Abstract
Craniofacial development requires a set of patterning codes that define the identities of postmigratory mesenchymal cells in a region-specific manner, in which locally expressed morphogens, including fibroblast growth factors (FGFs) and bone morphogenetic proteins (BMPs), provide instructive cues. Msx2, a bona fide target of BMP signaling, is a transcription factor regulating Runx2 and osterix (Osx), whose mutations are associated with cranial deformities in humans. Here we show that Msx2 defines osteo-chondro precursor cells in specific regions of the craniofacial mesenchyme at the postmigratory stage, particularly in the mandibular process and the posterior cranial vault. Analysis of Msx2-creER mice revealed that early mesenchymal cells in proximity to the BMP4-expressing mesenchyme were marked upon tamoxifen injection, and their descendants contributed to diverse types of mesenchymal cells in the later stage, such as chondrocytes and perichondrial cells of the transient cartilage, as well as osteoblasts and suture mesenchymal cells. By contrast, Osx-creER marked osteoblast precursors at the later stage, and their descendants continued to become osteoblasts well into the postnatal stage. Therefore, Msx2 marks spatially restricted populations of mesenchymal precursor cells with diverse differentiation potential, suggesting that extrinsic molecular cues can dictate the nature of postmigratory mesenchymal cells in craniofacial development.
Keywords: osteoblasts, maxillofacial development, cell differentiation, transcription factors, chondrocytes, cranial sutures
Introduction
Craniofacial development is a deliberate process requiring segmentation, migration, and colonization of neural crest– and mesoderm-derived mesenchymal cells into the primordium. These postmigratory cells are subsequently exposed to extrinsic molecular cues at their destinations so that they can evolve into regional precursor cells that later contribute to the bone, cartilage, and connective tissue in a region-specific manner (Minoux and Rijli 2010; Bhatt et al. 2013). Locally expressed morphogens/cytokines, including fibroblast growth factors (FGFs), bone morphogenetic proteins (BMPs), and hedgehog proteins (HHs), provide instructive cues to define the identity and the fate of these mesenchymal precursor cells in the adjacent region. Of these, ectodermally derived FGF8 and BMP4 induce expression of patterning code genes in the underlying mesenchyme (Shigetani et al. 2000). Signaling by the BMP pathway is particularly important in the morphogenesis and development of craniofacial structures, including the mandible and the midface (Stottmann et al. 2001; Komatsu et al. 2013; Hayano et al. 2015). Msx2 (muscle segment homeobox-like 2) is a bona fide target of BMP signaling (Brugger et al. 2004) and regulates the expression of Runx2 and Osx, which are the essential transcription factors for osteoblast differentiation (Lee et al. 2005; Hassan et al. 2006; Matsubara et al. 2008). Msx2 is transiently expressed in postmigratory mesenchymal cells in cranial neural crest–derived tissues and demonstrates the antagonism for Sox9 and chondrogenesis (Semba et al. 2000). In addition, mutations in MSX2 cause Boston-type craniosynostosis (CRS2, OMIM: 604757) (Jabs et al. 1993) and parietal foramina (PFM1, OMIM: 168500) (Wilkie et al. 2000), indicating that Msx2 plays an important role in the craniofacial osteoblast lineage.
In this study, we undertook a comprehensive lineage-tracing approach to reveal cell fates of Msx2-expressing early mesenchymal cells. We generated Msx2-creER bacterial artificial chromosome (BAC) transgenic mice for in vivo lineage-tracing experiments. We embedded a creER transgene within a ~180-kb BAC to recapitulate endogenous expression patterns of Msx2, in contrast to a previously reported line using a 0.5-kb promoter fragment (Kimmel et al. 2000). Moreover, the use of the cre-loxP system circumvents problems associated with the Tet system, including cytotoxicity and promiscuous target gene expression (Lin et al. 2009). In addition, we also took advantage of Osx-creER BAC transgenic mice (Maes et al. 2010) to trace fates of osteoblast precursors to highlight their difference with those of earlier mesenchymal cells. This approach revealed long-term fates of lineage-committed osteoblasts precursors, which had been impossible with previous DiI labeling experiments (Yoshida et al. 2008). Our findings collectively revealed that Msx2 defines osteo-chondro precursor cells in specific regions of the craniofacial mesenchyme at the postmigratory stage, particularly in the mandibular process and the posterior cranial vault, indicating that extrinsic molecular cues locally specify the nature of adjacent mesenchymal cells in a spatially restricted manner during craniofacial development.
Materials and Methods
Mice
Msx2-creER-SV40pA transgenic mice were generated by BAC transgenesis. Osteocalcin (Oc)-GFP (JAX13134) and Osx-creERT2 (Maes et al. 2010) mice have been described elsewhere. Rosa26-CAG-loxP-stop-loxP-tdTomato (Ai14: R26R-tdTomato, JAX7914) mice were acquired from the Jackson Laboratory. All procedures were conducted in compliance with the Guidelines for the Care and Use of Laboratory Animals approved by the University of Michigan’s Institutional Animal Care and Use Committee (IACUC), protocol PRO00005703 (Ono) and the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. For all experiments, male breeder mice (Msx2-creER or Osx-creER; R26RtdTomato/tdTomato) were mated to female CD1 mice (>8 wk old; Charles River Laboratories 022), and the vaginal plug was checked in the morning. Pregnant mice received 4 mg tamoxifen (Sigma T5648) and 4 mg progesterone (Sigma P3972) intraperitoneally at an indicated embryonic day in a home cage. Embryos or pups were used for analysis regardless of the sex. At least 3 independent biological samples of the indicated genotype were examined for each data shown in the figures.
