Abstract
Glioma is the most common primary malignant tumor of the central nervous system. Emerging evidence has demonstrated that long non-coding RNAs (lncRNAs) serve a major role of regulation in various types of human cancer, including glioma. However, the biological roles of thousands of lncRNAs remain unknown and require further identification. The present study investigated the functional role of lncRNA-HOXA10-AS in glioma. The present study examined the expression patterns of HOXA10-AS in glioma and normal brain tissues, as well as glioma cell lines and normal human astrocytes (HA) via reverse transcription-quantitative polymerase chain reaction. HOXA10-AS knockdown cells were generated using lentiviral short hairpin RNA against HOXA10-AS in A172 and U251 glioma cells. Cell growth was assessed by MTT assay, and a flow cytometer was used to investigate cell proliferation, cell cycle distribution and cell apoptosis. Western blot analysis was performed to analyze the expression levels of apoptosis-related proteins. HOXA10-AS was significantly upregulated in glioma tissues and cell lines, and increased HOXA10-AS expression levels were associated with higher grades of glioma. Knockdown of HOXA10-AS inhibited glioma cell proliferation and increased cell apoptosis rates compared with the control cells. HOXA10-AS markedly regulated the expression of the homeobox A10 (HOXA10) gene. Similarly, HOXA10 expression was increased with higher grades of glioma, and silencing of HOXA10 by small interfering RNA suppressed glioma cell proliferation and induced cell apoptosis. The results of the present study demonstrated that HOXA10-AS promoted cell growth and survival through activation of HOXA10 gene expression in glioma, which may potentially act as a novel biomarker and therapeutic target for clinical assay development.
Keywords: HOXA10-AS, glioma, cell proliferation, cell apoptosis, homeobox A10
Introduction
Glioma is the most frequent and aggressive primary malignant tumor of the central nervous system (CNS), and accounts for ~80% of primary CNS tumors. According to the 2016 World Health Organization (WHO) classification, gliomas are classified into four histopathological grades based on the degree of malignancy, including low-grade glioma (grade I and II) and high-grade glioma (grade III and IV), and glioblastoma (GBM) which is the highest grade of glioma (grade IV) (1). Despite significant improvements in the treatment of glioma over recent decades, including surgical resection, local radiotherapy and systemic chemotherapy, the prognosis of glioma patients remains poor, with a 5-year survival rate of 20–30% (2,3). Therefore, investigations into the molecular mechanisms and therapeutic targets of glioma have attracted extensive attention and have potential uses in the treatment of glioma (4,5).
With novel technologies having provided accelerating depths of RNA sequencing, recent studies have demonstrated that the human genome contains thousands of long non-coding RNAs (lncRNAs) (6). lncRNAs are >200 nucleotides in length and regulate gene expression by chromatin modification, at transcriptional and post-transcriptional levels (7,8). Functional studies for a number of lncRNAs have demonstrated that they participate in various aspects of cell biology, including cell proliferation, apoptosis, differentiation, invasion and metastasis, and potentially contribute to tumor initiation and malignant progression (9,10). lncRNAs have also been demonstrated to be involved in hematological malignancies (11) and multiple types of solid tumors, including hepatocellular carcinoma (12), breast (13), lung (14), gastric (15) and gynecological cancer (16), as well as melanoma (17). Downregulation or upregulation of lncRNA expression has been revealed to contribute to the transformed cancer phenotype, further affecting the clinicopathological appearance, prognosis and outcome of the cancer (18–20).
lncRNAs have been implicated in the oncogenesis of gliomas and are increasingly being considered as potential therapeutic targets. According to the function of lncRNAs in glioma, they are classified into oncogenes or tumor suppressors. HOTAIR (21,22), CRNDE (23), H19 (24,25) and XIST (26) are notable oncogenic lncRNAs, which can facilitate tumorigenesis. ADAMTS9-AS2 (27), CASC2 (28), MEG3 (29) and MALAT1 (30) are well-known tumor suppressor lncRNAs. Dysregulation of tumor suppressor lncRNAs leads to tumor formation. Therefore, advances in identifying glioma-related lncRNAs and elucidating their precise molecular mechanisms is critical for the diagnosis and treatment of glioma.
The present study identified a novel lncRNA, HOXA10-AS, which is transcribed from the antisense strand of the homeobox A10 (HOXA10) gene locus at chromosome 7p15.3. HOXA10-AS was markedly upregulated in glioma, and its increased expression levels were correlated with the malignancy of glioma. Furthermore, the present study addressed HOXA10-AS as a regulator of cell proliferation and apoptosis in glioma, and identified HOXA10 as a target of HOXA10-AS. Additionally, the oncogene HOXA10 had an important role in promoting the tumorigenesis of glioma.
Materials and methods
Patients and samples
A total of 59 glioma and 20 normal brain samples were obtained from the Department of Neurosurgery at the First Hospital of Jilin University (Changchu, China) from January to December of 2014. The 59 glioma patients (age range, 35–72 years; mean age, 46.6 years; 41 males and 28 females) consisted of 32 cases of low-grade glioma (WHO grade I and II) and 24 cases of high-grade glioma (WHO grade III and IV). All tissue samples were frozen in liquid nitrogen immediately after resection and stored in liquid nitrogen until use. All clinical pathological and biological data were available for these patients. The present study was approved by the Ethics Committee of The First Hospital of Jilin University and written informed consent was obtained from all patients. All the tumor tissues were obtained at primary resection, and none of the patients had undergone chemotherapy or radiation therapy prior to surgery.
