucleotide polymorphisms and SSRs). The alignment indicated that two single nucleotide polymorphisms and the loss of two 5-bp microsatellite motifs (GTTAT and TTTTA) were found in the “IRa” regions of Pinus koraiensis and Pinus contorta, respectively (Figure 5b). These could lead to variations in the size and sequence of short IRs in conifer CPGs, but they are not the main causes of large IR expansion and contraction. Large IRs can provide enhanced copy-correction activity because the rates of nucleotide substitution in IRs are several times slower than in SSC regions [5,8]. In addition, the loss of gene copies in large IRs could result in IR contraction, similar to how the loss of an ycf2 copy led to IR contraction in G. biloba [3]. Although the loss of large IRs has been confirmed in species of Cephalotaxaceae [12], Taxodiaceae [10,11] and Legumes [8], determining the main cause requires further study.
Figure 5.
Variations of inverted repeats (IRs) using multiple alignment. (a) Variations between P. massoniana and P. taiwanensis; (b) variations between “IRa” and “IRb” in P. koraiensis and P. contorta. Variations are in red frames: Single Nucleotide Polymorphisms (SNPs) are in yellow frames; microsatellites are in blue frames.
2.7. Phylogenetic Analysis
CPG sequences have been widely used in the phylogenetic analyses of land plants [5,12,26]. In particular, some CDSs of matK, rbcL and rpoB genes and non-CDSs of atpF-atpH, trnH-psbA and psbK-psbI have been used in phylogenetic studies [19,20]. However, few CDSs and non-CDSs in CPGs are not informative enough when studying closely related species and cultivars. Thus, whole CPG sequences that contain more variation could play important roles in phylogenetic analyses.
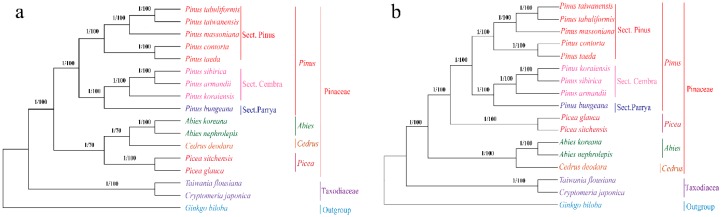
In the present study, phylogenetic analyses were performed based on the whole CPG sequences of 16 conifers, using G. biloba as an outgroup. We obtained a congruent phylogenetic tree with different support values using maximum likelihood (ML) and Bayesian inference (BI) approaches (Figure 6). All 16 conifers could be discriminated completely into two clades of Pinaceae and Taxodiaceae with high BI posterior probability and ML bootstrap support. Nine pine species were divided into two groups (sect. Pinus and sect. Cembra + sect. Parrya) (posterior probability = 1 and ML bootstrap support = 100), which supports the results of Parks et al [26]. In addition, P. massoniana is closely related to P. taiwanensis and P. tabuliformis, but not to P. taeda or P. contorta. These results corroborate previous studies [7,12,21]. However, the phylogenetic relationships based on whole CPG sequences between Pinus, Abies, Cedrus, and Picea in this study conflicted with previous studies. Here, Pinus was placed into a clade, while Abies, Cedrus, and Picea were placed into another clade, which is consistent with conventional plant taxonomy (Figure 6). In some previous studies, Pinus and Picea were placed into a clade, and Abies and Cedrus were placed into another clade, which is similar to our phylogenetic analyses based on 56 shared PCGs of 17 species (Supplementary Table S2). However, it is worth noting that the phylogenetic trees in previous studies were constructed based on shared PCGs [7,21] and low and high heterotachous data sets [12], while whole CPG sequences were used in our phylogenetic analyses. Thus, differences may be observed among the phylogenetic relationships established based on different CPG regions. As we all know, the chloroplast genome sequences are conserved and mainly embodied in the conservation of PCGs sequences. The result of average similarity scores of 56 shared PCGs sequences show that the similarity scores of shared PCGs sequences (average similarity score = 0.9592) in Pinaceae is much greater than that of Pinaceae-Ginkgoaceae (average similarity score = 0.8616) (Supplementary Table S3). Hence, particular CPG regions (like PCGs) that contain few variations among species are not suitable for establishing phylogenetic relationships of closely related species in Pinus, while whole CPG sequences may be powerful tools in phylogenetic analyses of closely related species or cultivars.
Figure 6.
