Abstract
Computationally designed transmembrane α-helical peptides (CHAMP) have been used to compete for helix–helix interactions within the membrane, enabling the ability to probe the activation of the integrins αIIbβ3 and αvβ3. Here, this method is extended towards the design of CHAMP peptides that inhibit the association of the α5β1 transmembrane (TM) domains, targeting the Ala–X3–Gly motif within α5. Our previous design algorithm was performed alongside a new workflow implemented within the widely used Rosetta molecular modeling suite. Peptides from each computational approach activated integrin α5β1 but not αVβ3 in human endothelial cells. Two CHAMP peptides were shown to directly associate with an α5 TM domain peptide in detergent micelles to a similar degree as a β1 TM peptide does. By solution-state nuclear magnetic resonance, one of these CHAMP peptides was shown to bind primarily the integrin β1 TM domain, which itself has a Gly–X3–Gly motif. The second peptide associated modestly with both α5 and β1 constructs, with slight preference for α5. Although the design goal was not fully realized, this work characterizes novel CHAMP peptides activating α5β1 that can serve as useful reagents for probing integrin biology.
Keywords: integrins, molecular modeling, protein design, Rosetta, transmembrane proteins
Introduction
Membrane proteins play crucial roles in many fundamental cellular processes including signal transduction, cell adhesion, and membrane trafficking. As a result, a battery of molecular tools has been developed to modulate the structural and functional states of membrane proteins, primarily through their water-soluble domains. However, oligomerization and induced conformational changes of their transmembrane (TM) α-helices often regulate these essential functions (Engelman et al., 2003). Thus, accessing critical functional states and signaling outputs of membrane proteins requires the development of agents that act by directly targeting TM domains as well (Yin and Flynn, 2016; Stone and Deber, 2017). Previous efforts have relied on natural sequences (Bennasroune et al., 2004; Poulsen and Deber, 2012; Fink et al., 2013) as dominant negative competitors or on large scale mutational scanning (Cammett et al., 2010; Freeman-Cook et al., 2004; Heim et al., 2015) of natural or artificial TM helices to generate agents for this purpose. However, recent work suggests that direct computational design of inhibitors of TM protein–protein interactions represents a promising strategy.
Our group has developed a method to computationally design peptides known as CHAMPs (Computed Helical Anti-Membrane Peptides), and used this approach to prepare peptides that were specifically directed against the TM domains of αIIb and αV, the α subunits of integrins αIIbβ3 and αVβ3 (Yin et al., 2007; Caputo et al., 2008). In the resting state, the TM helices of αIIbβ3 and αVβ3 interact, but they separate when the integrins are activated by pharmacological agonists. Accordingly we designed CHAMP peptides to determine the degree to which TM helix separation contributes to the process of integrin activation. Each CHAMP peptide was found to bind to its target TM domain with reasonable affinity in micelles by a FRET assay, in bacterial cell membranes via the TOXCAT assay, and in mammalian cell membranes by integrin activation assays. Importantly, the CHAMP peptides specifically enabled intact integrins to bind their cognate adhesion proteins when they were expressed in mammalian cells. More recently, the anti-αIIb CHAMP peptide was used to study the role of TM interactions in outside-in platelet signaling, and it was found to activate β3-associated Src and Syk, likely by inducing αIIbβ3 clustering (Fong et al., 2016).
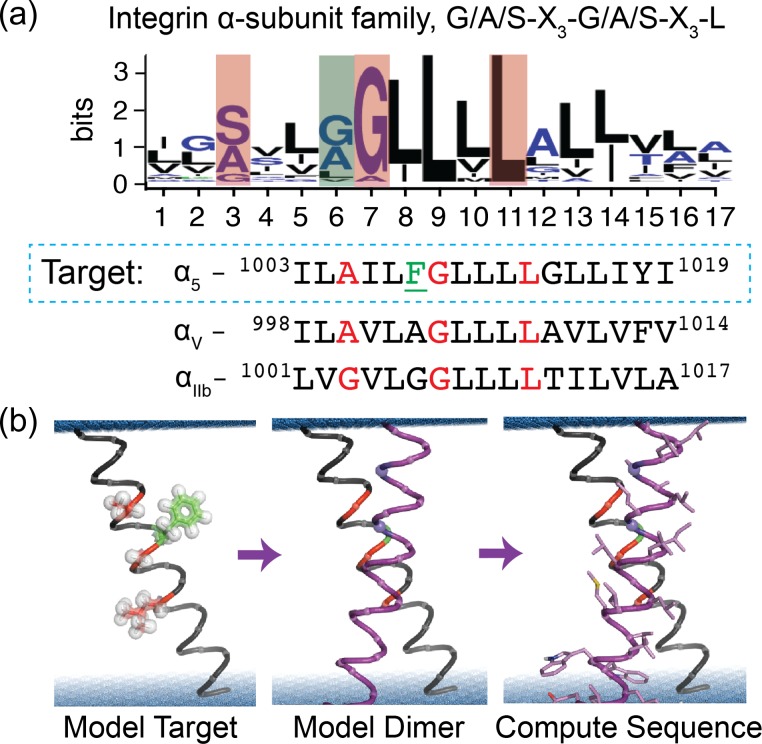
Here we sought to design a CHAMP peptide that selectively binds to the TM domain of α5, thereby causing the activation of the integrin α5β1. This would provide a reagent useful for deciphering the significantly overlapping biochemical signaling responses of integrins α5β1 versus αvβ3 in endothelial cell activation (Yurdagul et al., 2014; Chen et al., 2015; Yun et al., 2016). Because we expected it would be difficult to design CHAMPs with specificity for α5, given the sequence similarity within the integrin α-subunit family (Fig. 1a), we explored modifications on the original method that introduce computational advances that were made since the original development of the CHAMPs procedure.
Fig. 1.
CHAMP peptide design strategy. (a) First the intended interface of the target is defined, here the conserved A–xxx–G–xxx–L motif of the integrin α-subunit TM domain, colored red in the sequences and the weblogo for human isoforms. Integrin α5 has a unique large residue (Phe, green) in a residue position typically having a small residue. (b) Next an atomic model of the monomer is built, a second helix is selected from a database of pairs of associated helices extracted from natural membrane protein X-ray structures and modeled. Finally, the sequence of the CHAMP is sampled and selected by rotamer trials given a potential energy function.
The original CHAMP protocol proceeded in several discrete steps. First, potential interaction motifs were targeted in the sequence of the target TM domain. Here, we targeted the G/A/S–X3–G/A/S sequence motif of α5, a motif common to dimeric TM domain complexes interacting in a parallel right-handed geometry (GASright) (Senes et al., 2007; Teese and Langosch, 2015). Next, examples of 3D structures of these motifs were chosen from the Protein Data Bank (PDB) to guide modeling of a CHAMP peptide (Walters and DeGrado, 2006). The sequence of the target protein, here the α5 TM domain, was threaded in the appropriate register onto one of the two helices in the extracted helical pair, and the sequence of the neighboring CHAMP peptide was computationally designed using a sidechain packing algorithm. The exterior-facing residues of the CHAMP peptide were chosen based on the preference of specific sidechains to lie in distinct regions of a bilayer (Senes et al., 2007). Previously, we used this protocol to successfully design two distinct CHAMPs that were able to differentiate between the distinct, but homologous G/A/S–X3–G/A/S–X3–Leu motifs of the αIIb and αv TM helices.