Cell culture
The human glioma A172 and U251 cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and normal human astrocytes (HA) were obtained from ScienCell Research Laboratories (San Diego, CA, USA). A172 and U251 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), and HA cells were maintained in astrocyte medium (ScienCell Research Laboratories) at 37°C in humidified atmosphere with 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from tissues or cultured cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA quantity was determined using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA synthesis was performed with 1 µg total RNA, using the PrimeScript™ RT reagent kit (Takara Biotechnology, Dalian, China), according to the manufacturer's protocol. Real-time PCR was performed on the Takara system, using the SYBR® Premix Ex Taq™ II Kit (Takara Biotechnology). The cycling conditions were the following: 30 sec at 95°C, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. GAPDH was used as the endogenous control. The relative expression was calculated using the 2−ΔΔCq method (31). The primers of HOXA10-AS, HOXA10 and GAPDH are listed in Table I.
Table I.
Primers for real-time qPCR.
| Primer | Sequences |
|---|---|
| HOXA10-AS | F: CCCAGTAAGCCAAAGTCAAGCC |
| R: CTGAGGTCAATGGTGCAAAGG | |
| HOXA10 | F: CAACTGGCTCACGGCAAAGA |
| R: TTCAGTTTCATCCTGCGGTTC | |
| GAPDH | F: GCACCGTCAAGGCTGAGAAC |
| R: TGGTGAAGACGCCAGTGGA |
F, forward; R, reverse.
Lentiviral packaging and stable cell line establishment
A piLenti-shRNA-GFP lentiviral vector (Applied Biological Materials, Inc., Richmond, BC, Canada) was used for the HOXA10-AS-knockdown experiment. In brief, lentiviral particles expressing HOXA10-AS-specific or control scrambled shRNAs were co-transfected into 293T cells with the mixed set of packaging plasmids (SPAX2 and MD2G) using Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The medium was changed after 8 h and the supernatant containing lentiviruses was collected 48 h later. Produced lentiviruses were concentrated using the Centricon Plus-20 centrifugal filter device (EMD Millipore, Billerica, MA, USA). Lentiviral stock was titred and stored at −80°C. A172 and U251 cells were infected with the HOXA10-AS shRNA construct, and GFP+ cells were sorted using a flow cytometer (FACSAria II; Becton-Dickinson, Mountain View, CA, USA). Subsequently, the cells were cultured with regular complete medium. Finally, the cells were analyzed for mRNA expression by RT-qPCR. The following two targets were used for HOXA10-AS knockdown: Target 1, ACAGGAAACTACCTAAATCACCGACCAGT; and target 2, GTTCTGGTGCTGCCCGCGAAGGGCTGCCT.
MTT cell viability analysis
MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was performed to determine cell viability. A total of 1×103 cells were seeded into each well of 96-well plates and cultured in 200 µl growth medium. The cells were incubated with fresh complete medium every 2 days and after 6 days, 20 µg MTT was added to each well, followed by incubation at 37°C for 4 h. The reaction was stopped by adding 150 µl dimethyl sulfoxide (DMSO) to each well and incubating at 37°C for 10 min. The absorbance was then determined at 450 nm using a microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA).
BrdU cell proliferation assay
Cell proliferation was evaluated using BrdU Flow kit (BD Biosciences, San Diego, CA, USA). Cells were plated into 6-well plates with 2 ml complete growth medium/well, and prior to adding BrdU, the complete medium was replaced with serum-free medium. After 12 h, the serum-free medium was replaced with complete medium, and BrdU (1 mM/ml) was added to each well and incubated for 4 h. Subsequently, the cells were harvested, fixed and incubated with anti-BrdU and 7-AAD according to the manufacturer's protocol. Cell cycle distribution was determined using a flow cytometer (FACSAria II; BD Biosciences).
Cell apoptosis analysis
The Annexin V-PE assay kit (BD Biosciences) was used to analyze cell apoptosis in glioma cells. Cells were harvested at their exponential growth phase and were resuspended in 100 µl binding buffer at a density of 1×106 cells/ml. PE-conjugated Annexin V and 7-AAD reagent staining was performed at the concentrations and times recommended by the manufacturer. Stained cells were analyzed using a flow cytometer (FACSAria II; BD Biosciences), and data were analyzed using FlowJo V10 software (FlowJo LLC, Ashland, OR, USA).
Western blot analysis
Total protein was isolated from cells using radioimmunoprecipitation assay lysis buffer (Pulilai Gene Technology, Co., Ltd., Beijing, China). Protein concentrations were determined with a BCA protein assay kit (Pulilai Gene Technology, Co., Ltd.) and the mass of protein loaded per lane was 40 µg. Total proteins were separated by 12% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% (W/V) non-fat milk (Pulilai Gene Technology, Co., Ltd.) at room temperature for 1 h, and incubated with the following primary antibodies: Anti-caspase-3 (1:1,000; cat. no. 9662; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-cleaved caspase-3 (1:1,000; cat. no. 9664; Cell Signaling Technology, Inc.), anti-Bcl-2 (1:1,000; cat. no. 4223; Cell Signaling Technology, Inc.) and anti-β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.) at 4°C overnight and then incubated with secondary antibody (anti-rabbit IgG, HRP-linked antibody; 1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room temperature for 1 h. Bands were visualized using an enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.). β-actin was used as a loading control.