Histology and Immunohistochemistry
Samples were fixed in 4% paraformaldehyde for a proper period, typically ranging from 3 h to overnight at 4°C , then decalcified in 15% ethylenediaminetetraacetic acid (EDTA) for a proper period, typically ranging from 3 h to 14 d. Decalcified samples were cryoprotected in 30% sucrose/phosphate-buffered saline (PBS) solutions and then in 30% sucrose/PBS:OCT (1:1) solutions, each at least overnight at 4°C. Samples were embedded in an OCT compound (Tissue-Tek; Sakura). Frozen sections at 15 to 25 µm thickness were prepared using a cryostat (Leica CM1850) and adhered on positively charged glass slides (Fisherbrand ColorFrost Plus) and stained with anti-Sox9 polyclonal antibody (1:1,000; EMD-Millipore, AB5535), anti–osteopontin (OPN) polyclonal antibody (1:500; R&D, AF808), anti–periostin (POSTN) polyclonal antibody (1:2,000; EMD-Millipore, ABT280), rabbit anti–type I collagen (COL1) polyclonal antibody (1:500; Cederlane, CL50151AP), sheep anti–dentin matrix protein 1 (DMP1) polyclonal antibody (1:100; R&D, AF4386), goat anti–osteocalcin (OCN) polyclonal antibody (1:200; AbD Serotec/Bio-Rad, 7060-1815), and/or anti–E-cadherin (E-cad) monoclonal antibody (1:200; Abcam, ab11512) overnight at 4°C. Sections were subsequently stained with Alexa Fluor 488–conjugated donkey anti–rabbit Immunoglobulin G (IgG) (A21206), Alexa Fluor 647–conjugated donkey anti–goat IgG (A21082), or goat anti–rat IgG (A21094) (1:400; Invitrogen) for 3 h at 4°C, followed by DAPI (4′,6-diamidino-2-phenylindole, 5 µg/mL; Invitrogen D1306) staining. Sections were analyzed using an automated inverted fluorescence microscope with a structured illumination system (Zeiss Axio Observer Z1 with ApoTome.2 system). Images were typically tile-scanned with a motorized stage, Z-stacked, and reconstructed by a maximum-intensity projection (MIP) function. Differential interference contrast (DIC) was used for objectives higher than ×10. More detailed experimental procedures are available in the Appendix Materials and Methods.
RNAscope In Situ Hybridization
Samples were fixed in 4% paraformaldehyde for 3 h at 4°C and cryoprotected. Frozen sections at 15 µm were prepared on positively charged glass slides. In situ hybridization was performed with RNAscope 2.5 HD Reagent kit Brown (Advanced Cell Diagnostics 322300) using the following probes: Msx2 (421851), Bmp4 (401301), and Fgf8 (313411) according to the manufacturer’s protocol.
Statistical Analysis
Results are presented as mean values ± SD. Statistical evaluation was conducted using the unpaired t test. A P value of <0.05 was considered significant.
Results
Generation and Characterization of Msx2-creER BAC Transgenic Mice
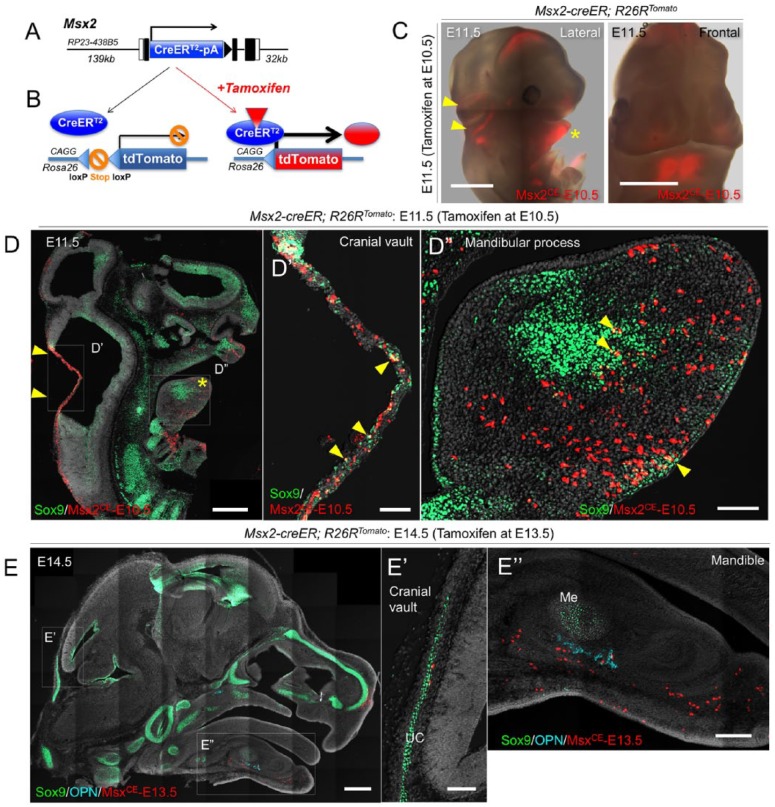
To study the fate of Msx2+ cells in vivo, we generated tamoxifen-inducible Msx2-creER BAC transgenic mice (Fig. 1A and Appendix Fig. 1A). We established 2 transgenic lines (Line1 and Line2) from 2 independent founders. Quantitative polymerase chain reaction (PCR) analysis on tail biopsies revealed that the Line2 expressed the creER gene at a significantly high level (~60-fold higher than Line1) (Appendix Fig. 1B). We further crossed these lines with an R26R-tdTomato reporter allele (Madisen et al. 2010) to test if recombination faithfully occurs in a tamoxifen-dependent manner (Fig. 1B). Importantly, virtually no tdTomato expression was observed without tamoxifen in both lines of Msx2-creER; R26R-tdTomato mice (Appendix Fig. 1C). Upon tamoxifen injection, the Line2 marked a great majority, whereas the Line1 marked only a small portion of hair follicles by tdTomato (Appendix Fig. 1C), indicating the Line2 induced recombination efficiently. We therefore focused on the Line2 for our subsequent analysis. We further took a closer look at early limb bud development to validate activities of the Line2. Msx2 is expressed both in the apical ectodermal ridge (AER) and in its underlying mesenchyme (Ferrari et al. 1998), whereas the 0.5-kb Msx2 promoter is specifically active in the AER (Kimmel et al. 2000). Recombination occurred both in the AER and its underlying mesenchyme at E11.5 upon tamoxifen injection at E10.5 (Appendix Fig. 1D). Therefore, the activity of the Msx2-creER BAC transgenic line that we have established here (Line2) seems to accurately reflect endogenous expression patterns of Msx2.