Despite its simplicity, to our knowledge, similar de novo TM peptide design methods have not been reported to date. To increase the accessibility of this method to the broader scientific community, we revised the protocol to make use of the ROSETTA molecular modeling suite (Alford et al., 2015) and to automate the scoring and ranking of modeled CHAMP peptides. To compare the original and revised methods, we designed anti-α5 peptides using both methods in parallel and assessed the ability of the resulting CHAMPs to cause integrin activation in a mammalian cell system. The ability of the peptides to associate with either α5 or β1 TM domain fragments was then tested by thiol-disulfide exchange equilibrium crosslinking (North et al., 2006; Zhang et al., 2009) and nuclear magnetic resonance (NMR) spectroscopy. This work highlights the promise and limitations of current computational approaches to engineer specific α-helical protein–protein interactions within the membrane in the background of a promiscuous small–X3–small motif. Furthermore, the designed peptides expand the set of reagents available to dissect the mechanisms and outcomes of integrin signaling through α5β1 and potentially other β1 integrins.
Materials and Methods
Computational CHAMP design
A schematic showing the general steps of the CHAMP design workflow is shown in Fig. 1b. Two approaches were taken in this study: protocol 1 mimics Yin et al. (2007); protocol 2 has pre-determined criteria to enable automated computer-based decision-making. In both cases, we first defined the approximate preferred position of the target integrin sequence in a bilayer. An atomic model for the α5 target was built by threading the α5 TM domain sequence onto an idealized alpha-helix and building side chain coordinates from a rotamer library. In protocol 1, side chains were built manually and oriented by the EZ-potential (Senes et al., 2007). In protocol 2, side chains were built through rounds of side chain repacking, rigid body motions, and Cartesian minimization, all relative to an implicit bilayer with a depth-dependent dielectric solvation model using the Rosetta membrane protein framework (RosettaMP) (Alford et al., 2015).
Next, a dimer model was built by extracting coordinates from a geometrically clustered database of pairs of α-helices derived from a non-redundant set of natural TM protein x-ray structures (Zhang et al., 2015). The CHAMP was designed to bind the target integrin TM domain at its conserved G/A/S–X3–G/A/S–X3–Leu motif. These sequence motifs commonly associate in a ‘GASright’ geometry with parallel right-handed crossing angle (−35°) and close inter-helical distance (8.3 Å) (Senes et al., 2000; Moore et al., 2008; Zhang et al., 2015).
In protocol 1, the helix dimer backbone coordinates were selected by choosing a subset of the helical pairs from the PDB with this geometry. Specifically, a subset of the parallel right-handed dimers from cluster #4 was chosen based on the criterion that they should have inter-helical distances less than one standard deviation from the mean inter-helical distance of all the members within the cluster (n = 109, mean = 8.3 Å) in order to facilitate close backbone packing and interaction within the helix dimer. All candidates were then culled to eliminate those with insufficient helix length, steric clashes with integrin α5 side chains, unusual conformations and non-α helical backbone hydrogen bonding patterns.
The protocol 1 sequence design was conducted by first fixing residues of non-interfacial positions to Val to simplify rotamer selection and interface positions to Gly as a starting sequence. Only residues at the dimer’s interface were sampled. Given that the dielectric environment within protein cores and the lipid bilayer are similar, rotamer trials were conducted using Rosetta with the Talaris2013 score function modified to utilize only the Lennard-Jones and rotamer energies, with equal weights (Leaver-Fay et al., 2011). Lowest energy designs were parsed and selected by visualization for inter-helical packing. Finally, the non-interfacial residues were selected semi-randomly: with a 0.6 chance of Leu and 0.1 chance of each Ala, Val, Phe and Ile. Terminal Lys residues were added or mutated to aid solubility, synthesis and purification. In protocol 2, the dimer model was selected by a series of automated criteria. First, the cluster (n = 109 members) was searched for helices with a sequence matching the pattern: [G/A/S]–X2–[L/F/Y/W/M]–[G/A/S]–X3–[L/F/V/I/M], where X is variable. This pattern was used to capture the sterics of the unique Phe1008 of the integrin α5 TM domain, which is usually a small residue within the α-subunit family (Fig. 1a) (Berger et al., 2010). Furthermore, it was required that template backbones were in van der Waals contact (Cα distances of <5.0 Å) at the positions of the small Ala and Gly residues. Also, the signature Phe residue’s Cα was required to be within 7 Å of a non-Ala/Gly Cα atom on the designed helix. Next, the coordinates of helical pairs matching the motif were superimposed onto the modeled TM domain of the target so that the query and target sequence motifs overlap. The helical pairs were extended by superimposing 3-residue fragments in an idealized alpha-helical conformation until both helices spanned the implicit bilayer (length = 34 Å). Helices that were angled such that they required >32 residues to span the membrane were discarded.
The protocol 2 sequence design was conducted by only selecting interfacial residues. However, here we applied RosettaMP and the standard TM full-atom energy function (Barth et al., 2007). Interface residues were selected through the FastRelax application implemented in RosettaScripts, (Fleishman et al., 2011) incorporating rounds of simulated annealing with rigid body re-orientation in the bilayer, rotamer trials and Cartesian minimization. For each dimer scaffold, 800 design trajectories were conducted. In addition to the calculated energy score, each model was evaluated for void volume in the helix interface by the Rosetta PackStat score as described previously (Sheffler and Baker, 2010). Additionally, the individual helices of each model were separated by 40 Å and re-scored after FastRelax trajectories to allow calculation of ∆EAssociation in Rosetta energy units:
| 1 |
in which EDimer is the Rosetta energy of the helices modeled as a dimer, and ECHAMP and Eα5 are the Rosetta energies of those monomeric α-helical peptides separated by 40 Å. Finally, unique sequences for each backbone were independently clustered hierarchically with complete linkage using a normalized BLOSOM sequence similarity score as the distance function with a matrix corresponding to the average pairwise sequence identity of all designs, ca. 80% (e.g. BLOSOM80), using BioPython (Henikoff and Henikoff, 1992; Cock et al., 2009). Of the four most populated clusters, the sequence with the highest PackStat score was selected for synthesis (additionally, sequences with a tryptophan in the apolar membrane region were not prepared as they might not insert properly into bilayers). Non-interfacial residues were randomized between A, L, I, V or F as in protocol 1. Flanking lysine residues were added to aid synthesis, purification, and solubility, and one tryptophan was added for spectroscopic detection.
