Abstract
A quantitative structure – antioxidant activity relationship (QSAR) study of 36 flavonoids was performed using the partial least squares projection of latent structures (PLS) method. The chemical structures of the flavonoids have been characterized by constitutional descriptors, two-dimensional topological and connectivity indices. Our PLS model gave a proper description and a suitable prediction of the antioxidant activities of a diverse set of flavonoids having clustering tendency.
Keywords: Antioxidants, free radicals, flavonoids, QSAR, chemometrics, partial least squares, PLS
Introduction
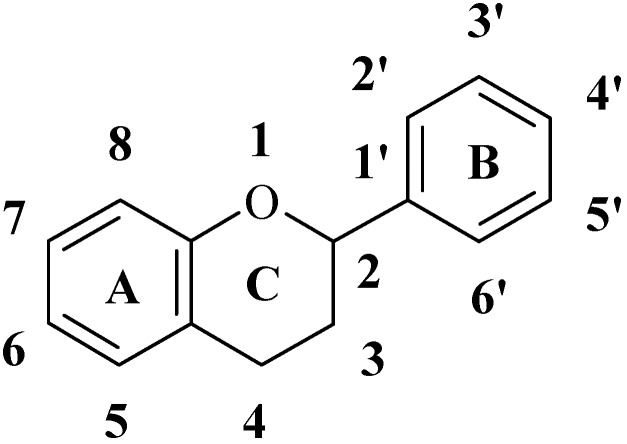
Flavonoids (see Figure 1), a group of polyphenolic compounds, can widely be found in fruits and vegetables. Numerous positive health effects of flavonoids have been described. They have been reported to exhibit anti-cancer [1,2,3], anti-viral [4] and anti-inflammatory [3,5,6] effects, and to reduce the risk of cardiovascular diseases [3,7,8,9]. These activities are generally associated with antioxidant or free radical-scavenging properties of flavonoids. The number of flavonoid derivatives is more than 4000 and their antioxidant properties are very different. It is a complex task to select the most effective antioxidants from a large amount of flavonoids. Quantitative structure-activity relationships (QSAR) have been often used to find correlations between biological activities and physicochemical properties of compounds. Because of their great number and positive biological effects, flavonoids are popular subjects of QSAR studies. Successful prediction of anti-HIV [10,11] and anti-tumor activities [12,13] of flavonoid compounds have been reported. Prediction of their inhibitory ability of cytochrome P-450 [14] and p5(lck) protein tyrosine kinase [15,16,17] has also been performed.
Figure 1.
General flavonoid structure
Many structure-activity relationship (SAR) investigations have been performed on the antioxidant activity of flavonoids, however, only a few were, in fact, quantitative. According to these studies, the antioxidant activity of flavonoids depends strongly on the number and position of hydroxyl groups in the molecule. Dihydroxylated B-ring (catechol structure), presence of unsaturation and of 4-oxo function in the C-ring are also presumed to increase the antioxidant capacity [18,19,20]. Lien et al. applied calculated parameters such as heat of formation, energy of highest occupied and lowest unoccupied molecular orbital, and number of OH groups for description of antiradical activity of flavonoids and other phenolic compounds [21]. Amić et al. defined indicator variables as the sum of the numbers and position of OH groups in the flavonoids [22]. It is noteworthy, that these models have been developed only for description of existing data. Our aims were (i) to build quantitative models for description and prediction of antioxidant activities of flavonoid compounds, as well as (ii) to classify them on the basis of biological activities and structural encoding.
Experimental
The antioxidant activity data of 36 flavonoids has been taken from reference [23]. These flavonoids can be grouped as follows: flavonols, flavones, flavanones, dihydroflavonols and their carbohydrate derivatives, biflavanones and isoflavones. Their antioxidant activities have been characterized by the ability to inhibit heat-induced oxidation in a β-carotene-linoleic acid-model-system [23]. Structures of flavonoids and antioxidant activity values are shown in Table 1
Table 1.