Figure 1.
Msx2-creER marks early mesenchymal cells in the mandibular process and cranial vault primordium. (A) Structure of the Msx2-creER-SV40pA bacterial artificial chromosome (BAC) transgene. Kozak-Msx2-creER-SV40pA-frt-NeoR-frt cassette containing ~300-bp homology arms was recombineered into BAC clone RP23-438B5 containing 139-kb upstream and 32-kb downstream genomic sequences of the Msx2 gene. (B) Diagram of lineage-tracing experiments based on a tamoxifen-inducible creER system. CreER recombinase is expressed by Msx2 promoter/enhancers. Left: in the absence of tamoxifen, creER is sequestered in the cytoplasm. Right: in the presence of tamoxifen, creER translocates to the nucleus and excises the stop sequences in the Rosa26 locus. Upon successful recombination, targeted cells permanently express tdTomato in a ubiquitously active CAG promoter-dependent manner. (C) Whole-mount image of Msx2-creER; R26RTomato mice at E11.5 (tamoxifen at E10.5). Left: lateral view; right: front view of the head. Arrowheads: tdTomato+ domain in the cranial vault primordium; asterisk: tdTomato+ domain in the mandibular process. Red: tdTomato. Scale bars: 1 mm. (D–D′′) Sagittal section of Msx2-creER; R26RTomato mice at E11.5 (tamoxifen at E10.5) stained for Sox9 and nuclei. D′: cranial vault; D′′: mandibular process. Arrowheads: Sox9+tdTomato+ cells. Green: Sox9-Alexa488; red: tdTomato; gray: 4′,6-diamidino-2-phenylindole (DAPI) and differential interference contrast (DIC). Scale bars: 500 µm (left panel) and 100 µm (center and right panel). (E–E′′) Sagittal section of Msx2-creER; R26RTomato mice at E14.5 (tamoxifen at E13.5) stained for Sox9, osteopontin (OPN), and nuclei. E′: cranial vault; E′′: mandibular process. UC, underlying cartilage. Light blue: OPN-Alexa647; green: Sox9-Alexa488; red: tdTomato; gray: DAPI and DIC. Scale bars: 500 µm (left panel), 100 µm (center panel), and 200 µm (right panel).
Msx2-creER Marks Early Mesenchymal Cells in the Mandibular Process and Cranial Vault Primordium
Next, we set out to map early mesenchymal cells that can be marked by Msx2-creER. For this purpose, we injected tamoxifen into Msx2-creER; R26R-tdTomato mice at embryonic day 10.5 (E10.5) and analyzed these mice after 24 h at E11.5, when postmigratory neural crest cells had already populated the primordium. Analysis of whole-mount embryos revealed robust tdTomato signal in the mandibular process of the first pharyngeal arch and in the cranial vault primordium (Fig. 1C). In both the cranial vault and the mandibular process, a small fraction of Msx2-creER–marked cells were positive for Sox9 (Fig. 1D′, D′′, arrowheads), while a majority of them were negative for Sox9. These Msx2-creER–marked cells were preferentially distributed in proximity to the Bmp4+ domain of the mesenchyme but were away from the Fgf8+ domain of the ectoderm (Appendix Fig. 2), as suggested from the previous study demonstrating ready responsiveness of the Msx2 promoter to BMP4 (Brugger et al. 2004). As expected, Msx2-creER–marked cells expressed Msx2 (Appendix Fig. 2). In addition, Msx2-creER–marked cells were also found in the ectoderm, as exemplified by positive staining for E-cadherin (Appendix Fig. 3B, arrows). Therefore, Msx2-creER can mark early cranial and mandibular postmigratory mesenchymal cells during a morphogenesis phase upon tamoxifen injection.
To further determine how the Msx2-creER activity changes over the course of development, we injected tamoxifen into Msx2-creER; R26R-tdTomato mice at later stages and analyzed these mice at 24 h after the pulse. An E11.5 pulse marked mesenchymal cells in the mandible and in the cranial vault, in a similar pattern to those arose from an E10.5 pulse (Appendix Fig. 3A, left panel). On coronal sections, tdTomato+ mesenchymal cells were particularly localized in proximity to the midline of the mandibular process (Appendix Fig. 3A, right panel). By contrast, an E13.5 pulse marked significantly fewer cells in the mandible and virtually no cells in the cranial vault (Fig. 1E, E′′), indicating that mesenchymal cells that can be marked by Msx2-creER dwindle prior to this stage. An E17.5 pulse marked dental papilla cells of the incisor (Appendix Fig. 3C′′, arrows) and whisker follicle cells (Appendix Fig. 3C′, arrowheads); however, other mesenchymal cells in the mandible and the calvaria were not labeled. We also injected tamoxifen into Msx2-creER; R26R-tdTomato mice at postnatal day 1 (P1) and analyzed these mice at P4. As a 5.2-kb Msx2-LacZ transgene is expressed in the calvarial suture (Liu et al. 1995), we expected that Msx2-creER could induce recombination in suture mesenchymal cells upon tamoxifen injection. Surprisingly, no tdTomato+ cells were observed in any portion of the calvaria, including the suture mesenchyme (Appendix Fig. 3D). In contrast, abundant tdTomato+ cells were observed in the dental papilla of molars (Appendix Fig. 3E), consistent with the previous study demonstrating Msx2 expression in the dental pulp (Yamashiro et al. 2003). Therefore, Msx2-creER is transiently expressed by postmigratory mesenchymal cells of the mandible and the posterior calvaria during an early phase of craniofacial development around E10.5 and E11.5 and becomes more restricted to dental papilla/pulp cells at the late fetal and postnatal stage.
