Abstract
Tumor recurrence hinders treatment of ovarian cancer. The present study aimed to identify potential biomarkers for ovarian cancer recurrence prognosis and explore relevant mechanisms. RNA-sequencing of data from the TCGA database and GSE17260 dataset was carried out. Samples of the data were grouped according to tumor recurrence information. Following data normalization, differentially expressed genes/micro RNAs (miRNAs)/long non-coding (lncRNAs) (DEGs/DEMs/DELs) were selected between recurrent and non-recurrent samples. Their correlations with clinical information were analyzed to identify prognostic RNAs. A support vector machine classifier was used to find the optimal gene set with feature genes that could conclusively distinguish different samples. A protein-protein interaction (PPI) network was established for DEGs using relevant protein databases. An integrated ‘lncRNA/miRNA/mRNA’ competing endogenous RNA (ceRNA) network was constructed to reveal potential regulatory relationships among different RNAs. We identified 36 feature genes (e.g. TP53 and RBPMS) for the classification of recurrent and non-recurrent ovarian cancer samples. Prediction with this gene set had a high accuracy (91.8%). Three DELs (WT1-AS, NBR2 and ZNF883) were highly associated with the prognosis of recurrent ovarian cancer. Predominant DEMs with their targets were hsa-miR-375 (target: RBPMS), hsa-miR-141 (target: RBPMS), and hsa-miR-27b (target: TP53). Highlighted interactions in the ceRNA network were ‘WT1-AS-hsa-miR-375-RBPMS’ and ‘WT1-AS-hsa-miR-27b-TP53’. TP53, RBPMS, hsa-miR-375, hsa-miR-141, hsa-miR-27b, and WT1-AS may be biomarkers for recurrent ovarian cancer. The interactions of ‘WT1-AS-hsa-miR-375-RBPMS’ and ‘WT1-AS-hsa-miR-27b-TP53’ may be potential regulatory mechanisms during cancer recurrence.
Keywords: ovarian cancer, recurrence, feature genes, lncRNA, miRNA, competing endogenous RNA
Introduction
Ovarian cancer is the sixth most frequent cancer worldwide (1). It is a major cause of cancer-related deaths among women. Based on global epidemiological data in 2008, 225,500 women were estimated to be diagnosed with ovarian cancer and 140,200 succumbed to the disease (2). In addition, the majority (>75%) of cases were in advanced stages requiring surgery and platinum-based chemotherapy. Although the standard treatment produces a high response rate of 40–60%, the 5-year survival rate is relatively poor (<25%) and recurrence occurs in >90% of patients after 18 months (3,4). Despite advancements in surgical and chemotherapeutic options, treatment of recurrent ovarian cancer is still a challenge.
A better understanding of the molecular mechanisms of ovarian cancer could help to develop more effective targeted therapies that contribute to improved prognosis. Upregulation of cluster of differentiation 44 (CD44) plays an important role in metastasis, recurrence, and drug resistance of ovarian cancer. Thus, CD44 is a potential target for prevention of recurrence in ovarian cancer (5). Human epididymis protein 4 (HE4) has been suggested as a serum biomarker for prognosis of epithelial ovarian cancer. Moreover, HE4 better predicts recurrence than the common marker carbohydrate antigen 125 (CA125) (6). Increased platelet-derived growth factor receptor-beta (PDGFR-β) and vascular endothelial growth factor receptor-2 (VEGFR-2) protein levels have been revealed to be associated with resistance to platinum-based chemotherapy and poorer outcome of ovarian cancer patients (7).
MicroRNAs (miRNAs/miRs) also play significant roles in the regulation of the disease recurrence. Loss of miR-200 family members has been revealed to be associated with relapse from early to advanced stages of epithelial ovarian cancer (8), suggesting that the expression of these miRNAs could be used as a target for prediction of recurrence. Moreover, miR-200 overexpression has been revealed to correspond with an advanced stage of ovarian cancer (9). Long non-coding RNAs (lncRNAs) are non-protein-coding RNA transcripts that control gene/miRNA expression and protein functions (10), which have been reported to be aberrantly expressed in ovarian carcinoma (11). LncRNAs also act as competing endogenous RNAs (ceRNAs) in the regulation of miRNA expression. Therefore, there is often a reverse expression between them (12). Recently, a study identified a six-lncRNA signature (RUNX1-IT1, MALAT1, H19, HOTAIRM1, LOC100190986 and AL132709.8) that was correlated with the recurrence of ovarian cancer (13).
The connection between lncRNAs with mRNAs or miRNAs in ovarian cancer is unclear. By constructing a functional lncRNA-mRNA co-expression network, Guo et al identified two immune-related lncRNA biomarkers (RP11-284N8.3.1 and AC104699.1.1) in the progression of malignant ovarian cancer (14). Although the biomarkers were reported to have crucial prognostic value on survival prediction at different stages of cancer, recurrence of the disease was not elucidated. Based on the ‘ceRNA hypothesis’, lncRNA-associated ceRNA networks were identified and ten lncRNA ceRNAs were proposed as potential candidates for ovarian cancer at different stages (15). In that study, miRNA-mediated ceRNA crosstalk between lncRNAs and mRNAs was evident but no information pertaining to recurrence was provided.
To provide more clarity concerning recurrence, we searched RNA-sequencing (RNA-seq) data in The Cancer Genome Atlas (TCGA) database and the Gene Expression Omnibus (GEO) database, which contain ovarian cancer samples with information about recurrence. We also explored potential regulations among lncRNAs, miRNAs, and mRNAs by establishing an integrated ceRNA network. A support vector machine (SVM) classifier with candidate feature genes was constructed to distinguish recurrent with non-recurrent ovarian cancer. These comprehensive analyses aimed to reveal novel lncRNA/miRNA/mRNA biomarkers of recurrent ovarian cancer and uncover the underlying regulatory mechanisms.
Materials and methods
Data resource and pretreatment
Data from TCGA database
The mRNA and miRNA expression profiles relevant to ovarian cancer were searched in the TCGA (https://gdc-portal.nci.nih.gov/) database. A total of 419 mRNA-sequencing profiles and 493 miRNA-sequencing profiles were obtained. These profiles were matched according to barcode numbers. Finally, 391 profiles with matched mRNA-sequencing and miRNA-sequencing data were generated. According to the clinical information, these 391 RNA-seq profiles were divided into a recurrence (n=220) and non-recurrence (n=171) group. The sequencing platform of all the samples was the HiSeq 2000 system (Illumina, Inc., San Diego, CA, USA).
All downloaded RNA-seq data were as files in the *.gene.quantification.txt format. Reads per kilobase of transcript per million mapped read (RPKM) values of expression of these RNAs were obtained. Since log2 (x+1) transformation had previously been performed, these data could be directly used for analysis in the present study.
Data from GEO database
The mRNA microarray data (accession no. GSE17260) was downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo). This dataset relevant to ovarian cancer consisted of 110 samples. These were also classified into a recurrence group (n=76) and non-recurrence group (n=34). The platform of the microarray data was the 014850 GPL6480 (Agilent Technologies, Inc., Santa Clara, CA, USA).
After downloading raw data from the GEO database, probe values corresponded to gene expressions based on annotation files. If more than one probe corresponded to the same gene, their values were averaged to calculate this gene expression. The expression of the genes were log2 transformed to reach an approximately normal distribution. Normalization was performed with the median method implemented in the Linear Models for Microarray Analysis (limma, http://www.bioconductor.org/packages/release/bioc/html/limma.html) package of R (16).
Analysis of the RNA-seq data
The mRNAs, miRNAs, and lncRNAs in 391 RNA-seq profiles downloaded from the TCGA database were identified according to 2,775 lncRNAs and 19,004 protein-coding gene annotation information recorded in the HUGO Gene Nomenclature Committee (HGNC, http://www.genenames.org/) (17). Low abundant mRNAs, miRNAs, and lncRNAs with an expressive abundance of <1, <5, and <5, respectively, were filtered out.
Differentially expressed genes/miRNAs (DEGs/DEMs) between recurrent and non-recurrent samples were selected using the edgeR package (version 3.0.1), a software in Bioconductor that adopts the over-dispersed Poisson model to differentiate biological and technical sources of variation (18). Notably, the edgeR package uses an empirical Bayes approach, which reduces overdispersion across different transcript samples and enhances analysis reliability (18). Α false discovery rate (FDR) <0.05 and |fold change (FC)|>1.5 were two criteria for DEG/DEM selection.
Relationships between DEGs and clinical features
All the clinical feature information of samples in the datasets was downloaded. The samples were divided into different groups based on the following dichotomous variables: Age at diagnosis (≥60 vs. <60 years), clinical stage (III+IV vs. I+II), neoplasm histological grade (G3+G4 vs. G1+G2), lymphatic invasion (Yes vs. No), and venous invasion (Yes vs. No). The expression of three types of RNAs (mRNAs, miRNAs, and lncRNAs) associated with different clinical features were selected using the edgeR package. Likewise, the cut-off values were FDR <0.05 and |FC|>1.5.