Small interfering RNA transfection
HOXA10-knockdown in glioma cells was performed by siRNA transfection. siRNA oligonucleotides targeting HOXA10 were designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China), and the non-specific siRNA oligonucleotides (Shanghai GenePharma Co., Ltd.) were used as a negative control. Cells were transfected with 150 pmol siRNA/well in 6-well plates, using Lipofectamine RNAiMax Reagent (Invitrogen.), according to the manufacturer's protocol. All siRNA oligonucleotide sequences are listed in Table II.
Table II.
siRNA oligonucleotides.
| siRNA | Sequences |
|---|---|
| siHOXA10-1 | S: GUCAGCCAGAAAGGGCUAUTT |
| A: AUAGCCCUUUCUGGCUGACTT | |
| siHOXA10-2 | S: CCAUAGACCUGUGGCUAGATT |
| A: UCUAGCCACAGGUCUAUGGTT | |
| siHOXA10-3 | S: CGCAGAACAUCAAAGAAGATT |
| A: UCUUCUUUGAUGUUCUGCGTT | |
| siControl | S: UUCUCCGAACGUGUCACGUTT |
| A: ACGUGACACGUUCGGAGAATT |
S, sense; A, antisense.
Statistical analysis
All data are presented as the mean ± standard error of the mean (SEM) and were analyzed using Student's t-test. Spearman's correlation analysis was performed to investigate the correlation between HOXA10 and HOXA10-AS. All experiments were performed at least three times. All statistical analyses were performed using SPSS 19.0 (IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
HOXA10-AS is overexpressed in human glioma samples and cell lines
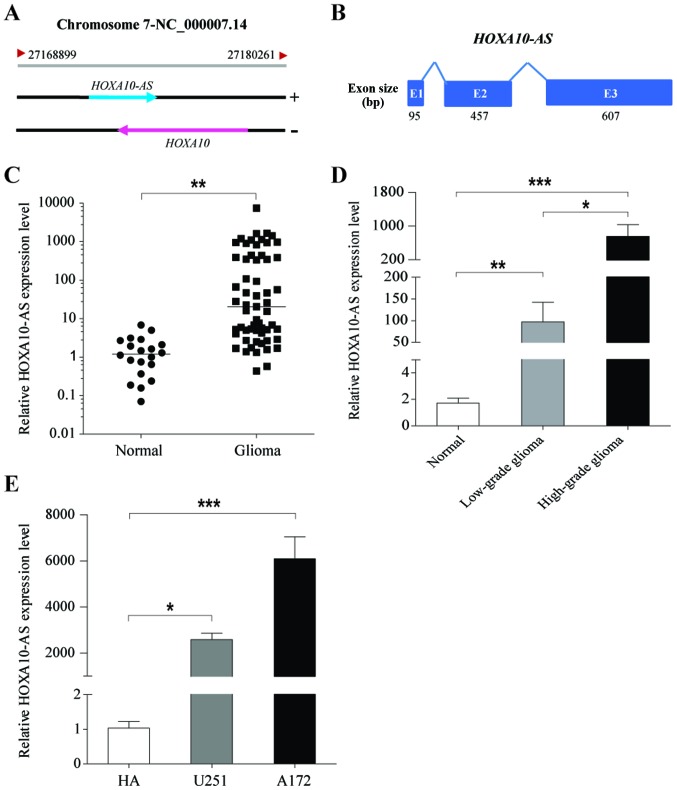
The human homeobox A (HOXA10) gene is located in the HOXA gene cluster at chromosome 7p15.3. A GeneBank search identified the gene HOXA10-AS in a tail-to-tail orientation relative to the HOXA gene, on the opposite strand of the HOXA10 gene (Fig. 1A). HOXA10-AS is a 1,159-bp long non-coding antisense transcript of HOXA10, consisting of three exons with a 3′ polyadenylation tail (Fig. 1B).
Figure 1.
Expression of HOXA10-AS in glioma tissues and cells. (A) Schematic representation of the HOXA10 and HOXA10-AS gene loci on human chromosome 7p15.3. (B) Exon composition of HOXA10-AS RNA. The relative HOXA10-AS expression was determined by quantitative polymerase chain reaction. GAPDH served as an internal control. (C) HOXA10-AS expression was higher in glioma tissues than in normal brain tissues. (D) HOXA10-AS expression in high-grade glioma (WHO III and IV) was higher, than in low-grade glioma (WHO I and II). (E) The relative HOXA0-AS expression was significantly upregulated in A172 and U251 cells, compared with the normal HA cells. Error bars represent the standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 by Student's t-test. WHO, World Health Organisation; HA, human astrocytes.
To investigate the expression levels of HOXA10-AS in glioma, tumor tissues were collected from 59 patients with glioma and normal brain tissues were obtained from 20 healthy patients, and the expression levels of HOXA10-AS were examined using qPCR. The results demonstrated that HOXA10-AS was significantly upregulated in glioma tissues compared with expression in normal brain tissues (Fig. 1C). Furthermore, it was demonstrated HOXA10-AS expression was associated with the malignancy grade of brain tumors, and that HOXA10-AS expression in cases of high-grade glioma (WHO III and IV, n=27) was significantly higher than in cases of low-grade glioma (WHO I and II, n=32; Fig. 1D). Additionally, HOXA10-AS expression was analyzed in two human glioma cell lines, A172 and U251 cells, and normal human astrocytes (HA). Data demonstrated that HOXA10-AS expression was significantly increased in glioma cell lines compared with that in HA (Fig. 1E). Therefore, these results indicated that HOXA10-AS was frequently upregulated in glioma, and may serve an important role in the progression of malignant glioma.