Msx2-creER+ Cells Become Diverse Types of Mesenchymal Cells in Fetal Development
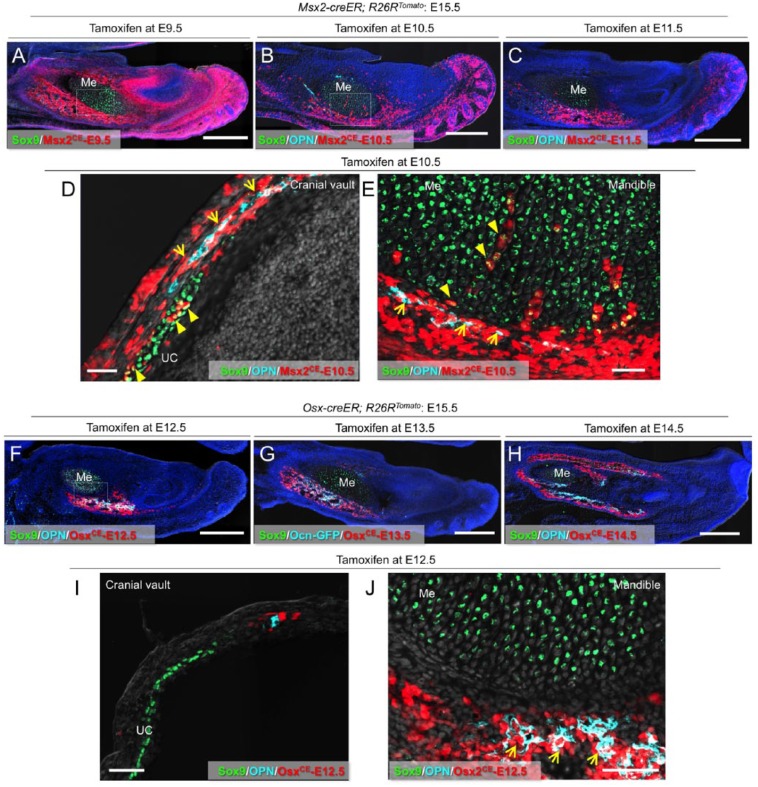
To define in vivo fates of Msx2-creER+ early mesenchymal cells during fetal craniofacial development, we analyzed Msx2-creER; R26R-tdTomato mice at E15.5 after being pulsed at various preceding time points (see the diagram in Appendix Fig. 4A). Cells marked by an E9.5 pulse to Msx2-creER; R26R-tdTomato mice (Msx2CE-E9.5 cells) contributed broadly to mesenchymal cells, including Sox9+ chondrocytes and perichondrial cells (Fig. 2A and Appendix Fig. 4B). In the mandible, Msx2CE-E9.5 cells on the innermost layer of the perichondrium of Meckel’s cartilage were positive for Sox9 (Appendix Fig. 4C′, arrowheads), whereas those on the outer layer of the perichondrium were positive for OPN, an extracellular matrix protein expressed by osteoblasts (Appendix Fig. 4C′′, arrows). In the posterior cranial vault, Msx2CE-E9.5 cells contributed to perichondrial cells and Sox9+ chondrocytes of the transient underlying cartilage (Appendix Fig. 4D, E). Cells marked by an E10.5 pulse (Msx2CE-E10.5 cells, as shown in Fig. 1C, D) contributed to E15.5 mesenchymal cells in a similar but more restricted pattern. Msx2CE-E10.5 cells became perichondrial cells and Sox9+ chondrocytes of the transient underlying cartilage (Fig. 2D, E, arrowheads) as well as OPN+ osteoblasts (Fig. 2D, E, arrows; see also Appendix Fig. 4F) in the mandible and the posterior cranial vault. A tamoxifen pulse at E11.5 resulted in further restricted domains of tdTomato+ cells toward the anterior portion in the mandible at E15.5 (Fig. 2C), while fewer tdTomato+ cells were observed in the posterior cranial vault (Appendix Fig. 4G). Therefore, Msx2-creER marks early mesenchymal precursors for diverse cell types such as perichondrial cells and chondrocytes at the late embryonic stage of E9.5 and E10.5, which contribute significantly to fetal craniofacial development. By contrast, at the fetal stage of E14.5 and later, Msx2-creER predominantly marks dental papilla cells of the incisor while failing to mark any mesenchymal cells in the cranial vault.
Figure 2.