Selection of prognostic mRNAs, miRNAs, and lncRNAs
The expression of DEG/DEM/differentially expressed lncRNAs (DEL) between recurrent and non-recurrent samples were extracted, accompanied with the survival information in each sample. The single factor Cox analysis using the survfit function implemented in the R survival package was utilized to perform prognostic analysis (19). The mRNAs, miRNAs, and lncRNAs with a threshold P-value <0.05 were considered as significantly related to the prognosis. The survival result was expressed as a Kaplan-Meier (KM) curve.
Identification of key feature genes relevant to recurrence
Construction of protein-protein interaction network of DEGs
Relationships of the DEGs were explored by integrating human gene interactions in three protein databases, BioGRID (version 3.4.140, http://thebiogrid.org/), HPRD (release 9.0, http://www.hprd.org/), and DIP (http://dip.doe-mbi.ucla.edu/). Overlapping interactions in the three databases were extracted to establish the protein-protein interaction (PPI) network of the DEGs. Cytoscape (http://cytoscape.org/) software was used to visualize the network.
Optimization of feature genes dependent on network betweenness centrality
After the PPI network of the DEGs was established, its topological structure was analyzed according to the node's degree and betweenness centrality (BC) algorithm, using the following formula:
where σst denotes the shortest path from s to t, σst (v) stands for the node numbers (v) from s to t. BC values are 0 to 1, and the closeness of a node's value to ‘1’ is strongly associated with the importance of the node. Based on this definition, the nodes whose BC values were ranked in the top 100 were selected as candidate feature genes.
Selection of optimal feature gene set
Following the identification of candidate DEGs between recurrent and non-recurrent samples, the unsupervised clustering method was used to validate the efficacy of the classification using this feature gene set. In brief, the top 100 candidate feature DEGs underwent the optimal feature combination selection with the recursive feature elimination (RFE) algorithm (20). Genes in the most optimal feature gene set were supposed to be representative, prominent and could be used for clinical diagnosis.
Construction of a support vector machine classifier utilizing the feature gene sets
The significant feature gene set was selected by optimizing the feature of genes. The SVM classifier model was constructed to classify and distinguish the samples according to the expression of these feature genes in each sample (21), which were defined as an eigenvalue of these feature genes. By evaluating the eigenvalue of these feature genes in each sample, the probability of each sample in a certain classification was determined. In this way, the recurrent and non-recurrent ovarian cancer samples were predicted.
Independent validation and assessment of SVM classifier performance
To confirm the robustness and reproducibility of this SVM classifier, the dataset of GSE17260 was used as the validation set. Performance of the SVM classifier was evaluated by assessing the following indicators: Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and area under the curve (AUC) of the receiver operating characteristic (ROC) curve.
Prediction of lncRNA/miRNA network
By integrating miRNAs with lncRNAs information in miRcode (version 11.0, http://www.mircode.org/) (22) and starBase databases (version 2.0) (23), potential lncRNA/miRNA interactions were predicted for DEMs and DELs.
Target prediction of miRNAs
The miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw) provides the newest and the most comprehensive miRNA-target interactions that have been experimentally validated (24,25). In the present study, we used the latest version of the database in 2016 (release 6.0) to search for potential target genes of the miRNAs. Combining these target genes with DEGs in the PPI network, a miRNA-target network of the DEGs was constructed and visualized using Cytoscape software.
Construction of ceRNA regulatory network
Integrating the lncRNA/miRNA network and miRNA-target network, a comprehensive ceRNA network, termed the lncRNA/miRNA/mRNA regulatory network, was constructed.
Functional and pathway enrichment analysis of genes in the ceRNA network
After establishing the ceRNA network, genes in this network underwent functional and pathway enrichment analyses, integrating gene information in the Gene Ontology (GO; http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genome (KEGG; http://www.genome.jp/kegg/pathway.html) databases with the threshold as P-value <0.05. Fisher's exact test was used, as indicated in the following formula:
where N denotes total gene numbers in the whole genome, M represents gene numbers in the pathway, K stands for the number of DEGs, and p indicates the possibility that at least ‘x’ of ‘K’ DEGs were enriched in a specific function or pathway category.
Results
Ovarian cancer recurrence-related mRNAs, miRNAs and lncRNAs
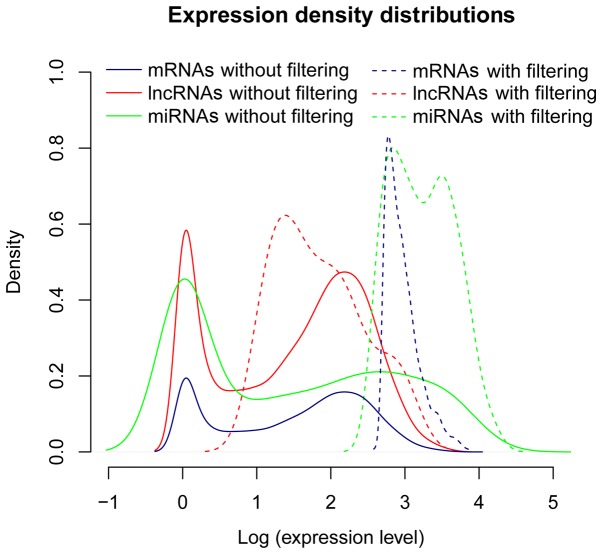
According to annotation information recorded in the HGNC, a set of 17,895 mRNAs that encodes proteins, 1,046 miRNAs, and 811 lncRNAs were identified in the 391 RNA-seq profiles downloaded from the TCGA database. Among these identified RNAs, those that had low abundance were filtered out. Cut-offs for low abundant mRNAs, miRNAs, and lncRNAs were expressed with an abundance <1, <5 and <5, respectively. After removing these low abundant RNAs, a group of 11,420 mRNAs, 169 miRNAs, and 398 lncRNAs remained. The expression distributions revealed that the peak values of expression density for the mRNAs, miRNAs and lncRNAs were evidently elevated after eliminating the low abundant ones (Fig. 1). Among these three types of RNAs, lncRNAs had an apparently lower expression density than the others.
Figure 1.
Expression density distributions of mRNAs, miRNAs and lncRNAs. The solid and dashed blue lines represent the expression density distributions of mRNAs before and after removal of the low abundant mRNAs, respectively. The solid and dashed red lines represent the expression density distributions of lncRNAs before and after removal of the low abundant lncRNAs, respectively. The solid and dashed green lines represent the expression density distribution of miRNAs before and after removal of the low abundant miRNAs, respectively. The peak values of expression density for the mRNAs, miRNAs, and lncRNAs after removal of the low abundant ones were evidently elevated.