HOXA10-AS promotes glioma cell proliferation
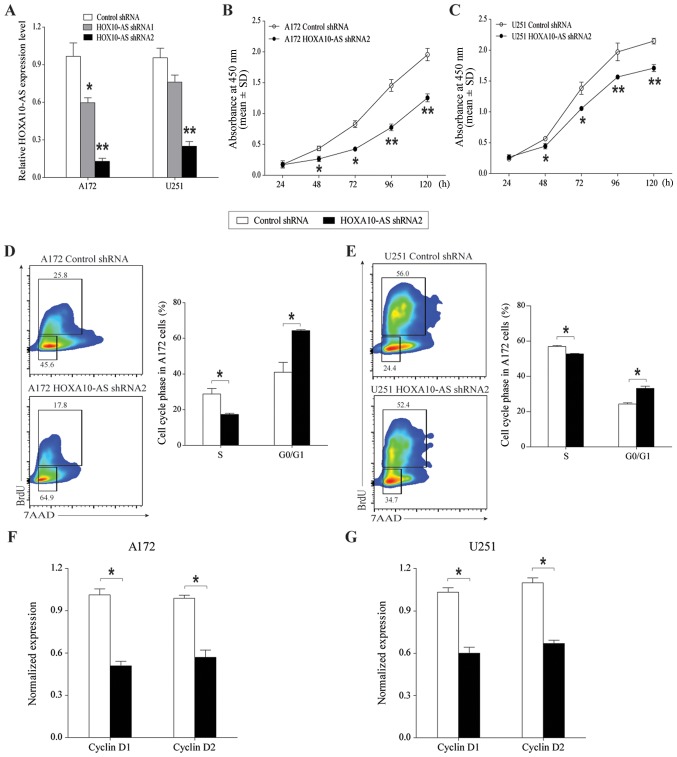
To investigate the involvement of HOXA10-AS in glioma cell growth, the changes in cell viability and proliferation following silencing of HOXA10-AS were analyzed. The effects of HOXA10-AS-knockdown were determined through lentivirus-mediated expression of shRNA targeting HOXA10-AS. These target sequences avoid overlap with the HOXA10 sequences, and do not cause silencing of the HOXA10 gene. The efficiency of HOXA10-AS downregulation was determined by RT-qPCR analysis. The results indicated that HOXA10-AS shRNA1 and shRNA2 induced significant reductions in the expression levels of HOXA10-AS, with a greater inhibitory effect with shRNA2 compared with shRNA1 in the two cell lines (Fig. 2A).
Figure 2.
Effects of HOXA10-AS-knockdown on glioma cell growth. (A) HOXA10-AS was stably silenced in A172 and U251 cells by transfection of shRNA. HOXA10-AS RNA expression was determined by quantitative polymerase chain reaction. GAPDH served as an internal control. (B and C) Following knockdown of HOXA10-AS, A172 and U251 cell growth curves were determined by MTT assay. The total number of control cells increased ~10-fold over 5 days, whereas the total number of cells with silenced HOXA10-AS increased ~5-fold over 5 days. (D and E) Representative flow cytometric cell-cycle profiles demonstrating a G0/G1 arrest and a decreased number of S-phase HOXA10-AS-knockdown cells, compared with control cells. BrdU was added to cells for 4 h, and then flow cytometry was used to analyze cell proliferation and cell cycle distribution. (F and G) Downregulation of cyclin D1 and cyclin D2 expression in HOXA10-AS-knockdown cells was determined by quantitative polymerase chain reaction. GAPDH served as an internal control. Error bars represent the standard error of the mean of three independent experiments. *P<0.05 and **P<0.01 by Student's t-test.
The growth inhibitory effects of HOXA10-AS-knockdown were confirmed by assessing the proliferation rate of cells for 5 consecutive days using MTT assays. In the scrambled control shRNA-infected cells, the number of cells increased by >10-fold over a 5-day culture period, whereas the number of HOXA10-AS-knockdown cells increased more slowly than that of the control cells. HOXA10-AS-knockdown cells cultured for 5 consecutive days exhibited statistically significantly lower proliferation rates than those of scrambled vector-containing cells (Fig. 2B and C).
To illustrate the underlining mechanisms by which HOXA10-AS regulated cell growth, the cell cycle of A172 and U251 cell lines was then investigated using a BrdU array. HOXA10-AS-knockdown markedly decreased the proportion of S phase cells and increased the proportion of G0/G1 phase cells (Fig. 2D and E). HOXA10-AS-knockdown led to arrest in the growth of cells (A172 cells: 56.7%; U251 cells: 36.6%), compared with the control cells. Additionally, RT-qPCR demonstrated that the RNA expression levels of cyclin D1 and cyclin D2, cell-cycle regulators, were decreased in HOXA10-AS-knockdown glioma cells (Fig. 2F and G). These data indicated that knockdown of HOXA10-AS inhibited glioma cell proliferation via G0/G1 phase arrest and a decrease in DNA synthesis.
Therefore, HOXA10-AS functions by promoting tumor cell growth in glioma and may be a tumor oncogene.