Msx2-creER+ cells become diverse types of mesenchymal cells in fetal development. (A) Sagittal sections of Msx2-creER; R26RTomato mandibles at E15.5 (tamoxifen at E9.5) stained for Sox9 and nuclei. Me, Meckel’s cartilage. Green: Sox9-Alexa488; red: tdTomato; blue: 4′,6-diamidino-2-phenylindole (DAPI); gray: differential interference contrast (DIC). Scale bars: 400 µm. (B) Sagittal sections of Msx2-creER; R26RTomato mandibles at E15.5 (tamoxifen at E10.5) stained for Sox9, osteopontin (OPN), and nuclei. Me, Meckel’s cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; blue: DAPI; gray: DIC. Scale bars: 400 µm. (C) Sagittal sections of Msx2-creER; R26RTomato mandibles at E15.5 (tamoxifen at E11.5) stained for Sox9, OPN, and nuclei. Me, Meckel’s cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; blue: DAPI; gray: DIC. Scale bars: 400 µm. (D) Sagittal sections of Msx2-creER; R26RTomato cranial vault at E15.5 (tamoxifen at E10.5) stained for Sox9, OPN, and nuclei. Arrows: OPN+tdTomato+ osteoblasts; arrowheads: Sox9+tdTomato+ chondrocytes. UC, underlying cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 50 µm. (E) Sagittal sections of Msx2-creER; R26RTomato mandible at E15.5 (tamoxifen at E10.5) stained for Sox9, OPN, and nuclei. Magnified view of the dotted area in (B). Me, Meckel’s cartilage. Arrows: OPN+tdTomato+ osteoblasts; arrowheads: Sox9+tdTomato+ chondrocytes and perichondrial cells. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 50 µm. (F) Sagittal sections of Osx-creER; R26RTomato mandibles at E15.5 (tamoxifen at E12.5) stained for Sox9, OPN, and nuclei. Arrows: OPN+tdTomato+ osteoblasts. Me, Meckel’s cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; blue: DAPI; gray: DIC. Scale bars: 400 µm. (G) Sagittal sections of Ocn-GFP; Osx-creER; R26RTomato mandibles at E15.5 (tamoxifen at E13.5) stained for Sox9 and nuclei. Me, Meckel’s cartilage. Light blue: Ocn-GFP; green: Sox9–Alexa 647; red: tdTomato; blue: DAPI; gray: DIC. Scale bars: 400 µm. (H) Sagittal sections of Osx-creER; R26RTomato mandibles at E15.5 (tamoxifen at E14.5) stained for Sox9, OPN, and nuclei. Me, Meckel’s cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 647; red: tdTomato; blue: DAPI; gray: DIC. Scale bars: 400 µm. (I) Sagittal sections of Osx-creER; R26RTomato cranial vault at E15.5 (tamoxifen at E12.5) stained for Sox9, OPN, and nuclei. UC, underlying cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 50 µm. (J) Sagittal sections of Osx-creER; R26RTomato mandibles at E15.5 (tamoxifen at E12.5) stained for Sox9, OPN, and nuclei. Magnified view of the dotted area in (F). Me, Meckel’s cartilage. Arrows: OPN+tdTomato+ osteoblasts. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 50 µm.
Osx-creER Marks Perichondrial Cells of the Transient Cartilage in the Fetal Stage
Osx-creER marks osteoblast precursors in the fetal osteogenic perichondrium (Maes et al. 2010), which have the capability to translocate into the marrow cavity and become osteoblasts and stromal cells. First, we set out to define the timing that Osx-creER+ precursors appear during craniofacial development. No tdTomato+ cells were observed in Osx-creER; R26R-tdTomato mice at E11.5 upon a tamoxifen pulse at E10.5 (Appendix Fig. 5A). However, a tamoxifen pulse at E12.5 marked a small group of tdTomato+ cells associated with the Meckel’s cartilage in the mandible (Appendix Fig. 5B, arrowheads), indicating that Osx-creER marks mesenchymal cells in the mandible at a later developmental stage than Msx2-creER does. Second, we analyzed Osx-creER; R26R-tdTomato mice at E15.5 after being pulsed at various preceding time points. In the mandible, cells marked by an E12.5 pulse to Osx-creER; R26R-tdTomato mice (OsxCE-E12.5 cells) became Sox9-negative perichondrial cells on the anterior-inferior portion of Meckel’s cartilage, some of which were positive for OPN (Fig. 2F, J). In the posterior cranial vault, OsxCE- E12.5 cells were found in a restricted domain of the perichondrium, without contributing to chondrocytes in the transient cartilage (Fig. 2I and Appendix Fig. 5C). Cells marked by a later pulse (E13.5 and E14.5) contributed to E15.5 mesenchymal cells in a broader domain. In the mandible, the superior-posterior portion of the perichondrium of Meckel’s cartilage was labeled (Fig. 2G, H and Appendix Fig. 5E), whereas in the cranial vault, a superior domain of the perichondrium was labeled (Appendix Fig. 5D, arrows). The expansion of the E15.5 tdTomato+ domain in E13.5/E14.5-pulsed mice implies that these cells started to express Osx-creER at a time later than E12.5, suggesting the existence of earlier mesenchymal cells contributing to Osx+ cells in these regions. As demonstrated above, descendants of Msx2-creER+ cells marked at an earlier day contributed to these mesenchymal cells. These findings suggest that Osx-creER marks perichondrial cells associated with the transient cartilage of the craniofacial complex, and Msx2-creER marks at least some of their precursors at an earlier stage.