Phylogenetic tree constructed by maximum likelihood (ML) and Bayesian inference (BI) methods based on whole chloroplast genome sequences and 56 shared protein-coding genes of 16 conifers. (a) Phylogenetic tree based on whole chloroplast genome sequences of 16 conifers; (b) Phylogenetic tree based on 56 shared protein-coding genes of 16 conifers chloroplast genomes; Ginkgo biloba as an outgroup; BI posterior probability/ML bootstrap support values were listed at each node.
3. Materials and Methods
3.1. DNA Source, Template Amplification
The needles of P. massoniana were collected from Longyan (Fujiang, China, N 25.25°, E 117.54°). Total DNA was extracted using the Plant Genprep DNA Kit (ZomanBio Inc., Beijing, China) and quantified using a NanoDrop 2000c (ThermoFisher Scientific, Wilmington, DE, USA). The entire CPG was amplified using PCR with 35 primer pairs [22]. Amplicons for these regions averaged ~3.6 kb, and fragments were obtained with PrimeSTAR Max DNA Polymerase (Takara Bio Inc., Dalian, China).
3.2. Sequencing, Assembly, PCR-Based Gap Filling and Annotation
PCR products were evaluated using electrophoresis on agarose gels and purified using a Gel Mini purification Kit (ZomanBio Inc., Beijing, China). All of the purified DNA products were sequenced using an ABI 3730 DNA sequencer. Sequence assemblies were performed using CAP3 [39] and manually confirmed against the reference CPG of Pinus taeda (KC427273.1).
The genome annotation was performed using a Dual Organellar GenoMe Annotator with default parameters [40], coupled with manual corrections of the start and stop codon positions. All of the tRNAs were identified using tRNAscan-SE v2.0 program (http://lowelab.ucsc.edu/tRNAscan-SE/) [41]. The boundaries of the exons and introns were verified using the BLASTn algorithm (2.6.0, National Center for Biotechnology Information, Bethesda, MD, USA, 2017) against other closely related pine species. The annotation map of the CPG was generated using Organellar Genome DRAW v1.2 program (http://ogdraw.mpimp-golm.mpg.de/) [42].
3.3. Simple Sequence Repeat Analysis
The microsatellite motifs were identified using the MIcroSAtellite identification tool (http://pgrc.ipk-gatersleben.de/misa/misa.html) with the following parameters [43]: the minimum repeats of SSRs were eight, five, four, three, three, and three mononucleotides (mono-), dinucleotides (di-), trinucleotides (tri-), tetranucleotides (tetra-), penta-nucleotides (penta-) and hexa-nucleotides, respectively.
3.4. Sequence Analysis
To highlight the differences in IRs between some species in Pinaceae and Taxodiaceae, a comparative analysis were conducted by aligning IR sequences using multiple sequence alignment software (MAFFT) [44]. Special attention was paid to the variations among the IR sequences. In addition, the loss of ndh genes was found through sequences alignments using MAFFT [44] based on the CPG annotation of P. massoniana. Whole-genome alignments were conducted to locate structural differences using MAUVE [45].
3.5. Phylogenetic Analysis
The whole CPG sequences of 16 conifers were used in a phylogenetic analysis with Ginkgo biloba as the outgroup (Supplementary Table S4). The phylogenetic analysis was carried out based on complete CPG sequences and 56 shared PCGs (Supplementary Table S2). The multiple alignments of the sequences mentioned above were performed on MAFFT [44]. The programs JModeltest2 [46] and Modelgenerator [47] were used to find an optimal substitution model for the subset by taking the Akaike Information Criterion (AIC) values into account [48]. The maximum likelihood (ML) tree of whole CPG sequences (Model: GTR+G) was conducted using RAxML 8.2.7 [49] with a bootstrap of 1000 replicates. A Bayesian inference (BI) analysis was run in MrBayes 3.2.6 with the setting of 1,000,000 generations with trees sampled every 1000 generations [50,51]. The first 25% of trees were discarded as burn-in to estimate the values of posterior probabilities.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No.31400575) and the Program Development of Jiangsu Higher Education Institutions (PAPD).
Supplementary Materials
Supplementary materials are available online.
Author Contributions
Z.N. and L.-A.X. conceived and designed the experiments; Z.N. performed the experiments and analyzed the data; Y.Y. contributed reagents/materials/analysis tools; Z.N. wrote the paper; T.B. and M.X. corrected the paper. Authorship must be limited to those who have contributed substantially to the work reported.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Sequence data of Pinus massoniana are not available from the authors.