Structures and antioxidant activity of tested flavonoids.
Flavonols and flavones
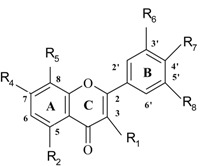
| No | Name | Code | Set** | R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | Antioxidant activity (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | kaempferol | kl | 1 | OH | OH | OH | H | H | H | OH | H | 65.3 |
| 2 | galangin | gl | 2 | OH | OH | OH | H | H | H | H | H | 64.9 |
| 3 | quercetin | qu | 1 | OH | OH | OH | H | H | OH | OH | H | 63.6 |
| 4 | robinetin | ro | 1 | OH | H | OH | H | H | OH | OH | OH | 61.7 |
| 5 | fisetin | fi | 1 | OH | H | OH | H | H | OH | OH | H | 61.6 |
| 6 | kaempferide | kd | 1 | OH | OH | OH | H | H | H | OMe | H | 60.0 |
| 7 | 3-hydroxy-flavone | h3 | 1 | OH | H | H | H | H | H | H | H | 59.4 |
| 8 | laricytrin | la | 2 | OH | OH | OH | H | H | OH | OH | OMe | 28.5 |
| 9 | laricytrin 3’- O-glucoside | l3 | 1 | OH | OH | OH | H | H | O-glu* | OH | OMe | 26.2 |
| 10 | myricetin | my | 1 | OH | OH | OH | H | H | OH | OH | OH | 18.4 |
| 11 | 3,5,7,3’,4’,5’-hexamethoxy-flavone | hm | 1 | OMe | OMe | OMe | H | H | OMe | OMe | OMe | 2.6 |
| 12 | 3,5,7,3’,4’-pentamethoxy-flavone | pm | 2 | OMe | OMe | OMe | H | H | O-glu | OMe | OH | 1.1 |
| 13 | larycitrin 3,3’- O-diglucoside | ld | 2 | O-glu | OH | OH | H | H | OH | OH | OMe | 1.1 |
| 14 | quercetin 3- O-glucoside-7-O-rhamnoside | qg | 1 | O-glu | OH | O-rha* | H | H | O-glu | OH | H | -6.2 |
| 15 | laricyrin 3,7,3’- O-triglucoside | lt | 1 | O-glu | OH | O-glu | H | H | OH | OH | OMe | -6.2 |
| 16 | rutin | ru | 1 | O-rut* | OH | OH | H | H | H | OH | H | -10.2 |
| 17 | morin | mo | 1 | OH | OH | OH | H | OH | H | OH | H | 63.5 |
| 18 | flavone | fl | 1 | H | H | H | H | H | H | H | H | -1.5 |
| 19 | 5-hydroxy-flavone | h5 | 2 | H | OH | H | H | H | H | H | H | -4.0 |
| 20 | 7-hydroxy-flavone | h7 | 1 | H | H | OH | H | H | H | H | H | 0.0 |
| 21 | chrysin | cr | 2 | H | OH | OH | H | H | H | H | H | -20.8 |
| 22 | 8-methoxy-flavone | m8 | 2 | H | H | OMe | H | H | H | H | H | -29.2 |
| 23 | apigenin 8- C-glucoside | a8 | 1 | H | OH | OH | glu | H | H | OH | H | -29.6 |
| 24 | luteolin 7- O-glucoside | lu | 2 | H | OH | O-glu | H | H | OH | OH | H | -25.3 |
Flavanones and dihydroflavonols
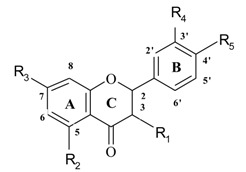
| No | Name | Code | Set** | R1 | R2 | R3 | R4 | R5 | Antioxidant activity |
|---|---|---|---|---|---|---|---|---|---|
| 25 | flavanone | fn | 1 | H | H | H | H | H | -23.0 |
| 26 | naringin | nh | 1 | H | OH | O-neohesp* | H | OH | 47.4 |
| 27 | hesperitin | he | 2 | H | OH | OH | OH | OMe | 4.7 |
| 28 | fustin | fu | 1 | OH | H | OH | OH | OH | -23.4 |
| 29 | taxifolin | ta | 1 | OH | OH | OH | OH | OH | -16.8 |
Biflavanones
| No | Name | Code | Set** | R1 | R2 | R3 | Antioxidant activity |
|---|---|---|---|---|---|---|---|
| 30 | GB-1 | gb | /1 | OH | OH | H | -30.1 |
| 31 | GB-1a | ga | /2 | H | OH | H | -16.9 |
Isoflavones

| No | Name | Code | Set** | R1 | R2 | R3 | Antioxidant activity |
|---|---|---|---|---|---|---|---|
| 32 | daidzein | da | 1 | H | OH | OH | 32.9 |
| 33 | formononetin | fm | 1 | H | OH | OMe | -20.4 |
| 34 | genistein | ge | 1 | OH | OH | OH | -24.6 |
| 35 | biochanin A | bi | 2 | OH | OH | OMe | -20.4 |

| No | Name | Code | Set | Antioxidant activity |
|---|---|---|---|---|