Peptide synthesis and purification
Peptides were synthesized at 0.1 mmol scale using a Biotage Initiator+ Alstra automated microwave peptide synthesizer on preloaded Rink-amide resin (Chem-Impex). Standard fluorenylmethyloxycarbonyl (Fmoc) deprotection was performed twice at 70°C for 5 min. Coupling reactions were performed twice for 5 min at 75°C using Fmoc-protected amino acids (5 equivalent, 0.5 M, Chem-Impex), O-(1 H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HCTU) (4.95 equivalent, 0.5 M) and N,N-diisopropylethylamine (DIEA) (10 equivalent, 0.5 M) in dimethylformide (DMF). N-terminal acetylation was carried out at room temperature in the presence of 10 equivalent of acetic anhydride and 20 equivalents of DIEA in DMF. Peptide cleavage was performed on the dried resin using trifluoroacetic acid (TFA)-triisopropylsilane (TIPS)-H2O (95:2.5:2.5) for 2 h at 22°C; in the presence of Cys or Met, the cleavage cocktail was changed to TFA-TIPS-H2O-1,2-ethanedithiol (EDT) (92.5:2.5:2.5:2.5). The crude peptide mixture was dried using N2 gas, precipitated with cold ether, dried again under N2 gas, and dissolved in a 1:1 mixture of solvents A and B (Solvent A: 0.1% TFA in H2O; solvent B: 0.1% TFA in 60% isopropanol, 30% CH3CN and 10% H2O). The mixture was purified by RP-high performance liquid chromatography (HPLC) (Higgens 300 Å C4 column, 10 μm, 10 × 50 mm2) with a flow rate of 5 ml/min over a linear gradient of 60–100% of buffer B over 40 min. Peptide mass was analyzed using MALDI (Shimadzu Axima) and electrospray ionization mass spectrometry (Qtrap 3200, ABSCI EX). Purity was determined by analytical HPLC. All peptides used were at least 95% pure. Organic solvents were purchased from Sigma.
Cell culture
Human aortic endothelial cells (HAEC) were purchased at passage 3 (Lonza) and cultured in MCDB131 media supplemented with 10% fetal bovine serum (FBS), 60 μg/ml heparin sodium, 25 μg/ml bovine brain extract (isolated from bovine hypothalamus, Pel-Freeze), 10 U/ml penicillin and 100 μg/ml streptomycin. Cells were used between passages 6 and 10, and experiments performed in low serum (0.5% FBS) media.
Integrin activation assay
A fusion protein composed of the 9th to 11th type III repeat of fibronectin (GST-FNIII9–11) and a glutathione S-transferase (GST) tag employed as a ligand mimetic for α5β1 as previously described (Orr et al., 2006). The activation state-sensitive anti-αvβ3 antibody His-WOW-1 (a gift of Sanford Shattil, University of California, San Diego) was used to measure αvβ3 activation. Briefly, cells were stimulated with CHAMP peptides in media containing either 20 μg/ml GST-FNIII9–11 or 20 μg/ml His-WOW-1 at 37°C for 30 min. The cells were then washed to remove unbound ligand, lysed in SDS sample buffer, and integrin activation assessed by Western blotting for GST (GST-FNIII9–11) or His (His-WOW-1).
Immunoblotting
Cell lysis and immunoblotting was performed as previously described (Orr et al., 2005). Antibodies included mouse anti-GST (Santa Cruz Biotechnology, Cat# sc-138, 1:1000 dilution), mouse anti-His (Thermo Fisher, Cat# R930-25, 1:1000 dilution), mouse anti-β actin (Cat# sc-47778, 1:2500 dilution) and rabbit anti-GAPDH (Cell Signaling Technology, Cat# 5174, 1:5000 dilution).
Disulfide exchange equilibrium in micelles
Sample preparation and thiol disulfide exchange were performed as previously described (Cristian et al., 2003; Zhang et al., 2009). For these experiments, peptides were synthesized with an N-terminal Cys residue followed by a three residue Gly linker (e.g. CGGG-R). Thiol-containing peptides and detergent dodecylphosphocholine (DPC) were co-dissolved in ethanol with a peptide to detergent/lipid ratio of 1:100, dried under N2 gas, and left under vacuum overnight. Samples were re-hydrated with thiol exchange buffer (100 mM Tris–HCl pH 8.6, 100 mM KCl and 1 mM ethylenediaminetetraacetic acid (EDTA)) and incubated for 4 h under reversible redox conditions in a glutathione buffer containing 0.45 mM oxidized (GSSG) and 1.05 mM reduced (GSH) glutathione before quenching with HCl (final 0.12 M). The mixture was separated by analytical reverse phase HPLC (Vydac™ 214TP C4 Column) (Fig. S2). Homo-dimer and target-CHAMP hetero-dimer peaks were identified by comparing the HPLC spectrum with samples containing only one peptide monomer. Overlapping peaks were analyzed with OriginPro 8 (OriginLab, Northampton, MA) multipeak fitting.
NMR spectroscopy
Construction of the α5 and β1 TM and cytoplasmic (TM-CT) domain fragment plasmids, expression, purification and NMR sample preparation were performed as previously described (Lu et al., 2016). The final NMR samples contained 20% (w/v) 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC)/1,2-dimyristoyl-sn-glycero-3-phosphocholine(DMPC) q = 0.3 bicelles, 1 mM EDTA and 10% v/v D2O in 50 mM NaPO4 at pH 6.5, in which the integrin concentration was 150 uM. The appropriate amount of 15 mM CHAMP peptide stock solution in the same NMR buffer was added to the NMR tube to make a series of mixtures of either integrin α5 or β1 with the CHAMP peptide in CHAMP:integrin molar ratios ranging from 0 to 8. An 1H15N-transverse relaxation-optimized spectroscopy (TROSY)-heteronuclear sequential quantum correlation (HSQC) spectrum was measured for each titration point at 45°C on a BRUKER 800 MHz spectrometer equipped with cryogenic triple-resonance probes with z-axis pulsed field gradients. The data were processed using NMRPIPE (Delaglio et al., 1995) and analyzed using NMRVIEWJ (Johnson, 2018). The observed decay in peak intensities for several resonances over the titration series were fit to a simple equimolar binding model as previously described (Lu et al., 2016).
Results
CHAMP design
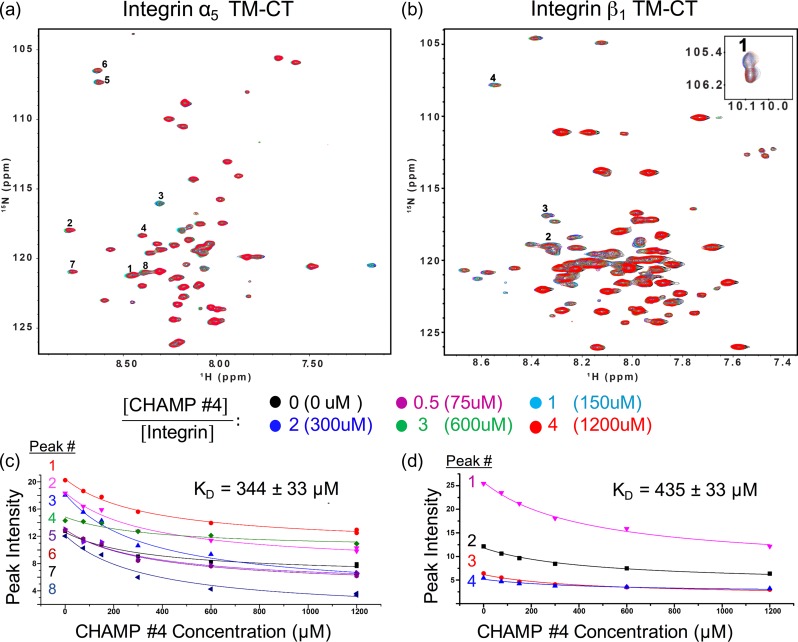
We performed structure-based protein design to generate sequences we expected to bind specifically to the TM domain of the integrin α5. The designs were achieved using two slightly different protocols, as summarized in Fig. 1b: the first protocol was previously described and used to generate sequences that bind specifically to the αIIb and αv TM domains (Yin et al., 2007) and the second relies on Rosetta’s membrane protein framework, RosettaMP (Alford et al., 2015). Both protocols began by selecting pairs of α-helices interacting in the GASRight geometry from our clustered database of helix pairs within natural TM proteins (Zhang et al., 2015), using their backbone coordinates to model the dimeric CHAMP–α5 TM complex. For protocol 1, we selected one pair of helices and the lowest energy sequence was synthesized (CHAMP #1). For protocol 2, we identified two backbone models from the 109 pairs in the GASRight cluster that contained a sequence motif similar to the α5 TM domain and also a distance restraint. After using RosettaMP for 800 sequence design trials, 159 and 26 unique sequences were generated for backbones 1 and 2, respectively. Sequences were clustered hierarchically and evaluated using an interfacial packing score and a relative association energy. The sequences in each cluster with the most favorable packing score were taken from the three most common clusters generated using backbone 1 (CHAMPs 2–4). Only one cluster from backbone 2 had a low Rosetta energy and packing score and we chose this sequence for experimental evaluation (CHAMP #5). The sequences for peptides generated using protocol 2 are shown in Table I.