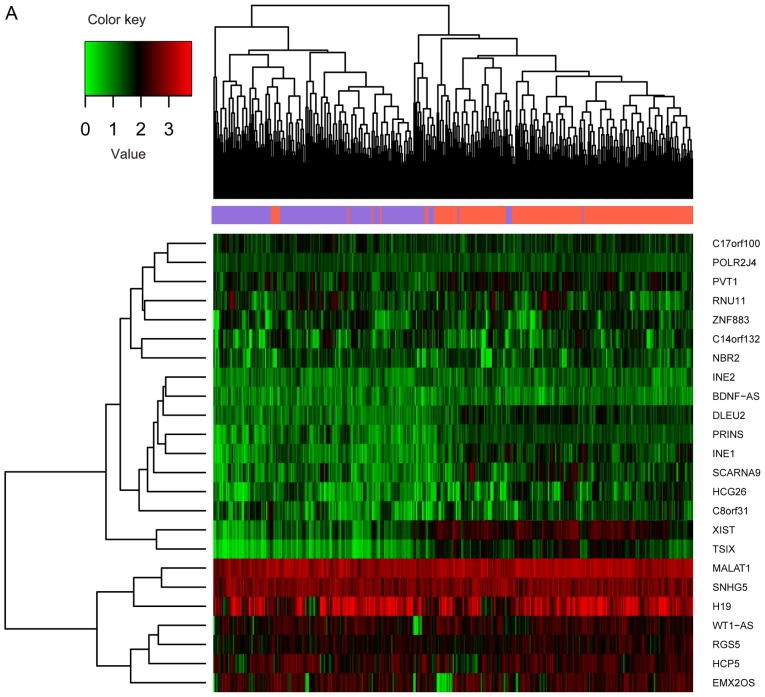
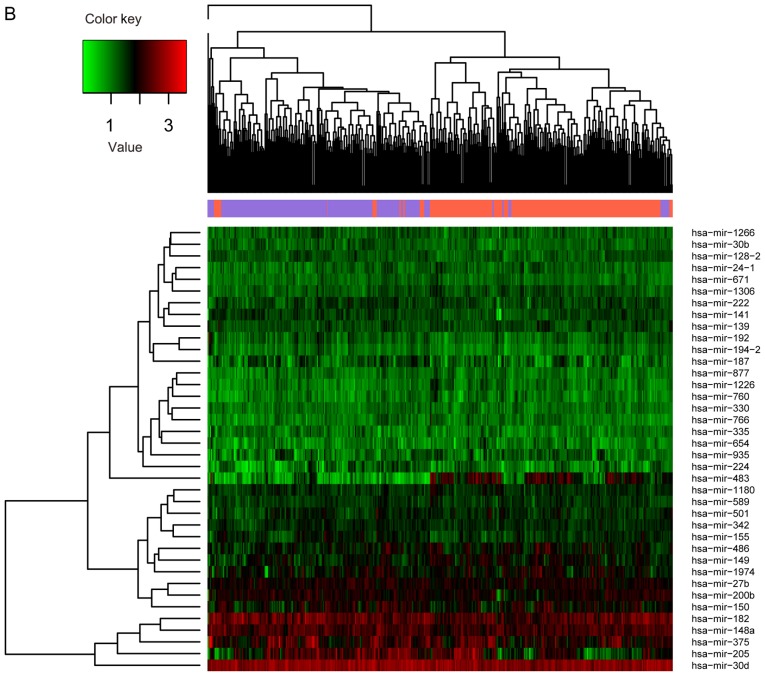
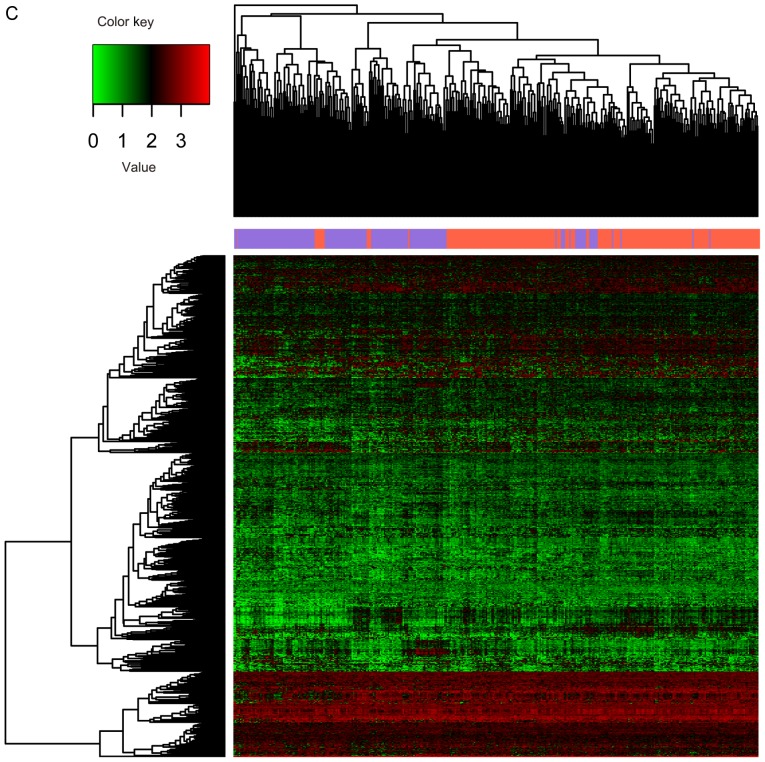
The 391 RNA-seq profiles were divided into recurrent (n=220) and non-recurrent (n=171) samples based on the clinical information. A total of 826 significant DEGs, 38 DEMs and 24 DELs between recurrent (n=220) and non-recurrent (n=171) samples were selected through differential analysis. Expression differences of these RNAs in different samples are displayed in the heat map of the clustering analysis (Fig. 2A-C). The three types of RNAs could clearly distinguish the recurrent ovarian cancer from the non-recurrent.
Figure 2.
Expression of different RNAs in different samples. (A) lncRNAs; (B) miRNAs. The y-axis represents RNAs; red denotes upregulation and green denotes downregulation. (C) mRNAs. The x-axis denotes the samples; the purple indicates the non-recurrent ovarian cancer, and the orange indicates the recurrent ovarian cancer. The y-axis represents RNAs; red denotes upregulation and green denotes downregulation.
Association between recurrence-related genes and clinical features
Using the five dichotomous variables (age, clinical stage, neoplasm histological grade, lymphatic invasion, and venous invasion) correlated with clinical information, the samples were classified into different groups. Upregulated or downregulated RNAs (mRNAs, miRNAs, and lncRNAs) between different groups within each comparison were selected (data not shown). Clinical features that the recurrence-related genes could reflect were revealed.
Prognosis-related mRNAs, miRNAs, and lncRNAs
Based on the expression of the DEGs, DEMs, and DELs, and the survival analysis (e.g. overall survival time and survival condition) of these RNAs, prognostic mRNAs, miRNAs, and lncRNAs were identified (Table I). Three upregulated lncRNAs were associated with prognosis including NBR2 (P=0.003), ZNF883 (P=0.016) and WT1-AS (P=0.014). has-miR-1974 (P=0.004), hsa-miR-155 (P=0.005), hsa-mir-1266 (P=0.006), hsa-mir-1306 (P=0.009), hsa-mir-935 (P=0.017) and hsa-mir-375 (P=0.028) were related to prognosis. A total of 58 DEGs (P<0.005) were significantly associated with prognosis.
Table I.
mRNAs, miRNAs and lncRNAs significantly related to prognosis.
| RNA | Type | Coef | Exp (coef) | Se (coef) | Z score | P-value | Regulation |
|---|---|---|---|---|---|---|---|
| NBR2 | lncRNA | −0.193 | 0.825 | 0.102 | −1.880 | 0.003 | Upregulated |
| WT1-AS | lncRNA | −0.070 | 0.932 | 0.064 | −1.100 | 0.014 | Upregulated |
| ZNF883 | lncRNA | 0.191 | 1.210 | 0.080 | 2.400 | 0.016 | Upregulated |
| hsa-miR-1974 | miRNA | −0.076 | 0.927 | 0.042 | −1.810 | 0.004 | Downregulated |
| hsa-miR-155 | miRNA | −0.087 | 0.917 | 0.052 | −1.680 | 0.005 | Upregulated |
| hsa-miR-1266 | miRNA | 0.098 | 1.100 | 0.063 | 1.540 | 0.006 | Downregulated |
| hsa-miR-1306 | miRNA | −0.091 | 0.913 | 0.066 | −1.370 | 0.009 | Upregulated |
| hsa-miR-935 | miRNA | 0.041 | 1.040 | 0.043 | 0.965 | 0.017 | Upregulated |
| hsa-miR-375 | miRNA | −0.018 | 0.982 | 0.031 | −0.582 | 0.028 | Downregulated |
| VEPH1 | mRNA | 0.341 | 1.410 | 0.108 | 3.160 | 0.002 | Downregulated |
| TSHZ3 | mRNA | −0.162 | 0.850 | 0.091 | −1.790 | 0.004 | Upregulated |
| SORBS2 | mRNA | 0.159 | 1.170 | 0.094 | 1.680 | 0.005 | Upregulated |
| NOTUM | mRNA | −0.080 | 0.923 | 0.051 | −1.590 | 0.006 | Downregulated |
| CASC1 | mRNA | 0.116 | 1.120 | 0.075 | 1.560 | 0.006 | Upregulated |
| CCDC65 | mRNA | 0.114 | 1.120 | 0.074 | 1.540 | 0.006 | Upregulated |
| ALDH1A2 | mRNA | −0.063 | 0.939 | 0.043 | −1.480 | 0.007 | Downregulated |
| REM1 | mRNA | −0.109 | 0.897 | 0.075 | −1.450 | 0.008 | Downregulated |
| PHOSPHO1 | mRNA | 0.073 | 1.080 | 0.055 | 1.320 | 0.010 | Downregulated |
| TBX3 | mRNA | −0.097 | 0.908 | 0.073 | −1.320 | 0.010 | Upregulated |
| OXGR1 | mRNA | 0.080 | 1.080 | 0.062 | 1.300 | 0.010 | Upregulated |
| C1orf194 | mRNA | 0.050 | 1.050 | 0.040 | 1.230 | 0.011 | Upregulated |
| INHA | mRNA | 0.098 | 1.100 | 0.082 | 1.190 | 0.012 | Upregulated |
| CLIC6 | mRNA | −0.079 | 0.924 | 0.067 | −1.170 | 0.012 | Upregulated |
| BNC2 | mRNA | 0.124 | 1.130 | 0.107 | 1.160 | 0.013 | Downregulated |
| CST6 | mRNA | −0.080 | 0.923 | 0.070 | −1.150 | 0.013 | Upregulated |
| PLCE1 | mRNA | 0.102 | 1.110 | 0.097 | 1.060 | 0.015 | Upregulated |
| MAT1A | mRNA | −0.080 | 0.923 | 0.078 | −1.020 | 0.016 | Downregulated |
| PHF7 | mRNA | 0.149 | 1.160 | 0.158 | 0.941 | 0.018 | Upregulated |
| HOXA3 | mRNA | −0.054 | 0.948 | 0.059 | −0.911 | 0.018 | Downregulated |
| WDR78 | mRNA | 0.108 | 1.110 | 0.128 | 0.842 | 0.020 | Upregulated |
| ZNF521 | mRNA | −0.052 | 0.949 | 0.062 | −0.847 | 0.020 | Upregulated |
| FAM155B | mRNA | 0.184 | 1.200 | 0.080 | 2.300 | 0.021 | Upregulated |
| SIGLEC14 | mRNA | 0.077 | 1.080 | 0.096 | 0.810 | 0.021 | Downregulated |