Knockdown of HOXA10-AS induces glioma cell apoptosis
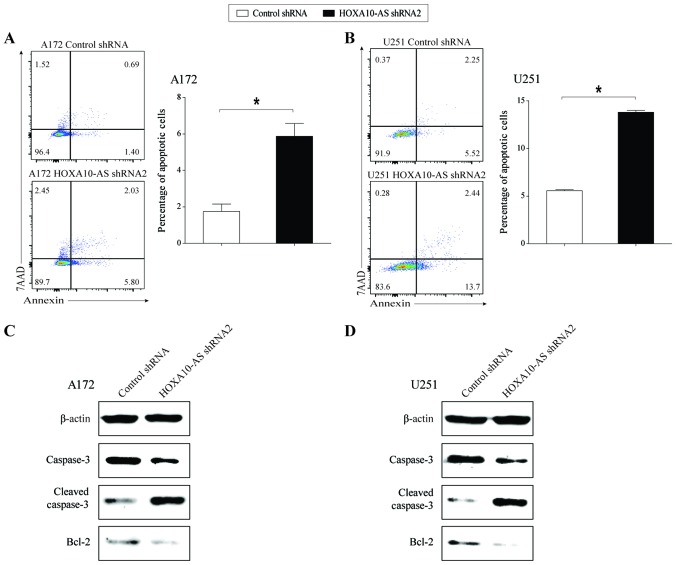
In order to examine the effect of HOXA10-AS-knockdown on tumor cell apoptosis, cell apoptosis analysis was performed using flow cytometry in HOXA10-AS-knockdown and shRNA-control glioma cell lines. Knockdown of HOXA10-AS resulted in a marked increase in the proportion of Annexin V-positive and 7AAD-negative cells in knocked down A172 and U251 cells, compared with that of the scrambled shRNA-expressing cells (Fig. 3A and B). This result demonstrated that the knockdown of HOXA10-AS induced glioma cell apoptosis.
Figure 3.
Effects of HOXA10-AS-knockdown on glioma cell apoptosis. HOXA10-AS was stably silenced in A172 and U251 cells by transfection of shRNA. (A and B) Flow cytometric analysis demonstrated that the cell apoptosis rate was increased in HOXA10-AS-knockdown glioma cells. Error bars represent the standard error of the mean of three independent experiments. *P<0.05 by Student's t-test. (C and D) Western blot analysis demonstrating the downregulation of total caspase-3 and Bcl-2 protein expression, and the upregulation of cleaved caspase-3 protein in HOXA10-AS-knockdown glioma cells. β-actin served as a loading control.
In order to confirm the effect of HOXA10-AS on cell apoptosis, the protein expression levels of apoptotic regulators, including caspase, cleaved-caspase and Bcl-2 were investigated in HOXA10-AS-knockdown cells. Western blot analysis demonstrated that total caspase-3 and Bcl-2 protein expression levels were decreased and the protein expression level of cleaved-caspase-3 was increased following knockdown of HOXA10-AS in glioma cells (Fig. 3C and D). The expression trends of three apoptosis-related proteins were consistent with the change in the cell apoptosis rate following knockdown of HOXA10-AS.
These findings indicated that HOXA10-AS may serve a role in promoting glioma cell survival.
HOXA10-AS activates HOXA10 gene expression
In order to investigate the molecular mechanisms underlying the tumor promoting effects of HOXA10-AS on glioma cells, the ability of HOXA10-AS to regulate the transcription activity of the HOXA10 gene was investigated in A172 and U251 cells.
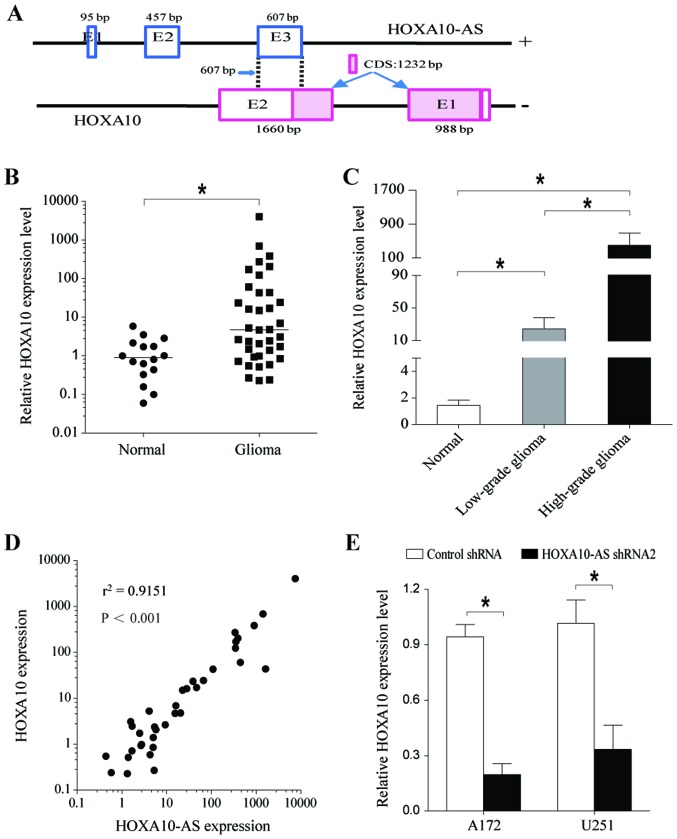
Sequence analysis demonstrated that exon 3 of HOXA10-AS overlaps with the second exon of HOXA10 by 607 bp in an antisense manner. The 607 bp of overlapping sequences is mostly located in 3′-UTR of HOXA10 mRNA, excluding 15 bp (Fig. 4A). Antisense lncRNAs have been implicated in the regulation of their overlapping sense transcripts (32,33). As a first step in determining whether HOXA10-AS regulates the expression of the HOXA10 gene, the present study analyzed the expression of HOXA10 in human glioma tissues and compared it with that in normal brain tissues. The results indicated that HOXA10 expression levels were significantly increased in glioma tissues (Fig. 4B). Additionally, HOXA10 mRNA expression levels in high-grade glioma tissues were higher than those in low-grade glioma tissues, and HOXA10 expression was associated with the degree of malignancy of the brain tumor (Fig. 4C). Further statistical analysis demonstrated a significant and positive correlation between HOXA10 and HOXA10-AS gene expression (Fig. 4D).