Table I.
Sequences of CHAMP peptides designed in this study
| Name | Sequence | Design pipeline | Backbone source |
|---|---|---|---|
| CHAMP #1 | KKAWSLVLGGLIGSLIAFAVFLLLWKK |
Original protocol | PDB: 2w2e |
| CHAMP #2 | KKLIMLVFLLFAALLGLILGLLLVFLWKK |
RosettaMembrane | PDB: 1kpl |
| CHAMP #3 | KKLVMVLLLLIAAILGIALGLVAVWLLKK |
RosettaMembrane | PDB: 1kpl |
| CHAMP #4 | KKVLMVLLLLLAALLGILLGLIMVLWVKK |
RosettaMembrane | PDB: 1kpl |
| CHAMP #5 | KKWFAFVLILAVMVFIALLVALLLMLFAILKK |
RosettaMembrane | PDB: 1r3j |
The CHAMP peptides activate α5β1 and not αVβ3
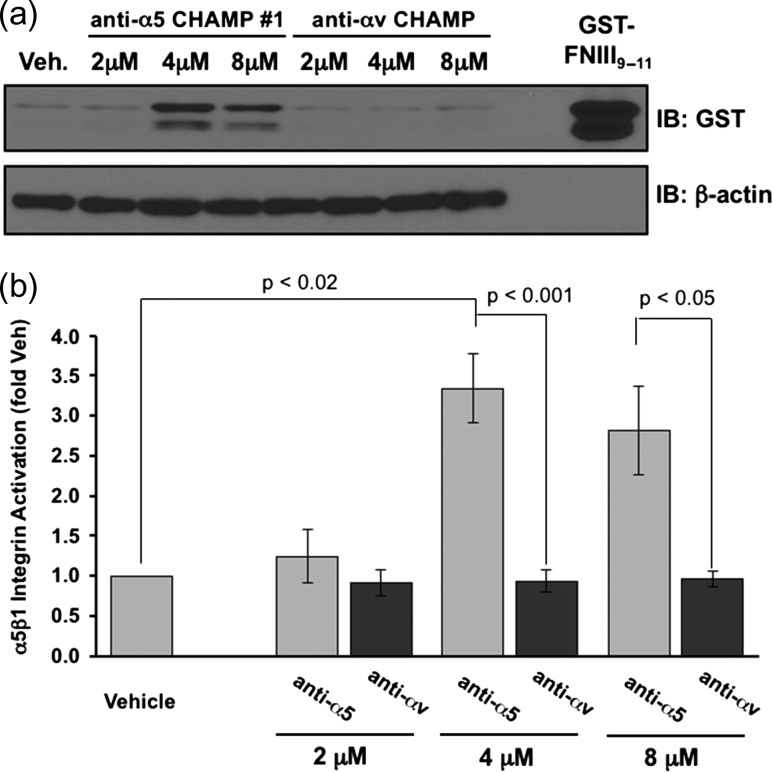
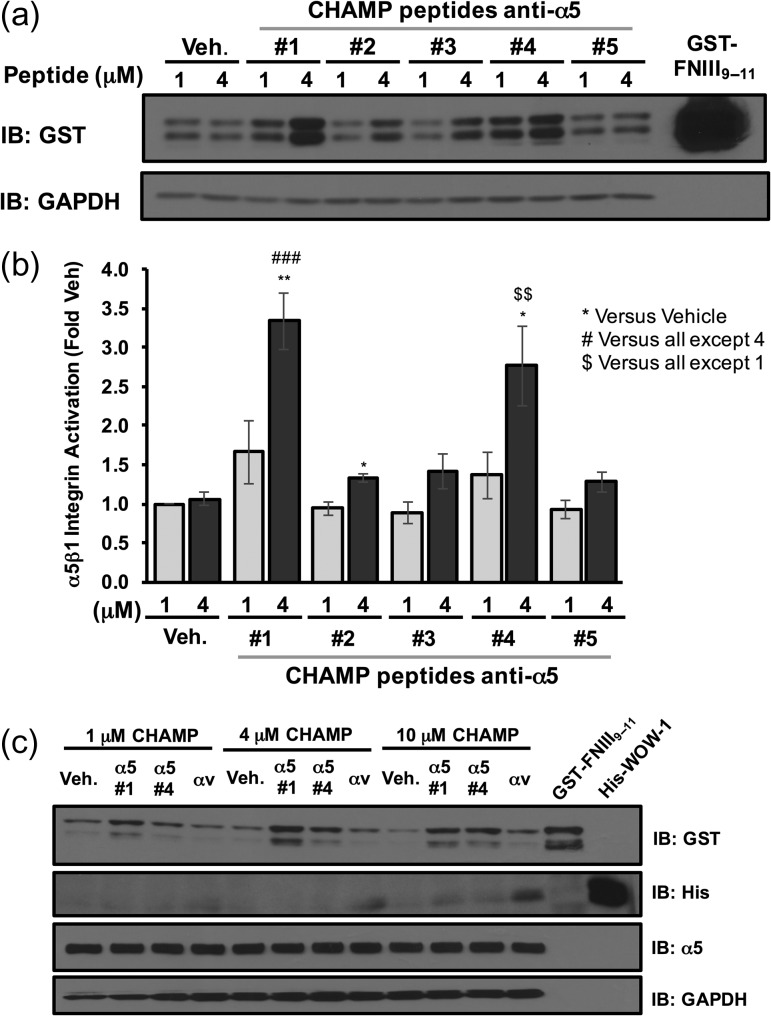
As a primary test, HAEC were incubated with increasing concentrations of either CHAMP #1 or the anti-αv CHAMP peptide. After which, α5β1 activation was assessed using the α5β1 ligand mimetic GST-FNIII9–11, as previously described (Hughes et al., 1997; Orr et al., 2006). Although GST-FNIII9–11 (a recombinant, GST-tagged fusion protein containing then 9th through 11th type III repeat of fibronectin) can interact with several integrins (Magnusson and Mosher, 1998), the soluble recombinant protein interacts specifically with activated α5β1 integrins, similar to endogenous fibronectin (Orr et al., 2006; Huveneers et al., 2008). CHAMP peptide #1 robustly induced GST-FNIII9–11 binding to HAEC at both 4 and 8 μM concentrations as assessed by Western blotting for the GST-tag (Fig. 2a and b). By contrast, anti-αv CHAMP peptide (8 μM) did not activate α5β1. Similarly, CHAMPs #4, generated using protocol 2, activated α5β1, although the magnitude of GST-FNIII9–11 induced by CHAMP #1 was consistently greater (Fig. 2a and b). Finally, CHAMP #2, CHAMP #3 and CHAMP #5 caused only a mild increase in GST-FNIII9–11 binding, with only that by CHAMP #2 reaching statistical significance (Fig. 3a and b).