| 36 | coumestrol | cu | 1 | 38.7 |
* O-glu: glucose, O-rha: rhamnose, O-rut: rutin, O-neohesp: neohesperidin; These sugars are connected via their glucoside OH group to the flavonoids.
** 1: training set of flavonoids, 2: test set of flavonoids
The two-dimensional structures were represented using the HyperChem 7.0 program package [24]. The independent variables used for description of the antioxidant activities were constitutional and 2D properties such as topological indices, connectivity indices and molecular walk counts. Using 2D descriptors instead of 3D properties reduced the calculation time significantly. Descriptors have been calculated using the Dragon program package [25]. The program omitted the constant and near constant descriptors, as well as one of the two (or more) variables automatically, which showed a correlation higher than 0.99. The number of calculated descriptors was 147. Model building has been performed by Statistica 5.5 program package [26] using the partial least squares projection of latent structures (PLS) method. The stability and validity of the models have been tested by leave-n-out cross-validation technique. Sixty-seven percent of the flavonoid molecules were chosen as a training- and thirty three percent as a test set. The training set was selected as follows: (i) the entire antioxidant activity scale has been covered, (ii) each flavonoid group was represented, including sugar containing and sugar free flavonoids. The real predictive power has been measured by the PRESS value (predicted error sum of squares). The number of PLS components in the model has been chosen on a minimum value of PRESS and maximum value of R2 (coefficient of determination). Further examination of model applicability was undertaken by reviewing the plot of PLS component scores and the plot of the predicted versus experimental antioxidant activity data of flavonoids.
Results and Discussion
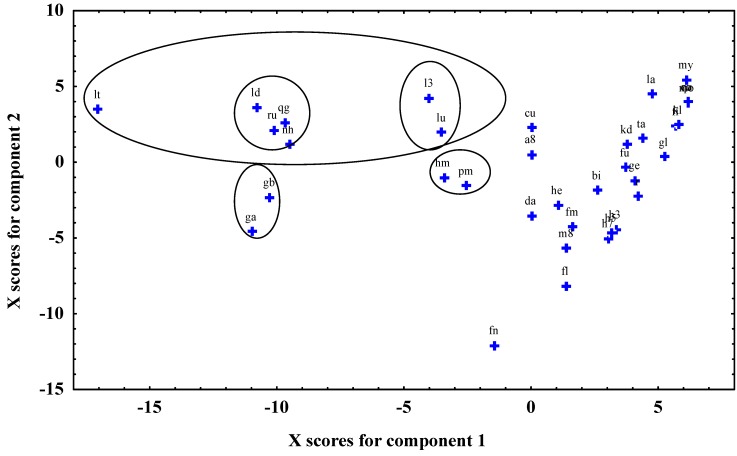
Retaining 13 PLS components in the model has resulted in a minimum of the PRESS value (PRESS=0.6253). The R2 value at the minimum value of Y PRESS was 0.9888. These statistical parameters show unambiguously that both the descriptive and the predictive power of the model is appropriate. The regression coefficient values of the independent variables show that connectivity indices play an important role in the description of antioxidant activities. The following descriptors have the highest regression coefficient in the PLS model: x3a (average connectivity index chi1), x2a (x3a (average connectivity index chi2), x3av (average valence connectivity index chi3), x5av (average valence connectivity index chi5), JGI5 (mean topological charge index of order 5), JGI8 (mean topological charge index of order 10). It is interesting, that constitutive descriptors (e.g. number of OH groups in the molecule) have no significant role in this PLS model. Figure 2 shows the PLS component scores. Several separated flavonoid groups can be observed on this plot. Carbohydrate derivatives of flavonoids (lt, ld, ru, qg, nh, l3, lu, for notation see Table 1) can be well distinguished from the other compounds. Biflavanones (gb, ga) and flavonoids with a lot of methoxy groups (hm, pm) are also clearly separated from other groups.