Figure 4.
HOXA10-AS promotes HOXA10 gene expression. (A) Exon composition of the HOXA10-AS transcript and organization of the overlapping exon with HOXA10. qPCR analysis of relative HOXA10 mRNA levels in normal brain tissues, low-grade glioma tissues and high-grade glioma tissues. (B) HOXA10 was upregulated in glioma tissues. (C) HOXA10 expression in high-grade glioma was greater than in low-grade glioma. (D) Spearman's correlation analysis demonstrated that the expression of HOXA10-AS and HOXA10 were positively correlated in glioma tissues. (E) HOXA10 expression was significantly decreased following knockdown of HOXA10-AS, compared with the control group in A172 and U251 cells by qPCR. GAPDH served as an internal control. Error bars represent the standard error of the mean of three independent experiments. *P<0.05 by Student's t-test. qPCR, quantitative polymerase chain reaction.
RT-qPCR analysis confirmed that HOXA10 expression was significantly decreased at the mRNA level in A172 and U251 cells transfected with shHOXA10-AS (Fig. 4E). These results indicated that HOXA10-AS regulated HOXA10 gene expression.
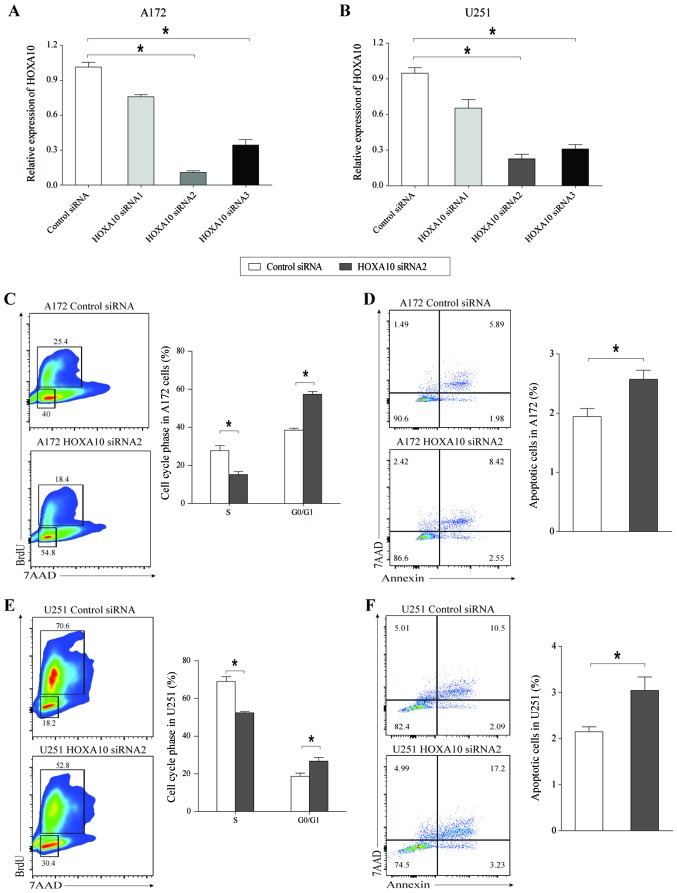
Knockdown of HOXA10 suppresses glioma cell proliferation and induces cell apoptosis
To further examine the effect of HOXA10 on glioma cell growth, the present study analyzed the changes in cell proliferation and apoptosis following silencing of HOXA10 using transcript-specific siRNAs in A172 and U251 cells. These siRNA sequences avoid overlap with the sequences of HOXA10-AS, and do not cause gene silencing of HOXA10-AS.
The present study first assessed HOXA10 mRNA expression levels in A172 and U251 cells treated with siHOXA10 for 48 h. The results indicated that siHOXA10-2 and siHOXA10-3 induced a significant and visible decrease in HOXA10 expression levels, with a greater inhibitory effect achieved with siHOXA10-2 compared with siHOXA10-3 in the two cell lines (Fig. 5A and B). Following knockdown of HOXA10 in A172 cells, the percentage of cells in the S phase was decreased and the percentage of cells in the G0/G1 phase was increased (Fig. 5C), and caused a significant increase in the rate of cell apoptosis (Fig. 5D). Similar results were also obtained in glioma cell line U251 (Fig. 5E and F). These results indicated that knockdown of HOXA10 inhibited glioma cell growth by inducing G0/G1 phase arrest and cell apoptosis. Therefore, we hypothesized that HOXA10-AS was able to promote tumor cell growth by targeting HOXA10 in human glioma cells.
Figure 5.
HOXA10-silencing inhibits the proliferative capacity of glioma cells and induces glioma cell apoptosis. (A and B) Quantitative polymerase chain reaction analysis of HOXA10 RNA levels following siHOXA10 treatment for 48 h in A172 and U251 cells. GAPDH served as an internal control. Following silencing of HOXA10, (C and E) cell proliferation and cell cycle distribution, and (D and F) cell apoptosis were analyzed by flow cytometry. Error bars represent the standard error of the mean of three independent experiments. *P<0.05 by Student's t-test.