Fig. 2.
CHAMP #1 induces activation of α5β1, increasing cell adhesion to GST-fibronectin ligand mimetic in endothelial cells. Human aortic endothelial cells were treated for 30 min with the indicated doses of anti-α5 CHAMP peptide #1, anti-αv CHAMP peptide or vehicle (ethanol) control in the presence of the α5β1 ligand mimetic GST-FNIII9–11. Retention of GST-FNIII9–11 was assessed by lysing cells, Western blotting for the GST-tag, and normalizing to β-actin. (a) Representative immunoblots are shown with recombinant GST-FNIII9–11 included as a positive control. (b) Quantification of the GST-FNIII9–11 retention. Results shown are mean ± S.E.M. n = 3–4.
Fig. 3.
CHAMP peptides selectively activate α5β1, but not αvβ3 in endothelial cells. Human aortic endothelial cells were treated for 30 min with the indicated doses of anti-α5 CHAMP peptides or vehicle (ethanol) control in the presence of the α5β1 ligand mimetic GST-FNIII9–11. Retention of GST-FNIII9–11 was assessed by lysing cells, Western blotting for the GST-tag, and normalizing to GAPDH. (a) Representative immunoblots are shown with recombinant GST-FNIII9–11 included as a positive control. (b) Quantification of the GST-FNIII9–11 retention. Results shown are mean ± S.E.M. n = 4. (c) Endothelial cells were treated with the indicated doses of CHAMP #1, CHAMP #4, anti-αv CHAMP or vehicle control for 30 min in the presence of both the α5β1 ligand mimetic GST-FNIII9–11 and His-WOW-1 antibody specific for the αvβ3 activated state. Western blotting for GST-tag (GST-FNIII9–11) or His-tag (His-WOW-1) was used to assess α5β1 and αvβ3 activation respectively. Representative immunoblots are shown with recombinant GST-FNIII9–11 and recombinant His-WOW-1 included as positive controls. n = 3.
To verify the specificity of anti-α5 CHAMP-induced α5β1 activation, HAEC were incubated with increasing concentrations of CHAMP #1, CHAMP #4 and anti-αv CHAMP. αvβ3 activation was measured using the ligand-mimetic antibody WOW-1. WOW-1 is an engineered αvβ3 ligand in which the 19 amino acid H-CDR3 of the αIIbβ3-specific activation-dependent PAC-1 Fab were replaced by the 50 amino acid αv binding domain of the adenovirus type 2 penton base, switching the specificity of the Fab from αIIbβ3 to the αv-containing integrins, αvβ3 and αvβ5 (Pampori et al., 1999). WOW-1 binding to the HAEC was only observed when the cells were incubated with anti-αv CHAMP peptides and then only at high peptide concentrations (Fig. 3c). Taken together, these data indicate that the CHAMP #1 and CHAMP #4 peptides strongly induce α5β1 activation in HAEC and do so without activating αvβ3.
CHAMP peptides #1 and #4 associate with the α5 TM domain in micelles
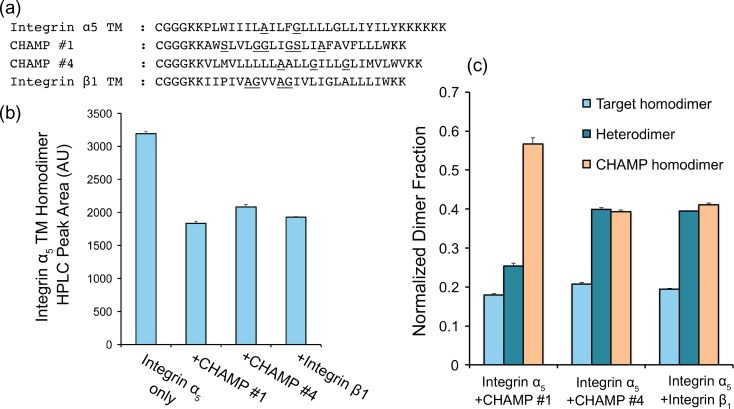
To measure the association of CHAMP peptides #1 and #4 with a peptide fragment of the α5 TM in vitro, we conducted thiol-disulfide exchange equilibrium. First, peptides were synthesized, including β1 and α5 TM fragments, with an N-terminal cysteine followed by a flexible triple glycine linker (Fig. 4a and Supplemental Scheme 1). As expected, the peptides were pre-dominantly helical in DPC micelles at a detergent to peptide ratio of 100:1 (Fig. S1). Mixtures of peptides in DPC micelles were brought to an equilibrium of monomeric and dimeric species in redox buffer in a two-step process of helix association followed by thiol oxidation (Supplemental Scheme 1). Covalent species were then separated and measured by analytical HPLC (Fig. S2) and the relative ratios of the monomers, as well as the homo-dimeric and hetero-dimeric species were quantified as shown previously (North et al., 2006; Zhang et al., 2009) (Supplemental Scheme 1, Fig. 4b). Under these conditions, if the peptides are primarily monomeric then the primary products are the free peptide thiol and a mixed disulfide between glutathione and the Cys-containing peptide. On the other hand, if the peptides dimerize in a geometry conducive to forming a disulfide, the primary product is a disulfide-bonded dimer. Additionally, if two different peptides are present in equimolar amounts, then the mole fraction of the homo- versus hetero-dimer provides an indication of the preference for homo- versus hetero-dimer formation. In the limit where the association constant for the homo- and hetero-dimers are the same, the ratio of the peaks would be 1:2:1. Deviations from this ratio indicate differences in the affinities of the individual homo- and hetero-dimeric species.
Fig. 4.
Disulfide exchange equilibrium assay indicated different dimerization potential of CHAMP peptides. (a) Sequences of Cys labeled peptides used; a cysteine was added to the N terminus of each peptide followed by a triple-glycine flexible linker to disulfide bond formation. (b) Integrin α5 TM homo-dimerization decrease due to a competitive binder. (c) Fraction of different dimers in equilibrium, indicating different preference of hetero-dimer formation over homo-dimer. Results shown are mean ± S.E.M. n = 3.
The β1 TM peptide had a slight propensity to form homo-dimers, in agreement with previous TOXCAT bacterial reporter assay experiments (Berger et al., 2010). Both CHAMP peptides had strong propensities for homo-dimerization as well. In particular, CHAMP #1 demonstrated a marked tendency to self-associate. Although the CHAMPs and β1 self-associated, they also formed a hetero-dimers with the α5 TM peptide. Moreover, the association of CHAMP #4 with the α5 TM hetero-dimers was similar to the association of α5 with β1, while all CHAMPs decreased the equilibrium concentration of the α5 to the same degree as β1. This behavior is consistent with a modest to weak association between the α5 TM fragment and either the CHAMPs or the β1 TM fragment.