| TMEM190 | mRNA | 0.040 | 1.040 | 0.051 | 0.789 | 0.022 | Upregulated |
| LMO3 | mRNA | 0.036 | 1.040 | 0.047 | 0.752 | 0.023 | Downregulated |
| FIGN | mRNA | −0.066 | 0.936 | 0.088 | −0.746 | 0.023 | Downregulated |
| FAM83E | mRNA | 0.047 | 1.050 | 0.063 | 0.741 | 0.023 | Upregulated |
| CLCN5 | mRNA | 0.090 | 1.090 | 0.124 | 0.727 | 0.024 | Downregulated |
| THBS4 | mRNA | −0.048 | 0.953 | 0.067 | −0.721 | 0.024 | Downregulated |
| HOXA5 | mRNA | −0.037 | 0.964 | 0.052 | −0.707 | 0.024 | Downregulated |
| HIST2H2BF | mRNA | 0.066 | 1.070 | 0.099 | 0.668 | 0.025 | Upregulated |
| PRR22 | mRNA | 0.060 | 1.060 | 0.097 | 0.619 | 0.027 | Downregulated |
| KCNH3 | mRNA | 0.036 | 1.040 | 0.062 | 0.575 | 0.029 | Downregulated |
| C16orf74 | mRNA | −0.043 | 0.958 | 0.088 | −0.492 | 0.031 | Upregulated |
| TGFA | mRNA | −0.034 | 0.967 | 0.072 | −0.475 | 0.032 | Upregulated |
| BHLHA15 | mRNA | 0.043 | 1.040 | 0.090 | 0.475 | 0.032 | Downregulated |
| FOXA2 | mRNA | −0.026 | 0.974 | 0.057 | −0.461 | 0.032 | Upregulated |
| NRL | mRNA | 0.093 | 1.100 | 0.204 | 0.456 | 0.033 | Downregulated |
| GEM | mRNA | −0.184 | 0.832 | 0.087 | −2.120 | 0.034 | Upregulated |
| FA2H | mRNA | −0.040 | 0.961 | 0.104 | −0.386 | 0.035 | Upregulated |
| ACAP1 | mRNA | 0.044 | 1.040 | 0.118 | 0.371 | 0.036 | Downregulated |
| SHC2 | mRNA | −0.026 | 0.975 | 0.072 | −0.354 | 0.036 | Upregulated |
| KRT16 | mRNA | 0.018 | 1.020 | 0.051 | 0.355 | 0.036 | Upregulated |
| TTC36 | mRNA | −0.039 | 0.962 | 0.113 | −0.345 | 0.037 | Downregulated |
| RBM11 | mRNA | 0.040 | 1.040 | 0.116 | 0.342 | 0.037 | Downregulated |
| ZNF569 | mRNA | −0.301 | 0.740 | 0.144 | −2.090 | 0.037 | Downregulated |
| LMTK3 | mRNA | 0.168 | 1.180 | 0.081 | 2.080 | 0.037 | Upregulated |
| ADAMDEC1 | mRNA | 0.018 | 1.020 | 0.060 | 0.306 | 0.038 | Downregulated |
| MACROD2 | mRNA | 0.022 | 1.020 | 0.076 | 0.292 | 0.039 | Downregulated |
| ZNF597 | mRNA | 0.031 | 1.030 | 0.102 | 0.298 | 0.039 | Upregulated |
| CD8A | mRNA | 0.018 | 1.020 | 0.071 | 0.249 | 0.040 | Downregulated |
| AGAP2 | mRNA | −0.033 | 0.968 | 0.137 | −0.238 | 0.041 | Downregulated |
| PRG4 | mRNA | 0.009 | 1.010 | 0.090 | 0.102 | 0.046 | Downregulated |
| GAL3ST3 | mRNA | −0.005 | 0.995 | 0.045 | −0.101 | 0.046 | Downregulated |
| CSPG5 | mRNA | −0.008 | 0.992 | 0.087 | −0.095 | 0.046 | Downregulated |
| SLAMF7 | mRNA | −0.004 | 0.996 | 0.057 | −0.078 | 0.047 | Downregulated |
| SP5 | mRNA | −0.002 | 0.998 | 0.052 | −0.043 | 0.049 | Downregulated |
Coef, coefficient; Se, standard error.
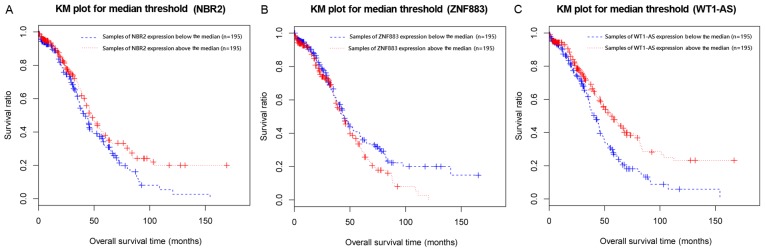
The median expression value of NBR2, ZNF883, or WT1-AS was used as the cut-off criterion for dividing the samples into two groups (below the median and above the median) based on the expression levels of these lncRNAs in the samples. KM survival analysis indicated the survival ratios of the below median and above median groups that were separated by the expression levels of NBR2 (P=0.0417), ZNF883 (P=0.0446), or WT1-AS (P=0.0131) were significantly different (Fig. 3A-C). This result indicated that these three lncRNAs could be used as prognostic predictors of recurrent ovarian cancer, particularly WT1-AS, since it had the lowest P-value.
Figure 3.
Kaplan-Meier (KM) curve of recurrent and non-recurrent ovarian cancer samples based on the expression levels of three differentially expressed lncRNAs. (A) KM plot for samples separated by the median threshold of NBR2; (B) KM plot for samples separated by the median threshold of ZNF883; (C) KM plot for samples separated by the median threshold of WT1-AS. The blue dash denotes samples with the expression levels below the median, and red denotes samples with the expression levels above the median.
Features of mRNAs related to recurrence
PPI network of the DEGs
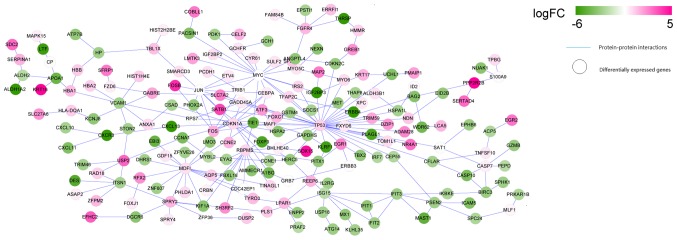
The search of the three protein databases identified protein interaction information for the DEGs. A total of 169, 221 and 234 protein interactions were identified from BioGRID, HPRD and DIP databases, respectively. Then, 299 overlapped interactions in three databases were extracted to establish the PPI network of the DEGs. As presented in Fig. 4, the network consisted of 234 nodes (the proteins encoded by the DEGs) and 299 edges (the protein interactions). Five predominant nodes in the PPI network were TP53 (degree=35), CDKN1A (degree=20), MYC (degree=17), MDFI (degree=16), and RBPMS (degree=13).
Figure 4.
Protein-protein interaction network of the differentially expressed genes. Pink denotes upregulated genes, and green denotes downregulated genes. The shade of color indicates different changes in expression; FC, fold change. The circle nodes denote genes and the lines denote the protein-protein interactions.