References
- 1.Asaf S., Waqas M., Khan A.L., Khan M.A., Kang S.-M., Imran Q.M., Shahzad R., Bilal S., Yun B.-W., Lee I.-J. The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species. Front. Plant Sci. 2017;8:304. doi: 10.3389/fpls.2017.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniell H., Lin C.-S., Yu M., Chang W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin C.-P., Wu C.-S., Huang Y.-Y., Chaw S.-M. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol. Evol. 2012;4:374–381. doi: 10.1093/gbe/evs021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cauz-Santos L.A., Munhoz C.F., Rodde N., Cauet S., Santos A.A., Penha H.A., Dornelas M.C., Varani A.M., Oliveira G.C.X., Bergès H., et al. The chloroplast genome of Passiflora edulis (passifloraceae) assembled from long sequence reads: Structural organization and phylogenomic studies in malpighiales. Front. Plant Sci. 2017;8:334. doi: 10.3389/fpls.2017.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu A., Guo W., Gupta S., Fan W., Mower J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016;209:1747–1756. doi: 10.1111/nph.13743. [DOI] [PubMed] [Google Scholar]
- 6.Wu C.-S., Chaw S.-M. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): Evolution towards shorter intergenic spacers. Plant Biotechnol. J. 2014;12:344–353. doi: 10.1111/pbi.12141. [DOI] [PubMed] [Google Scholar]
- 7.Zheng W., Chen J., Hao Z., Shi J. Comparative analysis of the chloroplast genomic information of Cunninghamia lanceolata (Lamb.) Hook with sibling species from the Genera Cryptomeria D. Don, Taiwania Hayata, and Calocedrus Kurz. Int. J. Mol. Sci. 2016;17:1084. doi: 10.3390/ijms17071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry A.S., Wolfe K.H. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J. Mol. Evol. 2002;55:501–508. doi: 10.1007/s00239-002-2333-y. [DOI] [PubMed] [Google Scholar]
- 9.Hirao T., Watanabe A., Kurita M., Kondo T., Takata K. Complete nucleotide sequence of the Cryptomeria japonica D. Don. Chloroplast genome and comparative chloroplast genomics: Diversified genomic structure of coniferous species. BMC Plant Biol. 2008;8:70. doi: 10.1186/1471-2229-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Gao L., Chen S., Tao K., Su Y., Wang T. Evolution of short inverted repeat in cupressophytes, transfer of accd to nucleus in Sciadopitys verticillata and phylogenetic position of Sciadopityaceae. Sci. Rep. 2016;6:20934. doi: 10.1038/srep20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi X., Gao L., Wang B., Su Y.-J., Wang T. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol. Evol. 2013;5:688–698. doi: 10.1093/gbe/evt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C.-S., Wang Y.-N., Hsu C.-Y., Lin C.-P., Chaw S.-M. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and Cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol. 2011;3:1284–1295. doi: 10.1093/gbe/evr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams K.L., Daley D.O., Qiu Y.-L., Whelan J., Palmer J.D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature. 2000;408:354–357. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- 14.Millen R.S., Olmstead R.G., Adams K.L., Palmer J.D., Lao N.T., Heggie L., Kavanagh T.A., Hibberd J.M., Gray J.C., Morden C.W., et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin C.-S., Chen J.J.W., Chiu C.-C., Hsiao H.C.W., Yang C.-J., Jin X.-H., Leebens-Mack J., de Pamphilis C.W., Huang Y.-T., Yang L.-H., et al. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 2017;90:994–1006. doi: 10.1111/tpj.13525. [DOI] [PubMed] [Google Scholar]
- 16.Chris Blazier J., Guisinger M.M., Jansen R.K. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae) Plant Mol. Biol. 2011;76:263–272. doi: 10.1007/s11103-011-9753-5. [DOI] [PubMed] [Google Scholar]
- 17.Ranade S.S., García-Gil M.R., Rosselló J.A. Non-functional plastid ndh gene fragments are present in the nuclear genome of Norway spruce (Picea abies L. Karsch): Insights from in silico analysis of nuclear and organellar genomes. Mol. Genet. Genomics. 2016;291:935–941. doi: 10.1007/s00438-015-1159-7. [DOI] [PubMed] [Google Scholar]
- 18.Martín M., Sabater B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 2010;48:636–645. doi: 10.1016/j.plaphy.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Gizaw A., Brochmann C., Nemomissa S., Wondimu T., Masao C.A., Tusiime F.M., Abdi A.A., Oxelman B., Popp M., Dimitrov D. Colonization and diversification in the African sky ‘islands‘: Insights from fossil-calibrated molecular dating of Lychnis (Caryophyllaceae) New Phytol. 2016;211:719–734. doi: 10.1111/nph.13937. [DOI] [PubMed] [Google Scholar]