Solution NMR titration of CHAMPs and α5 and β1 TM constructs in bicelles
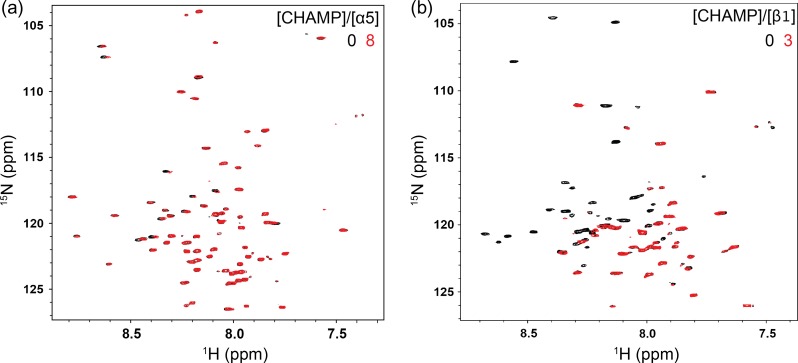
To determine whether the CHAMP #1 and #4 peptides bind to α5 or β1 during α5β1 activation, we titrated either 15N-labeled α5 or β1 TM and cytoplasmic domain (TM-CT) constructs in 20% (w/v) DHPC/DMPC bicelles with unlabeled CHAMP peptides and measured changes in previously characterized 1H-15N TROSY-HSQC spectra of the integrin (Lu et al., 2016). Figure 5a shows an overlay of the 1H-15N HSQC spectra of α5 in isolation and after the addition of 8 molar equivalents of unlabeled CHAMP #1. Even at the highest equivalent of CHAMP #1, only minor changes in either chemical shift or peak intensity can be observed for all resonances in the α5 TM-CT spectra, suggesting negligible interaction. The full titration series, including 0.5, 1, 2, 4, 6 and 8 molar equivalents of CHAMP #1, is shown in Fig. S3. However, drastic changes are observed in the β1 TM-CT 1H-15N HSQC spectra upon addition of 1 molar equivalent of CHAMP #1 (Fig. S3) which become more pronounced at 3 molar equivalent (Fig. 5b), indicative of a tight association. In the β1 spectra, the peak intensities of many resonances previously assigned to TM domain residues are markedly decreased, consistent with association-dissociation events between the β1 TM-CT and CHAMP #1 within the slow-exchange regime on the NMR time scale. This same behavior was observed upon mixing of integrin α5 TM-CT with β1 TM-CT (Lu et al., 2016). Thus, since CHAMP #1 binds more strongly to the integrin β1 TM domain than to integrin α5 TM-CT, it is likely that the CHAMP #1 causes α5β1 activation in cells by binding the β1 TM domain, thereby the disrupting the resting α5β1 TM hetero-dimer.
Fig. 5.
NMR titration experiments of integrin α5 and β1 TM-CT with CHAMP #1. (a) and (b), superimposed 800 MHz 1H-15N-TROSY-HSQC spectra from titrations of unlabeled CHAMP #1 into 15N-integrin α5 TM-CT or β1 TM-CT in bicelles, respectively. The concentrations of the integrin α5 and β1 TM-CT were fixed at 150 uM for all samples. Little change in the α5 TM-CT were observed upon addition of even 8 mol eqv. CHAMP#1, while significant peak intensity decay was observed for many resonances within the β1 TM domain with addition of CHAMP #1, 3 mol eqv. shown here.
The similar behavior of CHAMP #4 and #1 in the thiol-disulfide equilibrium assay suggests CHAMP #4 might also activate α5β1 in a similar manner. As shown in Fig. 6, there were slight decreases in peak intensity in the 1H-15N TROSY-HSQC of both α5 and β1 TM-CT with the addition of CHAMP #4 at 0.5, 1, 2, 3, 4 molar ratios, suggesting a modest association of the peptide to both α5 and β1 TM-CT constructs. For the β1 TM-CT, decay of peak intensities for the TM domain residues were plotted and globally fit to a simple equimolar binding model. This is shown in Fig. 6d for the W751 indole proton (peak 1) and resonances of L749, G744 and V736 amide protons. Similarly, eight resonances in the α5 spectra thought to be from TM domain residues were tracked and fit (Fig. 6c), although these peaks have not yet been assigned. The peak decay fits give equilibrium dissociation constants of 344 ± 33 μM (peptide/bicelle 1.6 mol%) and 435 ± 53 μM (2.03 mol%) for α5 TM-CT and β1 TM-CT respectively. This is an order of magnitude weaker than the dissociation constant of 0.17 ± 0.1 mol% similarly measured for the integrin α5 TM-CT and β1 TM-CT complex previously (Lu et al., 2016).
Fig. 6.
NMR titration experiments of integrin α5 and β1 TM-CT with CHAMP #4. (a) and (b), superimposed 800 MHz 1H-15N-TROSY-HSQC spectra from titration of 15N-integrin α5 transmembrane and cytoplasmic domain (TM-CT) construct or β1 TM-CT, respectively, in bicelles with unlabeled CHAMP #4 peptide. The concentrations of the integrin α5 and β1 TM-CT were fixed at 150 uM for all samples. (c) and (d) The intensity decay of several peaks are tracked for both spectra upon addition of CHAMP #4 at increased molar ratios, numbered consistently in the spectra (a) and (b) and the plots (c) and (d), respectively. The trace indicates the global fit of this data to an equimolar binding model, with listed dissociation constants: CHAMP#4-integrin α5, 344 ± 33 uM (1.6 mol%); CHAMP#4-integrin β1, 435 ± 53 uM (2.03 mol%). Trp 751 indol proton is shown in the inset of (b).
Discussion
Reagents that specifically bind to one integrin TM helix provide a way to interrogate the paradigm of integrin activation that is heavily based on the behavior of the platelet integrin αIIbβ3. However, the behavior of αIIbβ3 may not be typical of most integrins. For example, α1β1 and α2β1 activation may be less regulated and less dependent on a TM domain hetero-dimer to constrain their activity (Lu et al., 2016). Thus, CHAMP peptides that specifically bind to one integrin helix in situ provide a tool for studying the general role of TM domain interactions in regulating integrin function.
Our goal here was to specifically target the integrin α5β1 by designing CHAMP peptides that bind to the α5 TM domain. To this end, our design strategy was successful in that we generated and characterized two novel CHAMP peptides that specifically activate the α5β1 expressed by human aortic endothelial cells and not the co-expressed αvβ3. Nonetheless, subsequent biophysical characterization of the two most active CHAMP peptides indicated that they do not bind specifically to α5, but instead bind either to the β1 TM domain specifically or to both α5 and β1 TM domains. CHAMP #1 was found by solution NMR in bicelles to interact strongly with the β1 TM domain but had negligible interaction with α5 and it preferentially self-associated in the presence of α5 in DPC micelles. Thus, by binding the β1 TM domain, CHAMP #1 likely activates α5β1 by competitively dissociating the TM domain hetero-dimer of inactive α5β1. CHAMP #4 interacted modestly with both the α5 and β1 TM domains in the NMR titration assay with a slight preference for α5 in bicelles. In DPC micelles, it bound equally well to both the α5 and β1 TM domains. Thus, the molecular mechanism by which CHAMP #4 activates α5β1 is less clear. It is possible that at high concentrations, the peptide accumulates in the cell membrane relative to α5β1 and competitively inhibits the α5β1 TM hetero-dimer given its modest affinity. Nonetheless, because neither CHAMP peptide activates αVβ3, even a high concentrations, it is clear that their interaction with the α5 and β1 G/A/S–X3–G/A/S TM helix motifs are specific. However, the link between biological potency and binding affinity is convoluted and difficult to interpret given the many factors at play: peptide delivery and insertion into the membrane, peptide topology upon insertion, and promiscuous binding to other membrane proteins. Regardless, the CHAMPs designed here should be useful reagents to probe the biological role of TM domain interactions in integrin biology. In particular, the use of CHAMP #1 to specifically target β1-containing integrins across distinct cell types holds the most promise.