Verification of the SVM classifiers
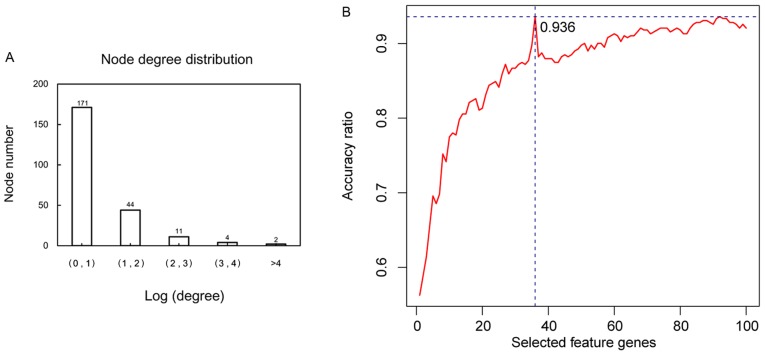
The node degree distribution of the genes in the PPI network was analyzed (Fig. 5A). The top 100 gene nodes in the PPI network ranked by their BC values were selected. The best prediction accuracy was up to 93.6% when the SVM classifier consisted of 36 specific feature genes (Fig. 5B) using the recursive feature elimination algorithm. These 36 specific feature genes (e.g. TP53, MYC, CDKN1A, RBPMS and JUN) are shown in Table II. This gene set was considered the optimal combination.
Figure 5.
Selection of the optimal feature gene set using recursive feature elimination algorithm. (A) Node degree distribution in the protein-protein interaction network. (B) Accuracy for sample classification using different feature gene combinations.
Table II.
Gene list of 36 feature genes in the optimal gene set identified by support vector machine classifier.
| Gene | BC | Degree | logFC | P-value | FDR | Gene | BC | Degree | logFC | P-value | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TP53 | 0.8549 | 36 | 1.1154 | 0.0034 | 0.0261 | GABRE | 0.2513 | 3 | 1.5580 | 0.0039 | 0.0303 |
| LIPH | 0.8000 | 3 | 2.1783 | 0.0035 | 0.0265 | KLRF1 | 0.2485 | 3 | −2.9154 | 0.0022 | 0.0166 |
| TNNT1 | 0.7000 | 2 | 0.8604 | 0.0049 | 0.0380 | SAT1 | 0.2477 | 3 | 0.5964 | 0.0062 | 0.0475 |
| MYC | 0.4611 | 17 | 0.6524 | 0.0064 | 0.0492 | SMARCD3 | 0.2354 | 2 | 0.9899 | 0.0042 | 0.0326 |
| VCAM1 | 0.3527 | 6 | −1.0917 | 0.0054 | 0.0414 | PLS1 | 0.2336 | 2 | 1.1975 | 0.0047 | 0.0364 |
| CDKN1A | 0.3446 | 12 | 1.0392 | 0.0044 | 0.0338 | EGR1 | 0.2312 | 4 | 1.8583 | 0.0005 | 0.0040 |
| RPS7 | 0.3278 | 3 | 0.6217 | 0.0064 | 0.0489 | SPC24 | 0.2207 | 2 | −0.8575 | 0.0061 | 0.0472 |
| HBA1 | 0.3031 | 3 | 1.3074 | 0.0040 | 0.0305 | HP | 0.2148 | 3 | −1.0778 | 0.0048 | 0.0368 |
| TIE1 | 0.3000 | 3 | −2.2494 | 0.0020 | 0.0150 | UCHL1 | 0.2140 | 3 | −1.6059 | 0.0013 | 0.0097 |
| JUN | 0.2881 | 10 | 0.6921 | 0.0057 | 0.0439 | SOCS1 | 0.2133 | 3 | −0.9843 | 0.0057 | 0.0437 |
| RBPMS | 0.2856 | 12 | 1.0742 | 0.0049 | 0.0377 | DGCR6 | 0.2104 | 2 | −1.4870 | 0.0046 | 0.0356 |
| APOA1 | 0.2813 | 4 | −2.0866 | 0.0003 | 0.0025 | FZD6 | 0.2104 | 2 | 0.8516 | 0.0056 | 0.0435 |
| HERC5 | 0.2803 | 4 | −1.2422 | 0.0046 | 0.0353 | RAD18 | 0.2086 | 2 | 1.1321 | 0.0054 | 0.0413 |
| BAG2 | 0.2712 | 3 | −1.5688 | 0.0030 | 0.0231 | HBB | 0.2080 | 2 | 1.1753 | 0.0037 | 0.0286 |
| BHLHE40 | 0.2651 | 3 | 0.8275 | 0.0051 | 0.0389 | SATB1 | 0.2075 | 3 | 2.4843 | 0.0014 | 0.0111 |
| CEP55 | 0.2538 | 2 | −0.8150 | 0.0064 | 0.0493 | FOXC1 | 0.2075 | 2 | 1.3750 | 0.0041 | 0.0314 |
| PSEN2 | 0.2534 | 5 | −1.1137 | 0.0049 | 0.0377 | PITX1 | 0.2055 | 2 | −1.3033 | 0.0045 | 0.0343 |
| ANGPTL4 | 0.2518 | 3 | −1.3853 | 0.0039 | 0.0298 | ANXA1 | 0.2047 | 2 | 1.0398 | 0.0030 | 0.0233 |
BC, betweenness centrality; FC, fold change; FDR, false discovery rate.
Data in the GSE17260 after normalization were used to validate the accuracy of the SVM classifier of the 36 feature genes. This SVM classifier could precisely distinguish 70 recurrent samples from 31 non-recurrent samples with an accuracy of 91.8%.
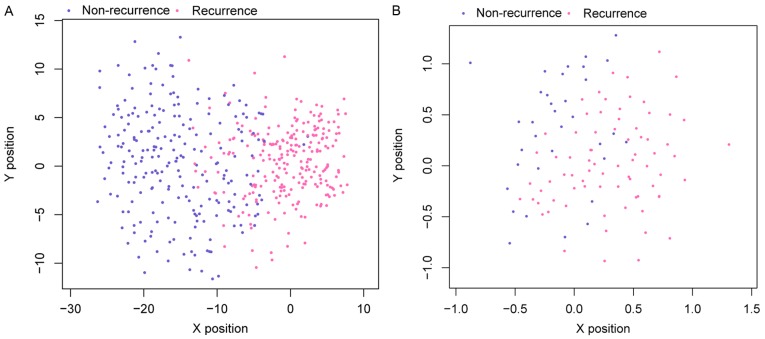
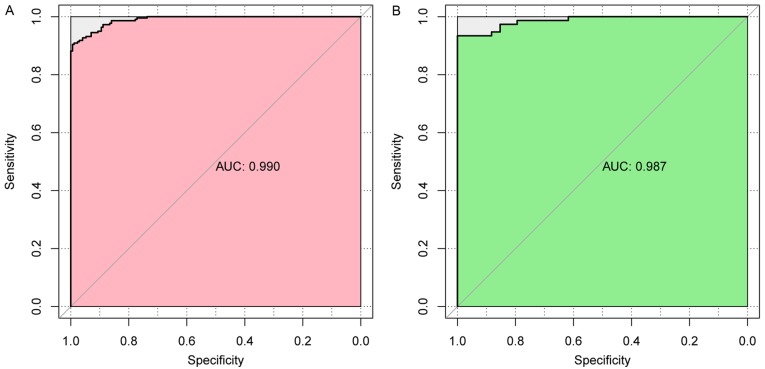
Scatter plots of sample classifications in validation dataset and data in TCGA are presented in Fig. 6. The findings indicated a good classification result. In addition, evaluation of the performance of this SVM classifier using five indicators (sensitivity, specificity, PPV, NPV, and AUC) further indicated that it had a high correct rate on classification (0.936 or 0.918), and most of the indicators had a high value of over 0.9 (Table III and Fig. 7).
Figure 6.
Scatter plot of samples using a support vector machine classifier. (A) TCGA training dataset; (B) GSE17260 validation dataset. The x-axis and y-axis represent the two-dimension position of the vector coordinates of the binary-class support vector machine. TCGA, The Cancer Genome Atlas.
Table III.
Performance evaluation of the SVM classifier in training dataset and validation dataset.
| Datasets | No. samples | Correct rate | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|---|---|
| TCGA | 391 | 0.936 | 0.897 | 0.942 | 0.923 | 0.921 | 0.990 |
| GSE13601 | 110 | 0.918 | 0.912 | 0.921 | 0.800 | 0.959 | 0.987 |
TCGA, The Cancer Genome Atlas; PPV, positive predictive value; NPV, negative predictive value; AUC, area under the curve of the receiver operating characteristic curve.
Figure 7.
Area under the curve of the receiver operating characteristic curve in different datasets. (A) TCGA training dataset; (B) GSE17260 validation dataset. AUC, area under the curve; TCGA, The Cancer Genome Atlas.