Figure 2.
Plot of PLS component scores (for abbreviations see code column in Table 1)
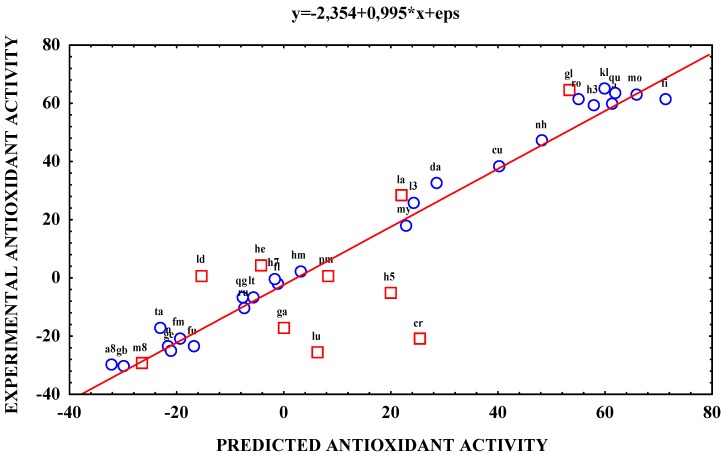
The plot of the predicted vs. experimental antioxidant activities is given in Figure 3. In order to give better comparison between the predicted and experimental data Table 2 shows both antioxidant activity values.
Figure 3.
Plot of predicted versus experimental antioxidant activities of flavonoids (for abbreviations see code column in Table 1)
Table 2.
Predicted and experimental antioxidant activities of flavonoids (for abbreviations see code column in Table 1)
| Antioxidant activity | Antioxidant activity | ||||
|---|---|---|---|---|---|
| Code | Experimental | Predicted | Code | Experimental | Predicted |
| a8 | -29.6 | 53.3 | hm | 2.6 | 22.7 |
| bi | -20.4 | -30.1 | kd | 60.0 | 48.1 |
| cr | -20.8 | -0.2 | kl | 65.3 | 8.1 |
| cu | 38.7 | -21.3 | la | 28.5 | 61.8 |
| da | 32.9 | -4.3 | l3 | 26.2 | -7.8 |
| fi | 61.6 | 57.7 | ld | 1.1 | 54.8 |
| fl | -1.5 | 19.8 | lt | -6.2 | -7.4 |
| fn | -23.0 | -1.9 | lu | -25.3 | -23.2 |
| fm | -20.4 | 2.9 | m8 | -29.2 | -26.7 |
| fu | -23.4 | 61.1 | mo | 63.5 | 65.8 |
| gl | 64.9 | 59.8 | my | 18.4 | 22.7 |
| gb | -30.1 | 22.0 | nh | 47.4 | 48.1 |
| ga | -16.9 | 24.2 | pm | 1.1 | 8.1 |
| ge | -24.6 | -15.5 | qu | 63.6 | 61.8 |
| he | 4.7 | -5.9 | qg | -6.2 | -7.8 |
| h3 | 59.4 | 6.1 | ro | 61.7 | 54.8 |
| h5 | -4.9 | -26.7 | ru | -10.2 | -7.4 |
| h7 | 0.0 | 65.8 | ta | -16.8 | -23.2 |
It shows that this model is adequate, the value of the slope is nearly equal to one and there is only a small additive error in the value of the intercept. The description of flavonoid antioxidant activities is correct and the model provides a suitable prediction in case of most flavonoids. A relatively large difference between the predicted and experimental values can be found in case of some flavonoids (e.g. cr and on a smaller extent lu, h5). This could be explained by the fact, that the flavonoid data set was diverse and 2D descriptors are not able to encode their chemical structures entirely.