Discussion
It is currently well-known that >75% of the human genome is functional and encodes a large number of ncRNAs (34). Based on the ENCODE project, it is estimated that the human genome encodes >28,000 distinct lncRNAs, a number of which continue to be discovered and are yet to be given a functional annotation (35). Therefore, revealing the biological function of lncRNAs and identifying the disease-related lncRNAs are one of the most investigated areas in the life sciences and medical research fields.
To the best of our knowledge, the present study was the first to report the role of HOXA10-AS in glioma. The original results of qPCR analysis revealed significantly increased HOXA10-AS expression in glioma tissues and cell lines, and demonstrated that it was strongly associated with the histological tumor grades of glioma tissues. We hypothesized that HOXA10-AS functions as a tumor oncogene in glioma. In order to verify this hypothesis, the tumor oncogenic roles of HOXA10-AS were investigated in glioma cell lines. The present study investigated the function of HOXA10-AS in glioma by HOXA10-AS-knockdown. Compared with the control group, cell proliferation was significantly suppressed and the cell apoptosis rate was increased in shHOXA10-AS-transfected glioma cells. These data indicated that HOX10-AS served a crucial role in glioma tumorigenesis.
Certain lncRNAs are cis-acting, affecting the expression of neighboring genes. HOXA10-AS is a 1,161-bp spliced and polyadenylated RNA, which transcripts the antisense strand of the HOXA10 gene with a tail-to-tail overlap with HOXA10 transcript. Based on the results obtained in the clinical sample analysis, the expression of HOXA10 was upregulated in glioma and the expression of HOXA10-AS was correlated with that of HOXA10.
HOXA10, a member of the HOXA gene cluster, is a pivotal transcriptional regulator of early embryonic development. Previous studies have demonstrated that high expression of HOXA10 in several types of cancer is involved in regulating cell proliferation, migration and invasion, and is correlated with a poor prognosis (36–40). For example, HOXA10 significantly promoted nasopharyngeal carcinoma cell proliferation and invasion (36). Overexpression of HOXA10 predicted a poor prognosis in a subgroup of patients with epithelial ovarian cancer (37) or gastric cancer (38). HOXA10 serves a role in the migration processes of medulloblastoma cells (39).
Furthermore, Kurscheid et al (41) identified high expression of HOXA10 in glioblastoma tissues, compared with non-cancerous brain tissues based on Affymetrix array data from TCGA, and demonstrated that HOXA10 levels were significantly higher in glioblastoma stem cells. The present study revealed that HOXA10 exerted effects on promoting the proliferation and inducing the apoptosis of glioma cells. It was further verified that the expression of HOXA10 was decreased in HOXA10-AS-knockdown glioma cells compared with the control group, indicating that HOXA10 may be activated by high HOXA10-AS expression levels.
However, further studies are required to elucidate the detailed mechanism of how HOXA10-AS regulates HOXA10. The results of the present study demonstrated that HOXA10-AS is overlapped with the 3′-UTR of HOXA10 mRNA. Therefore, the potential HOXA10-AS/HOXA10 RNA hybrid may act to stabilize the HOXA10 transcript. It has been confirmed that Wrap53, as a natural p53 antisense transcript, is required for p53 stabilization via Wrap53/p53 RNA interaction (42). In addition, previous studies have reported that DNA methylation at key regulatory CpGs in the promoters of HOXA10 was significantly associated with HOXA10-signature expression (41). Antisense lncRNAs have been proposed to cause DNA methylation (43,44). Therefore, we hypothesized that the transcriptional activity of the HOXA10 gene may be regulated in part by HOXA10-AS-directed DNA hypomethylation. Several miRNAs were also known to regulate HOXA10 expression in colorectal cancer (45), oral cancer (46), hepatocellular carcinoma (47) and breast cancer (48), suggesting that HOXA10-AS may function as a competing endogenous RNA (ceRNA) to regulate HOXA10 gene expression.
In conclusion, the results of the present study provided the first evidence of HOXA10-AS values in glioma tissues, which is a tumor oncogene. Increased expression of HOXA10-AS was associated with malignancy status in glioma and knockdown of HOXA10-AS may be a potential therapeutic strategy for glioma. In vitro experiments demonstrated that silencing of HOXA10-AS suppressed glioma cell proliferation and induced cell apoptosis. Furthermore, the present study identified HOXA10 as a target of HOXA10-AS and demonstrated that knockdown of HOXA10-AS downregulated the mRNA expression levels of HOXA10. Additionally, HOXA10 exhibited definitive oncogenic effects in glioma. Further evaluation of the interactions between HOXA10-AS and HOXA10 is required. The results of the present study indicated an important role of HOXA10-AS in the promotion of the proliferation and survival of glioma cells and that it may serve as a novel biomarker and therapeutic target for glioma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of Beijing City (7152098) and the Health Special Science Foundation of Jilin Province (grant no. 2018SCZWSZX-014).
Availability of data and materials
The datasets supporting the findings of this study are included within the article.