The cellular activity and the biophysical properties of our peptides emphasize key sequence-structure relationships within the G/A/S–X3-G/A/S motif critical to CHAMP design outcome. Principally, it is thought that the composition and sequence context of a G/A/S–X3–G/A/S motif provides limits on how strongly or weakly it interacts with complementary sequences. Motifs with strong homo- and hetero-dimerization propensities most commonly contain two Gly residues (G–X3–G) surrounded by either beta-branched or additional small residues (Russ and Engelman, 2000; Senes et al., 2000; Schneider and Engelman, 2004; Teese and Langosch, 2015; Anderson et al., 2017). It is also critical for the second small residue to have the potential to donate a side chain or Cα proton hydrogen bond across the helix (small–X3–G or G–X3–S) (Mueller et al., 2014). The α5 TM domain has a Leu-rich A–X3–G motif containing a bulky Phe residue unique within the α-subunit family (L-A-I-L-F-G-L, Figs 1a and 4a). Accordingly, we would expect close packing at this interface to be difficult, making it a difficult sequence to target. Consistent with this expectation, we found that the association between the α5 TM and CHAMP constructs to be weak. For example, CHAMP #5, which contained a single A–X3–A motif that could not donate a hydrogen bond in the GASRight geometry, was the only peptide that did not to induce detectable α5β1 activation (Table I). On the other hand, the β1 TM domain has features consistent with strong dimerization and potential promiscuity: a G–X3–G motif, preceded by additional small residues and flanked on each side by beta-branched amino acids (V-A-G-V-V-A-G-I). Likewise, CHAMP #1 has four small–X3–small motifs, including adjacent strong G–X3–G and G–X3–S motifs (L-G-G-L-I-G-S-L,). Thus, CHAMP #1 had a strong propensity to self-associate in micelles and bound to the β1 G–X3–G motif at near-stoichiometry. The unanticipated binding of CHAMP #1 and #4 to β1 suggest that much more rigorous modeling and energetic analysis will be required to overcome the inherent dimerization potential of strong small–X3–small motifs to achieve specificity for difficult targets like α5.
In addition to designing functional anti-α5 CHAMP peptides, our goal here was to test whether modifications to the de novo design protocol provided advantages over the original algorithm. Given that no specific anti-α5 peptide was identified, it is difficult to attribute changes in the computational algorithm to positive or negative biochemical outcomes. In the future, it will be important to implement changes in the potential function and introduce multi-state negative design. Here, we included the RosettaMP potential function, which was derived and trained on structure prediction a decade ago, for the design of CHAMPs #2–5 (Barth et al., 2007). Subsequent to designing our sequences, there was a report detailing the poor performance of RosettaMP and other potential functions on benchmark tests for TM proteins, basic tasks typically passed by standard Rosetta scoring functions for water-soluble proteins (Kroncke et al., 2016). However, others have supplemented RosettaMP with orientation restraints to achieve unprecedented success in TM homo-dimer structure prediction, utilizing protein family sequence analysis, residue entropy and co-evolution (Wang and Barth, 2015). Fortunately, efforts to revise and reparametrize the potential function used by RosettaMP has been recently reinvigorated. Alongside the imperative published by Kroncke et al. (2016), the Fleishman group recently focused on the depth-dependent insertion energy of amino acids in TM helices by deriving a statistical function from deep mutational scanning experiments with Glycophorin A and ErbB2 (Elazar et al., 2016a), which was subsequently benchmarked for topology prediction (Elazar et al., 2016b). Meanwhile, simpler potential functions, such as PREDIMER (Polyansky et al., 2012) and CATM (Mueller et al., 2014), capture critical structural features and achieve reasonable prediction of structure and energetics of TM homo-dimers utilizing just a polar surface complementation term (PREDIMER) or a two term function comprised of van der Waals and hydrogen bonds, including Cα-H donors (CATM). Despite limitations in modeling and energetic analysis of TM protein structure, exploring and benchmarking alternative potential functions holds great promise, especially for design applications such as CHAMP.
Given the current promiscuity of CHAMP binding, we hope to prevent unexpected interactions by implementing a multi-state modeling scheme to enable negative design. Specifically, we plan to explicitly model and evaluate the energy gap between ensembles of the desired state (e.g. CHAMP with α5) and alternative undesired states (i.e. CHAMP with β1 and CHAMP homo-dimers). Thereafter, mutations can be introduced to destabilize the undesired models without compromising the desired CHAMP hetero-dimer. The utility of this extra modeling stage can be directly evaluated by experiment. However, the accuracy and relevance of energy gaps calculated between models depends on the quality of the potential function, again highlighting the importance of potential functions that perform well for future TM protein modeling and design. Nonetheless, we anticipate that inclusion of these steps will advance the CHAMP algorithm towards routinely achieving specificity among difficult targets from closely related protein families such as integrins.
In summary, executing the CHAMP design protocol has provided structural insights into engineering protein–protein interaction in the membrane and the computational methodology to do so. Since pairwise interactions of α-helices serve as tertiary building blocks of TM proteins (Walters and DeGrado, 2006; Zhang et al., 2015), this type of de novo TM dimer design provides a forum for testing principles of TM protein folding. Likewise, CHAMP design, being the simplest TM protein design application, provides a stepping-stone towards the design of complex and functional multi-pass and higher-order oligomeric TM proteins.
Supplementary Material
Acknowledgements
We are grateful to T. Lemmin, R. Malmirchegini, S.Q. Zhang, R. Jung, H.T. Kratochvil and R. Newberry for helpful discussion, technical assistance and review of the article.
Funding
This work was supported by the National Institutes of Health [R01 GM54616 to W.F.D., R01 HL133497 to A.W.O and DK069921 to C.S.], the American Heart Association [15GRNT25560056 to A.W.O.], the China Scholarship Council Fellowship [to H.H.], University of California, San Francisco (UCSF) Discovery Fellowship [to M.M], and the Howard Hughes Medical Institute Gilliam Fellowship [to M.M.].