- 20.Group C.P.W. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C.-P., Huang J.-P., Wu C.-S., Hsu C.-Y., Chaw S.-M. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol. Evol. 2010;2:504–517. doi: 10.1093/gbe/evq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cronn R., Liston A., Parks M., Gernandt D.S., Shen R., Mockler T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai Z., Penaflor C., Kuehl J.V., Leebens-Mack J., Carlson J.E., de Pamphilis C.W., Boore J.L., Jansen R.K. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: Implications for the phylogenetic relationships of magnoliids. BMC Evol. Biol. 2006;6:77. doi: 10.1186/1471-2148-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuskan G.A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa(Torr. & Gray) Science. 2006;313:1596. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 25.Li Z.H., Zhu J., Yang Y.X., Yang J., He J.W., Zhao G.F. The complete plastid genome of bunge’s pine Pinus bungeana (Pinaceae) Mitochondr. DNA Part A. 2016;27:2971–2972. doi: 10.3109/19401736.2015.1060465. [DOI] [PubMed] [Google Scholar]
- 26.Parks M., Cronn R., Liston A. Separating the wheat from the chaff: Mitigating the effects of noise in a plastome phylogenomic data set from Pinus L. (Pinaceae) BMC Evol. Biol. 2012;12:100. doi: 10.1186/1471-2148-12-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks M., Cronn R., Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang M.F., Wang Y.J., Zu Y.M., Dong W.L., Wang R.N., Deng T.T., Li Z.H. The complete chloroplast genome of the taiwan red pine Pinus taiwanensis (Pinaceae) Mitochondr. DNA Part A. 2016;27:2732–2733. doi: 10.3109/19401736.2015.1046169. [DOI] [PubMed] [Google Scholar]
- 29.Jansen R.K., Wojciechowski M.F., Sanniyasi E., Lee S.-B., Daniell H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae) Mol. Phylogenet. Evol. 2008;48:1204–1217. doi: 10.1016/j.ympev.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni L., Zhao Z., Xu H., Chen S., Dorje G. Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine. Curr. Genet. 2017;63:241–252. doi: 10.1007/s00294-016-0631-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Iaffaldano B.J., Zhuang X., Cardina J., Cornish K. Chloroplast genome resources and molecular markers differentiate rubber dandelion species from weedy relatives. BMC Plant Biol. 2017;17:34. doi: 10.1186/s12870-016-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y., Woeste K.E., Zhao P. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front. Plant Sci. 2016;7:1955. doi: 10.3389/fpls.2016.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y., Dong W., Liu B., Xu C., Yao X., Gao J., Corlett R.T. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 2015;6:662. doi: 10.3389/fpls.2015.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M.L., Barkley N.A., Jenkins T.M. Microsatellite markers in plants and insects. Part I: Applications of biotechnology. Genes Genomes Genom. 2009;29:623–627. [Google Scholar]
- 35.Redwan R., Saidin A., Kumar S. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 2015;15:294. doi: 10.1186/s12870-015-0587-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braukmann T.W.A., Kuzmina M., Stefanović S. Loss of all plastid ndh genes in Gnetales and conifers: Extent and evolutionary significance for the seed plant phylogeny. Curr. Genet. 2009;55:323–337. doi: 10.1007/s00294-009-0249-7. [DOI] [PubMed] [Google Scholar]
- 37.Wakasugi T., Tsudzuki J., Ito S., Nakashima K., Tsudzuki T., Sugiura M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z.H., Qian Z.Q., Liu Z.L., Deng T.T., Zu Y.M., Zhao P., Zhao G.F. The complete chloroplast genome of armand pine Pinus armandii, an endemic conifer tree species to China. Mitochondr. DNA Part A. 2016;27:2635–2636. doi: 10.3109/19401736.2015.1041130. [DOI] [PubMed] [Google Scholar]
- 39.Huang X., Madan A. Cap3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 41.Lowe T.M., Chan P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohse M., Drechsel O., Kahlau S., Bock R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:W575–W581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiel T., Michalek W., Varshney R., Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare H.) Theor. Appl. Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 44.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darling A.C.E., Mau B., Blattner F.R., Perna N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keane T.M., Creevey C.J., Pentony M.M., Naughton T.J., Mclnerney J.O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goss E.M., Tabima J.F., Cooke D.E.L., Restrepo S., Fry W.E., Forbes G.A., Fieland V.J., Cardenas M., Grünwald N.J. The Irish potato famine pathogen Phytophthora infestans originated in central Mexico rather than the Andes. Proc. Natl. Acad. Sci. USA. 2014;111:8791–8796. doi: 10.1073/pnas.1401884111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 50.Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 51.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.