Predicted lncRNA/miRNA regulatory network
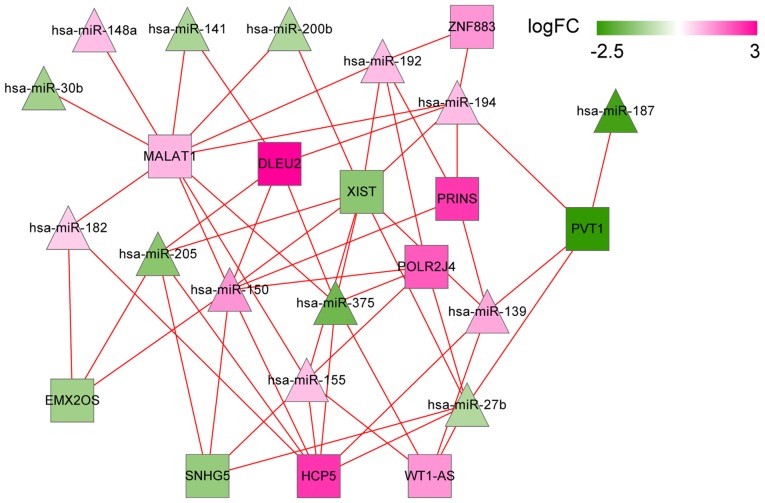
Using miRcode and starBase databases, a group of 469 and 396 lncRNA/miRNA interactions were identified in the two databases, respectively. Then, 562 overlapped interactions were extracted. Combining them with the DEMs and DELs, a set of 58 lncRNA/miRNA interactions were finally selected to construct the lncRNA/miRNA regulatory network. The network contained 11 DELs and 14 DEMs, such as DLEU2 (interplayed miRNAs: hsa-miR-141, hsa-miR-150 and hsa-miR-375), MALAT1 (interplayed miRNAs: hsa-miR-141, hsa-miR-150 and hsa-miR-375), and WT1-AS (interplayed miRNAs: hsa-miR-375, hsa-miR-155 and hsa-miR-27b; Fig. 8).
Figure 8.
DEL-DEM network in recurrent ovarian cancer. Diamonds denote miRNAs and squares denote lncRNAs. Pink denotes upregulation and green denotes downregulation. DEL, differentially expressed lncRNAs between recurrent and non-recurrent ovarian cancer; DEM, differentially expressed miRNAs between recurrent and non-recurrent ovarian cancer. The shade of color indicates different changes in expression; FC, fold change.
Predicted target genes of miRNAs
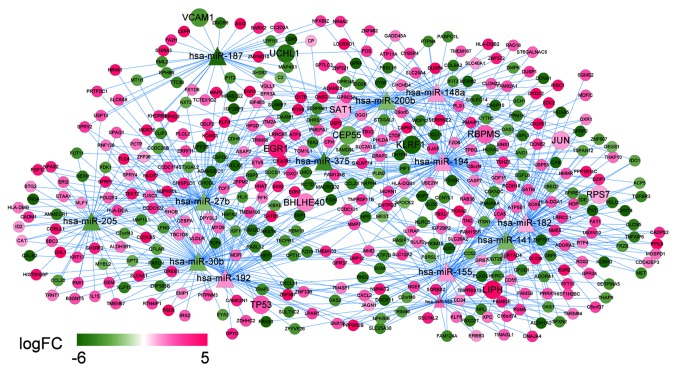
The 14 DEMs regulated by the DELs were mapped into the miRTarBase database to explore their target genes, accompanied with the DEGs. The predicted miRNA-target gene regulation network comprised of 426 nodes (13 miRNAs since hsa-miR-139 did not get any target information, and 413 mRNAs) and 743 interactions. In this network (Fig. 9), several nodes and interactions involving the feature genes in the optimal gene set that was identified by support vector machine classifier may be important, such as hsa-miR-375 (target genes: FOXC1, RBPMS and CCL28), hsa-miR-27b (target genes: TP53, PCTP, TOM1L1), and hsa-miR-141 (target genes: RBPMS, TINAGL1, and CCNE2).
Figure 9.
DEM-DEG network in recurrent ovarian cancer. Diamonds denote miRNAs, circles denote target genes and larger circles denote genes belonging to the optimal feature gene set. Pink denotes upregulation and green denotes downregulation. The shade of color indicates different changes in expression; FC, fold change.
Construction of ceRNA network
Integrated ceRNA network
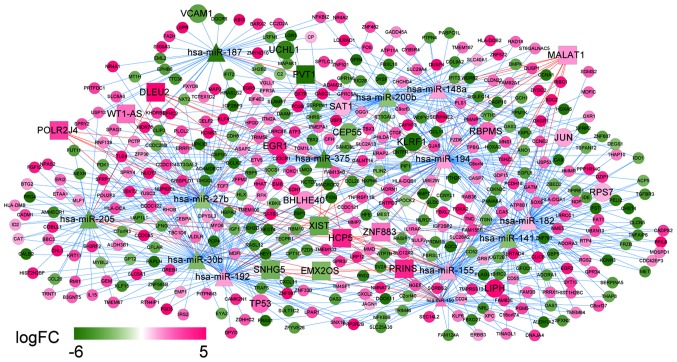
Integrating the lncRNA/miRNA interactions with the miRNA/mRNA interactions, a ceRNA network was constructed. The network was comprised of 437 nodes and 795 interactions. WT1-AS was a prognosis-related lncRNA with the best performance and hsa-miR-375 was also identified to be a prognosis-related miRNA. TP53 and RBPMS were the feature genes in the optimal gene set that were identified by the SVM classifier. In this integrated ceRNA network, two regulations ‘WT1-AS-hsa-miR-375-RBPMS’ and ‘WT1-AS-hsa-miR-27b-TP53’ may play important roles (Fig. 10).
Figure 10.
CeRNA network in recurrent ovarian cancer. Diamonds denote miRNAs, squares denote lncRNAs, circles denote target genes, and the larger circles denote genes belonging to the optimal feature gene set. Pink denotes upregulation and green denotes downregulation. The shade of color indicates different changes in expression. Pink lines denote lncRNA/miRNA interactions, and blue lines denote miRNA-target gene interactions; FC, fold change; ceRNA, competing endogenous RNA.
Enrichment analysis of target genes in the ceRNA network
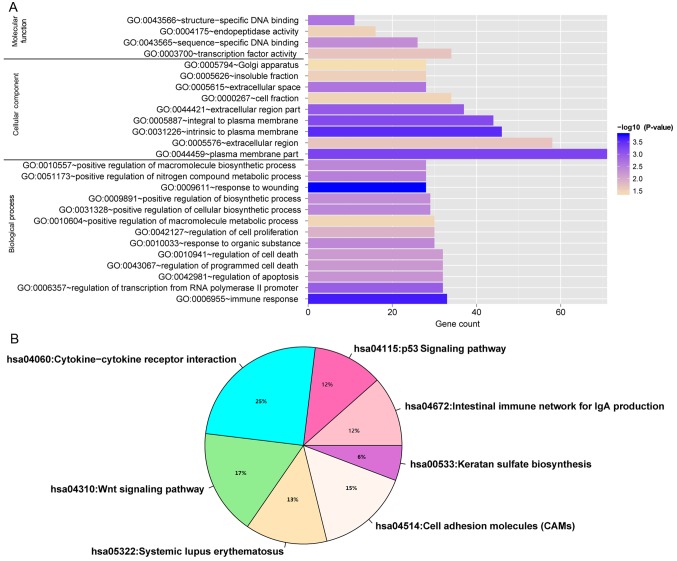
Enrichment analysis results indicated that target genes in this ceRNA network were significantly enriched in 26 GO functional categories (P<0.05; Fig. 11A) and seven KEGG pathway categories (P<0.05; Fig. 11B), including ‘immune response’, ‘response to wounding’, ‘intestinal immune network for IgA production’, ‘p53 signaling pathway’, ‘cytokine-cytokine receptor interaction’, and ‘Wnt signaling pathway’.
Figure 11.
Enriched pathways and functions of target genes in the ceRNA network. (A) Function categories; (B) Pathway categories. ceRNA, competing endogenous RNA; GO, Gene Ontology.