Authors' contributions
CYD, QL and JC performed the experiments. CYD, JC and DHL collected and analyzed the data. QL and XYH conceived and designed the study. QL and XYH wrote and edited the paper. CYD, JC and DHL reviewed and revised the paper. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work were appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Hospital of Jilin University and written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Omuro A, Deangelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 3.Taylor LP. Diagnosis, treatment, and prognosis of glioma: Five new things. Neurology. 2010;75(18 Suppl 1):S28–S32. doi: 10.1212/WNL.0b013e3181fb3661. [DOI] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 6.Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1930. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidle UH, Birzele F, Kollmorgen G, Rüger R. Long noncoding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez-Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Ye C, Xiong H, Shen Y, Lu Y, Zhou J, Wang L. Dysregulation of long non-coding RNA in breast cancer: An overview of mechanism and clinical implication. Oncotarget. 2017;8:5508–5522. doi: 10.18632/oncotarget.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie W, Yuan S, Sun Z, Li Y. Long noncoding and circular RNAs in lung cancer: Advances and perspectives. Epigenomics. 2016;8:1275–1287. doi: 10.2217/epi-2016-0036. [DOI] [PubMed] [Google Scholar]
- 15.Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016;142:2231–2237. doi: 10.1007/s00432-016-2183-7. [DOI] [PubMed] [Google Scholar]
- 16.Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16:107. doi: 10.1186/s12943-017-0671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richtig G, Ehall B, Richtig E, Aigelsreiter A, Gutschner T, Pichler M. Function and clinical implications of long non-coding RNAs in melanoma. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040715. pii: E715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. 2016;2016:9085195. doi: 10.1155/2016/9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, Yu SZ, Pu PY, Liu N, You YP, et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013;15:1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang B, Wei ZY, Wang BQ, Yang HC, Wang JY, Bu XY. Down-regulation of the long noncoding RNA-HOX transcript antisense intergenic RNA inhibits the occurrence and progression of glioma. J Cell Biochem. 2018;119:2278–2287. doi: 10.1002/jcb.26390. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y, Wu D, Wang Y, Zhuang Z, Xia H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg. 2016;124:129–136. doi: 10.3171/2014.12.JNS1426.test. [DOI] [PubMed] [Google Scholar]
- 25.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A. The oncofetal H19 RNA connection: Hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 27.Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu Y, Zhu W. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014;35:7935–7944. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 28.Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, Xue YX. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27:275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 30.Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, Sun T, Xie X, Zhou Y, Du Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016;7:e2123. doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 33.Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 34.Sanfilippo PG, Hewitt AW. Translating the ENCyclopedia of DNA elements project findings to the clinic: ENCODE's implications for eye disease. Clin Exp Ophthalmol. 2014;42:78–83. doi: 10.1111/ceo.12150. [DOI] [PubMed] [Google Scholar]
- 35.Tragante V, Moore JH, Asselberg FW. The ENCODE project and perspectives on pathways. Genet Epidemiol. 2014;38:275–280. doi: 10.1002/gepi.21802. [DOI] [PubMed] [Google Scholar]
- 36.Shen ZH, Zhao KM, Du T. HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci. 2017;21:945–952. [PubMed] [Google Scholar]
- 37.Eoh KJ, Kim HJ, Lee JY, Nam EJ, Kim S, Kim SW, Kim YT. Dysregulated expression of homeobox family genes may influence survival outcomes of patients with epithelial ovarian cancer: Analysis of data from The Cancer Genome Atlas. Oncotarget. 2017;8:70579–70585. doi: 10.18632/oncotarget.19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, Lee JS, Cho JY. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;19:7078–7088. doi: 10.3748/wjg.v19.i41.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonfim-Silva R, Melo Ferreira FU, Thomé CH, Abraham KJ, De Souza FAL, Ramalho FS, Machado HR, De Oliveira RS, Cardoso AA, Covas DT, et al. Functional analysis of HOXA10 and HOXB4 in human medulloblastoma cell lines. Int J Oncol. 2017;51:1929–1940. doi: 10.3892/ijo.2017.4151. [DOI] [PubMed] [Google Scholar]
- 40.Carrera M, Bitu CC, de Oliveira CE, Cervigne NK, Graner E, Manninen A, Salo T, Coletta RD. HOXA10 controls proliferation, migration and invasion in oral squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:3613–3623. [PMC free article] [PubMed] [Google Scholar]
- 41.Kurscheid S, Bady P, Sciuscio D, Samarzija I, Shay T, Vassallo I, Criekinge WV, Daniel RT, van den Bent MJ, Marosi C, et al. Chromosome 7 gain and DNA hypermethylation at the HOXA10 locus are associated with expression of a stem cell related HOX-signature in glioblastoma. Genome Biol. 2015;16:16. doi: 10.1186/s13059-015-0583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmoudi S, Henriksson S, Corcoran M, Méndez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 43.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 44.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 45.Sun S, Su C, Zhu Y, Li H, Liu N, Xu T, Sun C, Lv Y. MicroRNA-544a regulates migration and invasion in colorectal cancer cells via regulation of homeobox A10. Dig Dis Sci. 2016;61:2535–2544. doi: 10.1007/s10620-016-4186-2. [DOI] [PubMed] [Google Scholar]
- 46.Libório-Kimura TN, Jung HM, Chan EK. miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 2015;51:151–157. doi: 10.1016/j.oraloncology.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 47.Xiao ZD, Jiao CY, Huang HT, He LJ, Zhao JJ, Lu ZY, Liu LX. miR-218 modulate hepatocellular carcinoma cell proliferation through PTEN/AKT/PI3K pathway and HoxA10. Int J Clin Exp Pathol. 2014;7:4039–4044. [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Zhang J, Wang H, Zhao J, Xu C, Du Y, Luo X, Zheng F, Liu R, Zhang H, et al. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer. 2012;12:111. doi: 10.1186/1471-2407-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the findings of this study are included within the article.