Msx2-creER+ Early Mesenchymal Cells Exhibit Broader Differentiation Potential Than That of Osx-creER+ Osteoblast Precursors
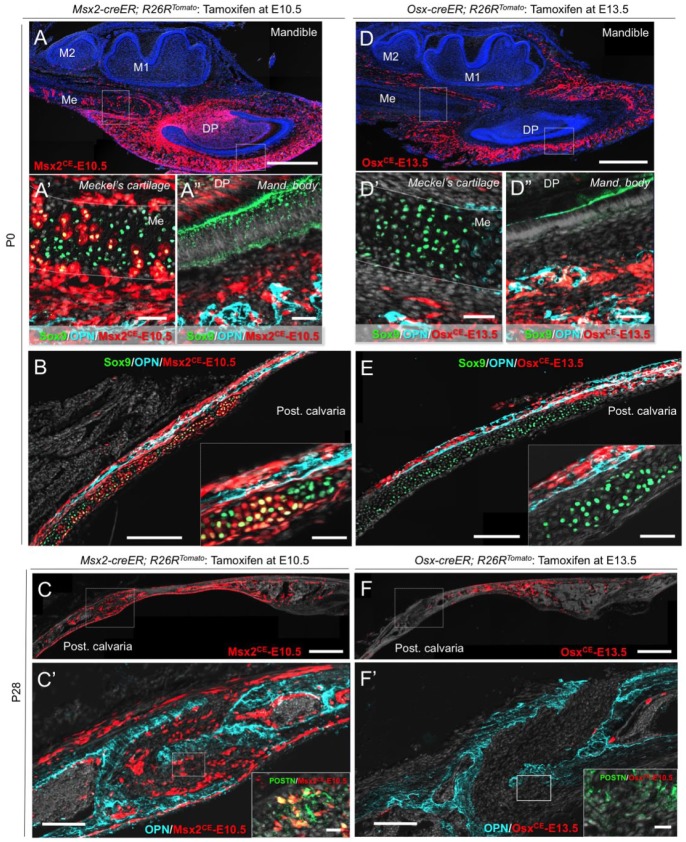
We next set out to test the hypothesis that Msx2+ early mesenchymal cells exhibit broader differentiation potential than that of Osx+ osteoblast precursors during postnatal craniofacial development. For this purpose, we pulsed Msx2-creER; R26R-tdTomato and Osx-creER; R26R-tdTomato mice at E10.5 and E13.5, respectively, to trace the fates of Msx2CE-E10.5 and OsxCE-E13.5 cells into postnatal days. Msx2CE-E10.5 cells contributed to Sox9+ chondrocytes in Meckel’s cartilage and OPN+ osteoblasts in surrounding bones, especially in the mandibular body at postnatal day 0 (P0) (Fig. 3A–A′′). Similarly, Msx2CE-E10.5 cells contributed to Sox9+ chondrocytes and perichondrial cells of the underlying cartilage and surrounding OPN+ osteoblasts, especially in the posterior portion of the calvaria (Fig. 3B). When transient cartilages had already disappeared at P21, Msx2CE-E10.5 cells continued to become OPN+ osteoblasts in the mandibular body (Appendix Fig. 6A). Msx2CE-E10.5 cells also became COL1+, DMP1+, and OCN+ osteoblasts (Appendix Fig. 6C), as well as bone marrow stromal cells in the marrow cavity (Appendix Fig. 6A, asterisk). In the posterior calvaria, Msx2CE-E10.5 cells became OPN+ osteoblasts and POSTN-positive suture mesenchymal cells of the lambdoid suture (Fig. 3C–C′). OsxCE- E13.5 cells contributed to OPN+ osteoblasts and osteocytes throughout the mandible at P0 (Fig. 3D–D′) and continued to provide these cells at P21 (Appendix Fig. 6B). In the calvaria, OsxCE-E13.5 cells robustly contributed to OPN+ osteoblasts and perichondrial cells but not to Sox9+ chondrocytes in the underlying cartilage at P0 (Fig. 3E). OsxCE-E13.5 perichondrial cells invaded into the transient cartilage and occupied the OPN+ ossification center, in a pattern similar to what Osx+ perichondrial precursors do in fetal endochondral bones (Appendix Fig. 6D, arrowheads; Maes et al. 2010). OsxCE-E13.5 cells continued to become OPN+ osteoblasts and osteocytes in the calvaria at P21 but not to POSTN+ suture mesenchymal cells (Fig. 3F–F′). Quantification of tdTomato+ cells revealed that Msx2CE-E10.5 cells contributed to a significantly higher number of Sox9+ chondrocytes in the calvarial transient cartilage at P0 than OsxCE-E13.5 cells did (tdTomato+Sox9+ cells: Msx2CE-E10.5; 1360 ± 149.3 cells, OsxCE-E13.5; 81.7 ± 51.2 cells per 250-µm thickness, n = 3 per group, P < 0.05, unpaired t test). At the postgrowth stage (P21–P28), Msx2CE-E10.5 cells contributed to a large number of POSTN+ suture mesenchymal cells, while virtually no OsxCE-E13.5 cells became these cells in the posterior calvaria (tdTomato+POSTN+ cells: Msx2CE-E10.5; 1555 ± 1252 cells, OsxCE-E13.5; 4.3 ± 1.2 cells per 250-µm thickness, n = 3 per group). Msx2CE-E10.5 contributed to OPN+ osteoblasts of the parietal bones to a larger extent than OsxCE-E13.5 cells did (tdTomato+OPN+ cells: Msx2CE-E10.5; 2026 ± 1410 cells, OsxCE-E13.5; 639.3 ± 72.1 cells per 250-µm thickness, n = 3 per group), although the difference was not statistically significant. These findings suggest that Msx2-creER marks spatially restricted populations of early mesenchymal cells with diverse differentiation potential to osteoblasts, chondrocytes, and suture mesenchymal cells, whereas Osx-creER marks lineage-committed precursors that can continue to provide osteoblasts well into postnatal craniofacial development (Fig. 4A, B).
Figure 3.