Conclusions
While the PLS model obtained this way could be easily constructed (there was no need for geometry optimization of flavonoids; variable selection can be similarly avoided), it provided appropriate description and reasonably accurate prediction for antioxidant activities of a very diverse set of flavonoids. The model can also be used for classification of different flavonoid groups.
Acknowledgements
This work was supported by the Hungarian Research Foundation: OTKA T 037684.
References
- 1.Block G. A role for antioxidants in reducing cancer risk. Nutr. Rev. 1992;50:207–213. doi: 10.1111/j.1753-4887.1992.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 2.Elangovan V., Sekar N., Govindasamy S. Chemoprotective potential of dietary bioflavonoids against 20-methylcholanthrene-induced tumorgenesis. Cancer Lett. 1994;87:278–284. doi: 10.1016/0304-3835(94)90416-2. [DOI] [PubMed] [Google Scholar]
- 3.Middleton E. J., Kandaswami C., Theoharides T. C. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000;52:673–651. [PubMed] [Google Scholar]
- 4.Selway J. W. Antiviral activity of flavones and flavans. Prog. Clin. Biol. Res. 1986;213:521–536. [PubMed] [Google Scholar]
- 5.Middleton E., Jr. Effect of plant flavonoids on immune and inflammatory cell function. Adv. Exp. Med. Biol. 1998;439:175–182. doi: 10.1007/978-1-4615-5335-9_13. [DOI] [PubMed] [Google Scholar]
- 6.Gabor M. Anti-inflammatory and anti-allergic properties of flavonoids. Prog. Clin. Biol. Res. 1986;213:471–480. [PubMed] [Google Scholar]
- 7.Facino R. M., Carini M., Aldini G., Berti F., Rossoni G., Bombardelli E., Morazzoni P. Diet enriched with procyanidins enhances antioxidant activity and reduces myocardial post-ischemic damage in rats. Life Sci. 1999;64:943–949. doi: 10.1016/S0024-3205(98)00605-5. [DOI] [PubMed] [Google Scholar]
- 8.Herteg M. G. L., Feskens E. J. M., Hollman P. C. H., Katan M. B., Kromhout D. Dietary flavonoids and risk of coronary heart disease. Lancet. 1993;342:1007–1011. doi: 10.1016/0140-6736(93)92876-u. [DOI] [PubMed] [Google Scholar]
- 9.Mazur A., Bayle D., Lab C., Rock E., Rayssiguier Y. Inhibitory effect of procyanidin-rich extracts on LDL-oxidation in vitro. Atherosclerosis. 1999;149:421–422. doi: 10.1016/s0021-9150(99)00115-x. [DOI] [PubMed] [Google Scholar]
- 10.Alves C. N., Pinheiro J. C., Camargo A. J., Ferreira M. M. C., Romero R. A. F., da Silva A. B. F. A multiple linear regression and partial least squares study of flavonoid compounds with anti-HIV activity. J. Mol. Struct. 2001;541:81–88. [Google Scholar]
- 11.Alves C. N., Pinheiro J. C., Camargo A. J., de Souza A. J., Carvalho R. B., da Silva A. B. F. A multiple linear regression and partial least squares study of flavonoid compounds with anti-HIV activity. J. Mol. Struct. 1999;491:123–131. doi: 10.1016/S0166-1280(99)00114-1. [DOI] [Google Scholar]