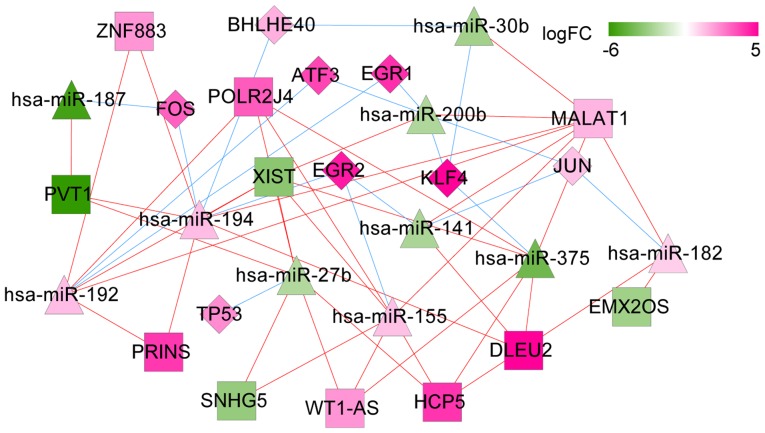
ceRNA network of transcription factors
To further detect regulations from transcription factors (TFs), two TF databases, Transcription Regulatory Regions Database (TRRD, http://wwwmgs.bionet.nsc.ru/mgs/gnw/trrd/) and JASPAR (http://jaspar.genereg.net/) were used. Integrating the TF information with the above ceRNA network, a sub ceRNA network relevant to TFs was extracted. In this subnetwork, eight important TFs were highlighted: KLF4, FOS, TP53, JUN, EGR1, EGR2, BHLHE40, and ATF3. In addition, TP53 was targeted by hsa-miR-27b and EGR2 was targeted by hsa-miR-141 (Fig. 12).
Figure 12.
CeRNA network of relevant transcription factors (TFs). Diamonds denote miRNAs, squares represent lncRNAs, and triangles denote TFs. Pink denotes upregulation and green denotes downregulation. The shade of color indicates different changes on expressions. Pink lines denote lncRNA/miRNA interactions, and blue lines denote miRNA-TF interactions; FC, fold change; ceRNA, competing endogenous RNA.
Discussion
In the present study, by analyzing the RNA-seq data in the GSE17260 dataset and the TCGA database, the optimal gene set containing 36 feature genes that could clearly distinguish recurrent with non-recurrent ovarian cancer was identified, including TP53 and RBPMS. These genes were also highlighted in the PPI network. We only identified three lncRNAs related to recurrent ovarian cancer: NBR2, ZNF883, and WT1-AS. Three predominant miRNAs with their target genes were also predicted: hsa-miR-375 (target: RBPMS), hsa-miR-27b (target: TP53), and hsa-miR-141 (target: RBPMS). Notably, ‘WT1-AS-hsa-miR-375-RBPMS’ and ‘WT1-AS-hsa-miR-27b-TP53’ interactions were striking in the ceRNA network.
The tumor suppressor tumor protein p53 (TP53) is mutated in the early stage of high-grade serous ovarian cancer, thus this gene mutation could act as a predictor for initiation of the disease (26). Notably, the TP53 mutation-regulated genomic instability induces the evolution of recurrence in epithelial ovarian cancer (27), indicating the close correlation between this gene expression and recurrence in ovarian cancer.
The protein encoded by RNA binding protein with multiple splicing (RBPMS) is a member of the RNA recognition motif family. It functions as a co-activator of transcriptional activity. Inhibition of miR-21-3p was revealed to significantly decrease proliferation and invasion in ovarian cancer, and RBPMS was confirmed as a target gene of miR-21-3p via luciferase reporter assays (28). For Korean patients with serous ovarian cancer at stage IIIC, RBPMS is a member of 27 genes located in chromosome 8p21.1-p12 regions with copy number loss, and it is enriched in ‘cellular macromolecule metabolic process’ involved in disease progression (29). However, no signs have indicated the relationship between this gene and recurrence in ovarian cancer.
EYA2 is identified as an oncogene in cervical carcinogenesis, while hsa-miR-375 is a tumor suppressor. EYA2 can also promote tumor growth of ovarian cancer (30). Considering the closeness of cervical cancer with ovarian cancer, hsa-miR-375 may also function as a tumor suppressor in ovarian cancer. Notably, hsa-miR-375 was revealed to be differentially expressed in ovarian serous carcinoma at stage I, and thus is a potential candidate miRNA signature for disease prediction (31). Alteration of hsa-miR-375 was highly correlated with recurrence in gastric cancer after surgery (32). However, the relationship between this miRNA and recurrence in ovarian cancer has not been reported.
hsa-miR-141 is a member of miR-200 family that has been revealed to be overexpressed in various cancer types, such as ovarian cancer, pancreatic ductal adenocarcinoma, and colorectal cancer (33). hsa-miR-141-5p is one of the ten miRNA signatures that may predict ovarian cancer development (34). High expression of hsa-miR-141 was related to poor prognosis of the disease (35). The collective data indicated the important role of hsa-miR-141 in ovarian cancer progression. Several miR-200 family members have been implicated in the correlation with recurrence. For instance, miR-429 was increased in metastatic ovarian cancer cells, and it was revealed to be a candidate therapeutic target that could reduce ovarian cancer metastasis and tumor recurrence (36). Another family member, miR-200b, was significantly associated with ovarian cancer recurrence (37). Whether the miR-200 family member hsa-miR-141 is involved in the recurrence of ovarian cancer is still unclear.
Based on our results, both of hsa-miR-375 and hsa-miR-141 were involved in different regulation networks, indicating that they participate in the process of ovarian cancer recurrence, or that their dysregulation accounts for the disease recurrence. Notably, RBPMS was the predicted target of both hsa-miR-375 and hsa-miR-141, indicating that these two miRNAs function by targeting this gene. The targeting relationships require validation by luciferase reporter assays.
Reportedly, the expression of miR-27 was associated with metastasis of ovarian cancer (38), and miR-27a and miR-27b were implicated in the control of drug resistance in ovarian cancer (39). No clues at present have linked miR-27 to recurrence in ovarian cancer. However, in our study, hsa-miR-27b was identified as an important miRNA for recurrent ovarian cancer, indicating it may be a novel signature. In human embryonal carcinoma cells, overexpressed miR-27 resulted in an increase of TP53 (40). Additionally, the TP53 gene transcript contains miR-27 binding sites (41). These indicate potential targeting regulations between miR-27 and TP53. Based on our study, TP53 was the predicted downstream target gene of hsa-miR-27b.
The Wilms tumor 1 (WT1) gene is frequently expressed in epithelial ovarian cancer (42). The lncRNA WT1 Antisense RNA (WT1-AS) encoded gene is located upstream of WT1. The two genes are bi-directionally transcribed from the same promoter region. Reportedly, interaction between WT1-AS and WT1 sense RNA resulted in the upregulation of the WT1 protein (43). In acute myeloid leukemia, alternative splicing of WT1-AS was reported (44). In gastric cancer, downregulation of WT1-AS promoted tumor cell proliferation and invasion (45). However, no correlations were indicated in ovarian cancer. Our results indicated that this lncRNA is a critical lncRNA in the ceRNA network, and the interactions of ‘WT1-AS-hsa-miR-375-RBPMS’ and ‘WT1-AS-hsa-miR-27b-TP53’ indicated that WT1-AS is a biomarker for prognosis of ovarian cancer recurrence, and participates in the aforementioned regulations during the process.
Despite the fact that our results provide many potential biomarkers and relevant regulations for ovarian cancer recurrence, there are several limitations in the study. The expression of these important genes, miRNAs and lncRNAs, as well as the predictive targeting relationships require further validation.