Msx2-creER+ early mesenchymal cells exhibit broader differentiation potential than Osx-creER+ osteoblast precursors. (A–A′′) Sagittal sections of Msx2-creER; R26RTomato mandibles at P0 (tamoxifen at E10.5) stained for Sox9, osteopontin (OPN), and nuclei. A′: Meckel’s cartilage; A′′: mandibular body. DP, dental pulp; M1, mandibular first molar; M2, mandibular second molar; Me, Meckel’s cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: 4′,6-diamidino-2-phenylindole (DAPI) and differential interference contrast (DIC). Scale bars: 500 µm (A) and 50 µm (A′–A′′). (B) Sagittal sections of Msx2-creER; R26RTomato posterior cranial vault at P0 (tamoxifen at E10.5) stained for Sox9, OPN, and nuclei. Inset: magnified view of the transitional area. UC, underlying cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 200 µm and 50 µm (inset). (C–C′) Sagittal sections of Msx2-creER; R26RTomato posterior cranial vault at P28 (tamoxifen at E10.5) stained for OPN, periostin (POSTN), and nuclei. Inset: magnified view of the dotted area of the suture. Light blue: OPN–Alexa 647; green: POSTN–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 500 µm and 20 µm (inset). (D–D′′) Sagittal sections of Osx-creER; R26RTomato mandibles at P0 (tamoxifen at E13.5) stained for Sox9, OPN, and nuclei. DP, dental pulp; M1, mandibular first molar; M2, mandibular second molar; Me, Meckel’s cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 500 µm (D) and 50 µm (D′–D′′). (E) Sagittal sections of Osx-creER; R26RTomato posterior cranial vault at P0 (tamoxifen at E13.5) stained for Sox9, OPN, and nuclei. Inset: magnified view of the transitional area. UC, underlying cartilage. Light blue: OPN–Alexa 647; green: Sox9–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 200 µm and 50 µm (inset). (F–F′) Sagittal sections of Osx-creER; R26RTomato posterior cranial vault at P28 (tamoxifen at E13.5) stained for OPN, POSTN, and nuclei. Inset: magnified view of the dotted area of the suture. Light blue: OPN–Alexa 647; green: POSTN–Alexa 488; red: tdTomato; gray: DAPI and DIC. Scale bars: 500 µm and 20 µm (inset).
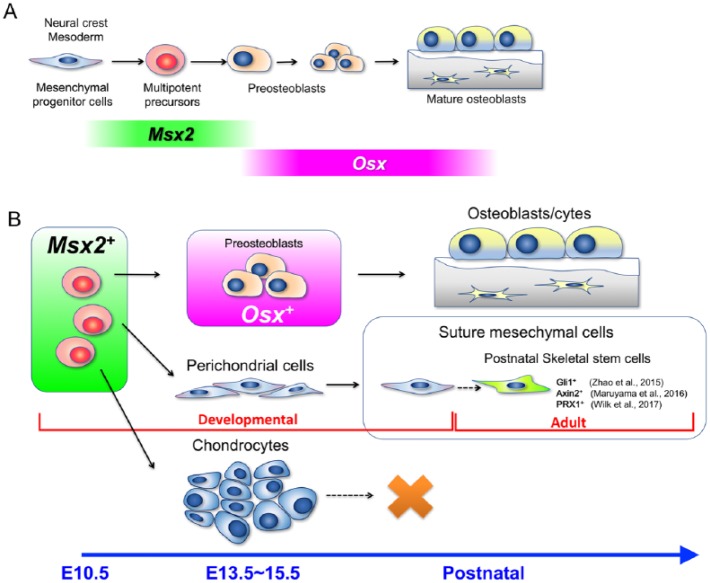
Figure 4.
Msx2 marks early mesenchymal precursors in craniofacial bone development. (A) Proposed model for craniofacial skeletal cell lineage development. Regardless of the origin of mesenchymal stem cells in the mesoderm or neural crest, Msx2 is transiently expressed by multipotent mesenchymal precursor cells at specific portions of the craniofacial primordium, such as the mandibular process and the posterior cranial vault. Osx starts to be expressed by lineage-restricted osteoblast precursors only after Msx2 is no longer expressed in the lineage. (B) Msx2-expressing early mesenchymal cells possess broad differentiation potential into diverse cell types during development, such as preosteoblasts, perichondrial cells, and chondrocytes of the transient cartilage of the craniofacial complex. Although these chondrocytes eventually disappear during postnatal growth, Msx2-derived perichondrial cells can further become suture mesenchymal cells. The cranial suture is a known niche for postnatal skeletal stem cells. Thus, Msx2-expressing early mesenchymal cells are likely to be one of the precursors for these adult skeletal stem cells.
Discussion
Our data collectively indicate that Msx2 functions as a craniofacial patterning code gene that defines osteo-chondro precursor cells specifically in the mandibular process and the posterior cranial vault at the postmigratory stage. Msx2 expression is induced by BMP4 derived from the specific domain of the ectoderm and the mesenchyme, indicating that extrinsic molecular cues can dictate the nature of postmigratory mesenchymal cells in craniofacial development. Msx2 is a bona fide target of BMP signaling, containing consensus binding sites for BMP-restricted Smads (Brugger et al. 2004). Consistent with the well-described function of BMPs that robustly induce osteoblast differentiation of undifferentiated mesenchymal cells, our data demonstrate that Msx2+ cells are endowed with the ability to become osteoblasts and chondrocytes. One hypothesis is that Msx2 determines the cell lineage of early mesenchymal cells based on its function as a transcription factor that regulates Runx2 and Osx expression (Lee et al. 2005; Hassan et al. 2006; Matsubara et al. 2008). Intriguingly, Msx2 expression becomes progressively restricted during development, while Osx becomes widely expressed by lineage-restricted osteoblast precursors and mature osteoblasts in later stages. Therefore, we speculate that Msx2 maintains broad differentiation potential and certain self-renewability of the population, although the more detailed function of Msx2 needs to be investigated by further experimentation.
We discovered that some of suture mesenchymal cells were derived from Msx2-creER–marked early mesenchymal cells. Interestingly, it was only until P28 before Msx2-creER+ descendants made a discernable contribution to suture mesenchymal cells (Fig. 3C). Therefore, suture mesenchymal cells might not be the principal source of cranial osteoblasts, at least during active calvarial bone growth during the fetal and early postnatal stage. The calvarial suture in adult mice hosts mesenchymal stem cells that contribute to a much slower remodeling process and response to injury (Zhao et al. 2015; Maruyama et al. 2016; Wilk et al. 2017). These findings indicate that some of adult mesenchymal stem cells in the cranial suture might uniquely develop from Msx2+ early mesenchymal precursors.