- 12.Moriani M. Z., Galati G., O’Brien P. J. Comparative quantitative structure toxicitz relationships for flavonoids evaluated in isolated rat hepatocytes and HeLa tumor cells. Chem. Bio. Interact. 2002;139:251–264. doi: 10.1016/S0009-2797(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 13.Sergediene E., Jönsson K., Szymusiak H., Tyrakowska B., Rietjens I. M. C. M., Cenas N. Prooxidant toxicity of polyphenolic antioxidants to HL-60 cells: description of quantitative structure-activity relationships. FEBS Letters. 1999;462:392–396. doi: 10.1016/s0014-5793(99)01561-6. [DOI] [PubMed] [Google Scholar]
- 14.Moon T., Chi M. H., Kim D. H., Yoon C. N., Choi Y. K. Quantitative structure-activity relationships (QSAR) study of flavonoid derivatives for inhibition of cytochrome P450 1A2. Quant. Struct. Act. Relat. 2000;19:257–263. [Google Scholar]
- 15.Nikolovska-Coleska Z., Suturkova L., Dorevski K., Krbavcic A., Solmajer T. Quantitative structure-activity relationship of flavonoid inhibitors of p56(lck) protein tyrosine kinase: A classical/quantum chemical approach. Quant. Struct. Act. Relat. 1998;17:7–13. [Google Scholar]
- 16.Oblak M., Randic M., Solmajer T. Quantitative structure-activity relationship of flavonoid analogues. 3. Inhibition of p56(lck) protein tyrosine kinase. J. Chem. Inf. Comput. Sci. 2000;40:994–1001. doi: 10.1021/ci000001a. [DOI] [PubMed] [Google Scholar]
- 17.Stefanic-Petek A., Krbavcic A., Solmajer T. QSAR of flavonoids: 4. Differential inhibition of aldose reductase and p56(lck) protein tyrosine kinase. Croat. Chem. Act. 2000;5:517–529. [Google Scholar]
- 18.Rice-Evans C. A., Miller N. J., Paganga G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free. Rad. Biol. Med. 1996;20:933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 19.Heim K. E., Tagliaferro A. R., Bobilya D. J. Flavonoid antioxidants: chemistry, metabolism and sructure-activity relationships. J. Nutr. Biochem. 2002;13:572–584. doi: 10.1016/s0955-2863(02)00208-5. [DOI] [PubMed] [Google Scholar]
- 20.Harborne J. B., Williams C. A. Advances in flavonoid research since 1992. Phytochem. 2000;55:481–504. doi: 10.1016/s0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- 21.Lien E. J., Ren S. J., Bui H. Y. H., Wang R. B. Quantitative structure-activity relationship analysis of phenolic antioxidants. Free. Rad. Biol. Med. 1999;26:285–297. doi: 10.1016/s0891-5849(98)00190-7. [DOI] [PubMed] [Google Scholar]
- 22.Amić D., Davidovic-Amić D., Bešlo D., Trinajstić N. Structure-radical scavenging activity relationships of flavonoids. Croat. Chem. Act. 2003;76:55–61. [Google Scholar]
- 23.Burda S., Oleszek W. Antioxidant and antiradical activities of flavonoids. J. Agric. Food Chem. 2001;49:2774–2779. doi: 10.1021/jf001413m. [DOI] [PubMed] [Google Scholar]
- 24.HyperCube Inc.; Toronto, Canada: 2002. HyperChem 7.0. [Google Scholar]
- 25.Todeschini R., Consonni V., Pavan M. 2002. Dragon Software version 2.1. [Google Scholar]
- 26.StatSoft Inc.; Tulsa, OK, USA: Statistica 5.5 software package. [Google Scholar]