References
- Alford R.F., Leman J.K., Weitzner B.D., Duran A.M., Tilley D.C., Elazar A. and Gray J.J. (2015) PLoS Comput. Biol., 11, e1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.M., Mueller B.K., Lange E.J. and Senes A. (2017) J. Am. Chem. Soc., 139, 15774–15783. 10.1021/jacs.7b07505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P., Schonbrun J. and Baker D. (2007) Proc. Natl. Acad. Sci. USA, 104, 15682–15687. 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasroune A., Fickova M., Gardin A., Dirrig-Grosch S., Aunis D., Crémel G. and Hubert P. (2004) Mol. Biol. Cell, 15, 3464–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B.W., Kulp D.W., Span L.M., DeGrado J.L., Billings P.C., Senes A., Bennett J.S. and DeGrado W.F. (2010) Proc. Natl. Acad. Sci. USA, 107, 703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammett T.J., Jun S.J., Cohen E.B., Barrera F.N., Engelman D.M. and DiMaio D. (2010) Proc. Natl. Acad. Sci. USA, 107, 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo G.A., Litvinov R.I., Li W., Bennett J.S., DeGrado W.F. and Yin H. (2008) Biochemistry, 47, 8600–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Green J., Yurdagul A. Jr., Albert P., McInnis M.C. and Orr A.W. (2015) Am. J. Pathol., 185, 2575–2589. 10.1016/j.ajpath.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.J., Antao T., Chang J.T. et al. (2009) Bioinformatics, 25, 1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristian L., Lear J.D. and DeGrado W.F. (2003) Proc. Natl. Acad. Sci. USA, 100, 14772–14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J. and Bax A. (1995) J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Elazar A., Weinstein J., Biran I., Fridman Y., Bibi E. and Fleishman S.J. (2016. a) Elife, 5 10.7554/eLife.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazar A., Weinstein J.J., Prilusky J. and Fleishman S.J. (2016. b) Proc. Natl. Acad. Sci. USA, 113, 10340–10345. 10.1073/pnas.1605888113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D.M., Chen Y., Chin C.-N. et al. (2003) FEBS Lett., 555, 122–125. [DOI] [PubMed] [Google Scholar]
- Fink A., Reuven E.M., Arnusch C.J., Shmuel-Galia L., Antonovsky N. and Shai Y. (2013) J. Immunol., 190, 6410–6422. [DOI] [PubMed] [Google Scholar]
- Fleishman S.J., Leaver-Fay A., Corn J.E. et al. (2011) PLoS One, 6, e20161 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K.P., Zhu H., Span L.M., Moore D.T., Yoon K., Tamura R., Yin H.H., DeGrado W.F. and Bennett J.S. (2016) J. Biol. Chem. 10.1074/jbc.M116.716613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman-Cook L.L., Dixon A.M., Frank J.B., Xia Y., Ely L., Gerstein M., Engelman D.M. and DiMaio D. (2004) J. Mol. Biol., 338, 907–920. 10.1016/j.jmb.2004.03.044. [DOI] [PubMed] [Google Scholar]
- Heim E.N., Marston J.L., Federman R.S., Edwards A.P., Karabadzhak A.G., Petti L.M., Engelman D.M. and DiMaio D. (2015) Proc. Natl. Acad. Sci. USA, 112, E4717–E4725. 10.1073/pnas.1514230112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. and Henikoff J.G. (1992) Proc. Natl. Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes P.E., Renshaw M.W., Pfaff M., Forsyth J., Keivens V.M., Schwartz M.A. and Ginsberg M.H. (1997) Cell, 88, 521–530. [DOI] [PubMed] [Google Scholar]
- Huveneers S., Truong H., Fassler R., Sonnenberg A. and Danen E.H. (2008) J. Cell Sci., 121, 2452–2462. 10.1242/jcs.033001. [DOI] [PubMed] [Google Scholar]
- Johnson B.A. (2018) Methods Mol. Biol., 1688, 257–310. 10.1007/978-1-4939-7386-6_13. [DOI] [PubMed] [Google Scholar]
- Kroncke B.M., Duran A.M., Mendenhall J.L., Meiler J., Blume J.D. and Sanders C.R. (2016) Biochemistry, 55, 5002–5009. 10.1021/acs.biochem.6b00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A., Tyka M., Lewis S.M. et al. (2011) Methods Enzymol., 487, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Mathew S., Chen J. et al. (2016) Elife, 5 10.7554/eLife.18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson M.K. and Mosher D.F. (1998) Arterioscler. Thromb. Vasc. Biol., 18, 1363–1370. [DOI] [PubMed] [Google Scholar]
- Moore D.T., Berger B.W. and DeGrado W.F. (2008) Structure, 16, 991–1001. 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B.K., Subramaniam S. and Senes A. (2014) Proc. Natl. Acad. Sci. USA, 111, E888–E895. 10.1073/pnas.1319944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B., Cristian L., Stowell X.F., Lear J.D., Saven J.G. and DeGrado W.F. (2006) J. Mol. Biol., 359, 930–939. [DOI] [PubMed] [Google Scholar]
- Orr A.W., Ginsberg M.H., Shattil S.J., Deckmyn H. and Schwartz M.A. (2006) Mol. Biol. Cell, 17, 4686–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A.W., Sanders J.M., Bevard M., Coleman E., Sarembock I.J. and Schwartz M.A. (2005) J. Cell Biol., 169, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampori N., Hato T., Stupack D.G., Aidoudi S., Cheresh D.A., Nemerow G.R. and Shattil S.J. (1999) J. Biol. Chem., 274, 21609–21616. [DOI] [PubMed] [Google Scholar]
- Polyansky A.A., Volynsky P.E. and Efremov R.G. (2012) J. Am. Chem. Soc., 134, 14390–14400. 10.1021/ja303483k. [DOI] [PubMed] [Google Scholar]
- Poulsen B.E. and Deber C.M. (2012) Antimicrob. Agents Chemother., 56, 3911–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ W.P. and Engelman D.M. (2000) J. Mol. Biol., 296, 911–919. 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Schneider D. and Engelman D.M. (2004) J. Mol. Biol., 343, 799–804. 10.1016/j.jmb.2004.08.083. [DOI] [PubMed] [Google Scholar]
- Senes A., Chadi D.C., Law P.B., Walters R.F., Nanda V. and Degrado W.F. (2007) J. Mol. Biol., 366, 436–448. 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Senes A., Gerstein M. and Engelman D.M. (2000) J. Mol. Biol., 296, 921–936. 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- Sheffler W. and Baker D. (2010) Protein Sci., 19, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone T.A. and Deber C.M. (2017) Biochim. Biophys. Acta, 1859, 577–585. 10.1016/j.bbamem.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Teese M.G. and Langosch D. (2015) Biochemistry, 54, 5125–5135. 10.1021/acs.biochem.5b00495. [DOI] [PubMed] [Google Scholar]
- Walters R.F. and DeGrado W.F. (2006) Proc. Natl. Acad. Sci. USA, 103, 13658–13663. 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. and Barth P. (2015) Nat. Commun., 6, 7196 10.1038/ncomms8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H. and Flynn A.D. (2016) Annu. Rev. Biomed. Eng., 18, 51–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Slusky J.S., Berger B.W. et al. (2007) Science, 315, 1817–1822. [DOI] [PubMed] [Google Scholar]
- Yun S., Budatha M., Dahlman J.E., Coon B.G., Cameron R.T., Langer R., Anderson D.G., Baillie G. and Schwartz M.A. (2016) Nat. Cell Biol., 18, 1043–1053. 10.1038/ncb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurdagul A. Jr., Green J., Albert P., McInnis M.C., Mazar A.P. and Orr A.W. (2014) Arterioscler. Thromb. Vasc. Biol., 34, 1362–1373. 10.1161/ATVBAHA.114.303863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Q., Kulp D.W., Schramm C.A., Mravic M., Samish I. and DeGrado W.F. (2015) Structure, 23, 527–541. 10.1016/j.str.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kulp D.W., Lear J.D. and DeGrado W.F. (2009) J. Am. Chem. Soc., 131, 11341–11343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.