In conclusion, our study provides the first evidence that Msx2 marks spatially restricted populations of early mesenchymal precursors during a morphogenesis phase. Further studies are needed to reveal more detailed molecular mechanisms governing the specification of diverse mesenchymal precursor cells in craniofacial development.
Author Contributions
N. Sakagami, S. Syklawer-Howle, contributed to conception, design, data acquisition, analysis, and interpretation, drafted the manuscript; Y. Matsushita, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript; H.M. Kronenberg, W. Ono, contributed to conception and design, critically revised the manuscript; N. Ono, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034518771014 for Msx2 Marks Spatially Restricted Populations of Mesenchymal Precursors by N. Sakagami, Y. Matsushita, S. Syklawer-Howle, H.M. Kronenberg, W. Ono, and N. Ono in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported by National Institutes of Health grants DE026666 and DE022564 to N.O., University of Michigan MCubed 2.0 Grant to N.O. and W.O., and Non-traditional Dean’s Fellowship in Organogenesis to N.S.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: W. Ono  https://orcid.org/0000-0002-0358-1897
https://orcid.org/0000-0002-0358-1897
References
- Bhatt S, Diaz R, Trainor PA. 2013. Signals and switches in mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol. 5(2). pii: a008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger SM, Merrill AE, Torres-Vazquez J, Wu N, Ting MC, Cho JY, Dobias SL, Yi SE, Lyons K, Bell JR, et al. 2004. A phylogenetically conserved cis-regulatory module in the Msx2 promoter is sufficient for BMP-dependent transcription in murine and drosophila embryos. Development. 131(20):5153–5165. [DOI] [PubMed] [Google Scholar]
- Ferrari D, Lichtler AC, Pan ZZ, Dealy CN, Upholt WB, Kosher RA. 1998. Ectopic expression of Msx-2 in posterior limb bud mesoderm impairs limb morphogenesis while inducing BMP-4 expression, inhibiting cell proliferation, and promoting apoptosis. Dev Biol. 197(1):12–24. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. 2006. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 281(52):40515–40526. [DOI] [PubMed] [Google Scholar]
- Hayano S, Komatsu Y, Pan H, Mishina Y. 2015. Augmented BMP signaling in the neural crest inhibits nasal cartilage morphogenesis by inducing p53-mediated apoptosis. Development. 142(7):1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs EW, Müller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB. 1993. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 75(3):443–450. [DOI] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. 2000. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 14(11):1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Komatsu Y, Yu PB, Kamiya N, Pan H, Fukuda T, Scott GJ, Ray MK, Yamamura K, Mishina Y. 2013. Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J Bone Miner Res. 28(6):1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS, Wozney JM, Chi XZ, Bae SC, Choi KY, et al. 2005. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J Biol Chem. 280(42):35579–35587. [DOI] [PubMed] [Google Scholar]
- Lin C, Yin Y, Chen H, Fisher AV, Chen F, Rauchman M, Ma L. 2009. Construction and characterization of a doxycycline-inducible transgenic system in Msx2 expressing cells. Genesis. 47(5):352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Kundu R, Wu L, Luo W, Ignelzi MA, Snead ML, Maxson RE. 1995. Premature suture closure and ectopic cranial bone in mice expressing Msx2 transgenes in the developing skull. Proc Natl Acad Sci USA. 92(13):6137–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13(1):133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM. 2010. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 19(2):329–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Jeong J, Sheu TJ, Hsu W. 2016. Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nat Commun. 7:10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. 2008. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 283(43):29119–29125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. 2010. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 137(16):2605–2621. [DOI] [PubMed] [Google Scholar]
- Semba I, Nonaka K, Takahashi I, Takahashi K, Dashner R, Shum L, Nuckolls GH, Slavkin HC. 2000. Positionally-dependent chondrogenesis induced by BMP4 is co-regulated by Sox9 and Msx2. Dev Dyn. 217(4):401–414. [DOI] [PubMed] [Google Scholar]
- Shigetani Y, Nobusada Y, Kuratani S. 2000. Ectodermally derived FGF8 defines the maxillomandibular region in the early chick embryo: epithelial-mesenchymal interactions in the specification of the craniofacial ectomesenchyme. Dev Biol. 228(1):73–85. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Anderson RM, Klingensmith J. 2001. The BMP antagonists Chordin and Noggin have essential but redundant roles in mouse mandibular outgrowth. Dev Biol. 240(2):457–473. [DOI] [PubMed] [Google Scholar]
- Wilk K, Yeh SA, Mortensen LJ, Ghaffarigarakani S, Lombardo CM, Bassir SH, Aldawood ZA, Lin CP, Intini G. 2017. Postnatal calvarial skeletal stem cells expressing PRX1 reside exclusively in the calvarial sutures and are required for bone regeneration. Stem Cell Reports. 8(4):933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO, Tang Z, Elanko N, Walsh S, Twigg SR, Hurst JA, Wall SA, Chrzanowska KH, Maxson RE. 2000. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat Genet. 24(4):387–390. [DOI] [PubMed] [Google Scholar]
- Yamashiro T, Tummers M, Thesleff I. 2003. Expression of bone morphogenetic proteins and Msx genes during root formation. J Dent Res. 82(3):172–176. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. 2008. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 125(9–10):797–808. [DOI] [PubMed] [Google Scholar]
- Zhao H, Feng J, Ho TV, Grimes W, Urata M, Chai Y. 2015. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat Cell Biol. 17(4):386–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034518771014 for Msx2 Marks Spatially Restricted Populations of Mesenchymal Precursors by N. Sakagami, Y. Matsushita, S. Syklawer-Howle, H.M. Kronenberg, W. Ono, and N. Ono in Journal of Dental Research