In conclusion, several biomarkers for ovarian cancer recurrence were identified. These included TP53, RBPMS, hsa-miR-375, hsa-miR-141, hsa-miR-27b, and WT1-AS. The interactions of ‘WT1-AS-hsa-miR-375-RBPMS’ and ‘WT1-AS-hsa-miR-27b-TP53’ may be potential regulatory mechanisms during this process.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
XW and LH performed data analyses and wrote the manuscript. LZ and LW significantly contributed in data analyses and manuscript revision. LMZ conceived and designed the study. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
In the original article of the datasets, the trials were approved by the local institutional review boards of all participating centers, and informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Diaz-Gil D, Fintelmann FJ, Molaei S, Elmi A, Hedgire SS, Harisinghani MG. Prediction of 5-year survival in advanced-stage ovarian cancer patients based on computed tomography peritoneal carcinomatosis index. Abdom Radiol. 2016;41:2196–2202. doi: 10.1007/s00261-016-0817-5. [DOI] [PubMed] [Google Scholar]
- 2.Lowe KA, Chia VM, Taylor A, O'Malley C, Kelsh M, Mohamed M, Mowat FS, Goff B. An international assessment of ovarian cancer incidence and mortality. Gynecol Oncol. 2013;130:107–114. doi: 10.1016/j.ygyno.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 3.Kehoe S, Hook J, Nankivell M, Jayson GC, Kitchener H, Lopes T, Luesley D, Perren T, Bannoo S, Mascarenhas M, et al. Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (CHORUS): An open-label, randomised, controlled, non-inferiority trial. Lancet. 2015;386:249–257. doi: 10.1016/S0140-6736(14)62223-6. [DOI] [PubMed] [Google Scholar]
- 4.Schmid BC, Oehler MK. New perspectives in ovarian cancer treatment. Maturitas. 2014;77:128–136. doi: 10.1016/j.maturitas.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y, Foster R, Yang X, Feng Y, Shen JK, Mankin HJ, Hornicek FJ, Amiji MM, Duan Z. Up-regulation of CD44 in the development of metastasis, recurrence and drug resistance of ovarian cancer. Oncotarget. 2015;6:9313–9326. doi: 10.18632/oncotarget.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen WT, Gao X, Han XD, Zheng H, Guo L, Lu RQ. HE4 as a serum biomarker for ROMA prediction and prognosis of epithelial ovarian cancer. Asian Pac J Cancer Prev. 2014;15:101–105. doi: 10.7314/APJCP.2014.15.1.101. [DOI] [PubMed] [Google Scholar]
- 7.Avril S, Dincer Y, Malinowsky K, Wolff C, Gündisch S, Hapfelmeier A, Boxberg M, Bronger H, Becker KF, Schmalfeldt B. Increased PDGFR-beta and VEGFR-2 protein levels are associated with resistance to platinum-based chemotherapy and adverse outcome of ovarian cancer patients. Oncotarget. 2017;8:97851–97861. doi: 10.18632/oncotarget.18415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng X, Muller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H. Circulating cell-free miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Adv Exp Med Biol. 2016;924:3–8. doi: 10.1007/978-3-319-42044-8_1. [DOI] [PubMed] [Google Scholar]
- 9.Koutsaki M, Libra M, Spandidos DA, Zaravinos A. The miR-200 family in ovarian cancer. Oncotarget. 2017;8:66629–66640. doi: 10.18632/oncotarget.18343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulitsky I. Evolution to the rescue: Using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 11.Luo P, Liu XF, Wang YC, Li ND, Liao SJ, Yu MX, Liang CZ, Tu JC. Prognostic value of abnormally expressed lncRNAs in ovarian carcinoma: A systematic review and meta-analysis. Oncotarget. 2017;8:23927–23936. doi: 10.18632/oncotarget.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, Xue YX. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. 2015;27:275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Yang K, Hou Y, Li A, Li Z, Wang W, Xie H, Rong Z, Lou G, Li K. Identification of a six-lncRNA signature associated with recurrence of ovarian cancer. Sci Rep. 2017;7:752. doi: 10.1038/s41598-017-00763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q, Cheng Y, Liang T, He Y, Ren C, Sun L, Zhang G. Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression. Sci Rep. 2015;5:17683. doi: 10.1038/srep17683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou M, Wang X, Shi H, Cheng L, Wang Z, Zhao H, Yang L, Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–12611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8:485–499. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- 17.Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ. The HUGO gene nomenclature database, 2006 updates. Nucleic Acids Res. 2006;34:D319–D321. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Lun ATL, Smyth GK. Differential expression analysis of complex RNA-seq experiments using edgeR. Springer International Publishing; Springer, Cham: 2014. pp. 51–74. [Google Scholar]
- 19.Yue S. Clinical trial data analysis using R. J Statistical Software. 2011;43 [Google Scholar]
- 20.Baur B, Bozdag S. A feature selection algorithm to compute gene centric methylation from probe level methylation data. PLoS One. 2016;11:e0148977. doi: 10.1371/journal.pone.0148977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavroforakis ME, Theodoridis S. A geometric approach to support vector machine (SVM) classification. IEEE Trans Neural Netw. 2006;17:671–682. doi: 10.1109/TNN.2006.873281. [DOI] [PubMed] [Google Scholar]
- 22.Jeggari A, Marks DS, Larsson E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chien J, Sicotte H, Fan JB, Humphray S, Cunningham JM, Kalli KR, Oberg AL, Hart SN, Li Y, Davila JI, et al. TP53 mutations, tetraploidy and homologous recombination repair defects in early stage high-grade serous ovarian cancer. Nucleic Acids Res. 2015;43:6945–6958. doi: 10.1093/nar/gkv111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M, Zhuang G, Sun X, Shen Y, Wang W, Li Q, Di W. TP53 mutation-mediated genomic instability induces the evolution of chemoresistance and recurrence in epithelial ovarian cancer. Diagn Pathol. 2017;12:16. doi: 10.1186/s13000-017-0605-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Báezvega PM, Vargas Echevarría IM, Valiyeva F, Encarnación-Rosado J, Roman A, Flores J, Marcos-Martínez MJ, Vivas-Mejía PE. Targeting miR-21-3p inhibits proliferation and invasion of ovarian cancer cells. Oncotarget. 2016;7:36321–36337. doi: 10.18632/oncotarget.9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon JY, Seo YR, Ahn WS. Recognition of potential predictive markers for diagnosis in Korean serous ovarian cancer patients at stage IIIc using array comparative genomic hybridization with high resolution. Mol Cell Toxicol. 2011;7:77–86. doi: 10.1007/s13273-011-0011-3. [DOI] [Google Scholar]
- 30.Bierkens M, Krijgsman O, Wilting SM, Bosch L, Jaspers A, Meijer GA, Meijer CJ, Snijders PJ, Ylstra B, Steenbergen RD. Focal aberrations indicate EYA2 and hsa-miR-375 as oncogene and tumor suppressor in cervical carcinogenesis. Genes Chromosomes Cancer. 2013;52:56–68. doi: 10.1002/gcc.22006. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, Zhang X, Bi T, Ding Y, Zhao J, Wang C, Jia T, Han D, Guo G, Wang B, et al. MiRNA expression signature for potentially predicting the prognosis of ovarian serous carcinoma. Tumour Biol. 2013;34:3501–3508. doi: 10.1007/s13277-013-0928-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Yan Z, Zhang J, Gong L, Li W, Cui J, Liu Y, Gao Z, Li J, Shen L, Lu Y. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol. 2011;22:2257–2266. doi: 10.1093/annonc/mdq758. [DOI] [PubMed] [Google Scholar]
- 33.Xu L, Li Q, Xu D, Wang Q, An Y, Du Q, Zhang J, Zhu Y, Miao Y. hsa-miR-141 downregulates TM4SF1 to inhibit pancreatic cancer cell invasion and migration. Int J Oncol. 2014;44:459–466. doi: 10.3892/ijo.2013.2189. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Zhu MJ, Ren AM, Wu HF, Han WM, Tan RY, Tu RQ. A ten-microRNA signature identified from a genome-wide microRNA expression profiling in human epithelial ovarian cancer. PLoS One. 2014;9:e96472. doi: 10.1371/journal.pone.0096472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waltering KK, Porkka KP, Jalava SE, Urbanucci A, Kohonen PJ, Latonen LM, Kallioniemi OP, Jenster G, Visakorpi T. Androgen regulation of micro-RNAs in prostate cancer. Prostate. 2011;71:604–614. doi: 10.1002/pros.21276. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol Oncol. 2011;121:200–205. doi: 10.1016/j.ygyno.2010.12.339. [DOI] [PubMed] [Google Scholar]
- 37.Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Park YT, Jeong JY, Lee MJ, Kim KI, Kim TH, Kwon YD, Lee C, Kim OJ, An HJ. MicroRNAs overexpressed in ovarian ALDH1-positive cells are associated with chemoresistance. J Ovarian Res. 2013;6:18. doi: 10.1186/1757-2215-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Hu S, Wang J, Cai J, Xiao L, Yu L, Wang Z. MiR-27a modulates MDR1/P-glycoprotein expression by targeting HIPK2 in human ovarian cancer cells. Gynecol Oncol. 2010;119:125–130. doi: 10.1016/j.ygyno.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Fuchs H, Theuser M, Wruck W, Adjaye J. miR-27 negatively regulates pluripotency-associated genes in human embryonal carcinoma cells. PLoS One. 2014;9:e111637. doi: 10.1371/journal.pone.0111637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halytskiy V. Shifts in microRNA expression pattern can facilitate the cancer cell stemness. Eur J Cancer. 2014;50:174. doi: 10.1016/S0959-8049(14)70662-8. [DOI] [Google Scholar]
- 42.Hylander B, Repasky E, Shrikant P, Intengan M, Beck A, Driscoll D, Singhal P, Lele S, Odunsi K. Expression of Wilms tumor gene (WT1) in epithelial ovarian cancer. Gynecol Oncol. 2006;101:12–17. doi: 10.1016/j.ygyno.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 43.Dallosso AR, Hancock AL, Malik S, Salpekar A, King-Underwood L, Pritchard-Jones K, Peters J, Moorwood K, Ward A, Malik KT, Brown KW. Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. RNA. 2007;13:2287–2299. doi: 10.1261/rna.562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, Mrózek K, Bucci D, Carroll AJ, Baer MR, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci USA. 2014;111:18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du T, Zhang B, Zhang S, Jiang X, Zheng P, Li J, Yan M, Zhu Z, Liu B. Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochim Biophys Acta. 2016;1862:12–19. doi: 10.1016/j.bbadis.2015.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author upon reasonable request.