Abstract
Coevolution of the human host and its associated microbiota has led to sophisticated interactions to maintain a delicate homeostasis. Emerging evidence suggests that in addition to small molecules, peptides, and proteins, small regulatory noncoding RNAs (sRNAs) might play an important role in cross-domain interactions. In this study, we revealed the presence of diverse host transfer RNA–derived small RNAs (tsRNAs) among human salivary sRNAs. We selected 2 tsRNAs (tsRNA-000794 and tsRNA-020498) for further study based on their high sequence similarity to specific tRNAs from a group of Gram-negative oral bacteria, including Fusobacterium nucleatum, a key oral commensal and opportunistic pathogen. We showed that the presence of F. nucleatum triggers exosome-mediated release of tsRNA-000794 and tsRNA-020498 by human normal oral keratinocyte cells. Furthermore, both tsRNA candidates exerted a growth inhibition effect on F. nucleatum, likely through interference with bacterial protein biosynthesis, but did not affect the growth of Streptococcus mitis, a health-associated oral Gram-positive bacterium whose genome does not carry sequences bearing high similarity to either tsRNA. Our data provide the first line of evidence for the modulatory role of host-derived tsRNAs in the microbial-host interaction.
Keywords: tsRNAs, cross-domain interactions, oral microbiome, antimicrobials, microbial-host interaction, sRNAs
Introduction
The human microbiota exists in a delicate balance with its host, and this equilibrium is maintained via multiple mechanisms of cross-domain communication and modulation (Artis 2008; Peterson and Artis 2014). A variety of host-derived mobile molecules, including antimicrobial peptides, immunoglobulins, and the complex complement network, can serve as interdomain modulators. These molecules often operate with other mechanisms, such as microbe-associated molecular pattern recognition systems, to maintain microbial-host homeostasis (Ganz 2003; Markiewski and Lambris 2007; Gallo and Hooper 2012; Chu and Mazmanian 2013).
Small regulatory noncoding RNAs (sRNAs) are a class of regulatory elements that have been identified in prokaryotes and eukaryotes and implicated in gene regulation (Shimoni et al. 2007; Waters and Storz 2009; Duran-Pinedo et al. 2015). In eukaryotic cells, sRNAs are RNA molecules that are 19 to 31 nucleotides in length and mainly classified per their biogenesis as microRNA, small interfering RNA, or PIWI-interacting RNA (piRNA; Carthew and Sontheimer 2009). These sRNAs were shown to be involved in the regulation of gene expression, and some of them were implicated in inflammation and cancer (Volinia et al. 2006; Tili et al. 2013), thus potentially serving as biomarkers for cancer screening and diagnosis. For example, while miR-371 acts as a tumor suppressor via regulating SLC7A11 in oral squamous cell carcinoma (Wu et al. 2017), oncogenic miR-155 promotes oral squamous cell carcinoma cell proliferation (Rather et al. 2013). Recent studies further revealed the potential role of sRNAs in interspecies and cross-domain interactions (Knip et al. 2014; Weiberg et al. 2015). These “social RNAs” could serve as gene silencing and modulating molecules in recipient cells across domains, a phenomenon called cross-domain RNAi (Sarkies and Miska 2013).
A novel class of sRNA was recently described: transfer RNA (tRNA)–derived small sRNA (tsRNA; Lee et al. 2009). Depending on the size and mechanism of biogenesis, tsRNAs can be classified into 2 types: the 5′ and 3′ tRNA halves and the shorter, tRNA-derived fragments (tRFs; Dhahbi 2015). tsRNAs are present in most organisms: some are constitutively produced; others are exclusively generated in response to certain stress conditions (Ivanov et al. 2011; Peng et al. 2012; Loss-Morais et al. 2013; Maute et al. 2013). Increasing evidence indicates that endogenous tsRNAs can function as modulators of gene expression in prokaryotic and eukaryotic organisms (Gebetsberger et al. 2012; Goodarzi et al. 2015).
Host-derived sRNAs, including tsRNAs, can also be found in human body fluids, including blood, tears, and saliva, either freely or contained within exosomes (Lasser et al. 2011; Williams et al. 2013). These secreted sRNAs have been extensively studied for diagnostic applications (Majem et al. 2015); however, their biological functions remain elusive. As a key component of host defense against oral infection, saliva contains innate antimicrobial proteins and adaptive immune mediators. Considering the complexity of commensal microbiota residing within oral cavity and the abundant host-derived sRNAs detected in saliva, it is intriguing to hypothesize that these sRNAs are potentially involved in interdomain interaction. In this study, we provide evidence that host-derived salivary tsRNAs may mediate microbial-host interaction through targeted modulation of oral bacterial growth.
Materials and Methods
Identification of Host-Derived Salivary tsRNAs and Determination of Homology to Bacterial tRNA
Salivary RNA sequences were initially trimmed for adapters and low-quality sequences. To identify host-derived salivary tsRNAs, salivary sRNA sequences were first mapped to the human genome (hg19) with Bowtie (version 1.1.2) and the commands “–v 2 –k 100 –best strata” (Langmead et al. 2009). Since tsRNAs are known to have 3′ modifications, unmapped reads were subjected to trimming one 3′ nucleotide and remapped with bowtie command “–v 1 –k 100 –best strata.” Trimming and remapping of the unmapped reads was performed 3 times to account for 3′ CCA extensions found in the tRF 3 series. Mapped reads were overlapped with annotated human (hg19) tRF coordinates (http://genome.bioch.virginia.edu/trfdb). Valid overlaps were defined by requiring the mapped read alignment to start either 3 bases before or 2 bases after the tsRNA-annotated start site and ending 2 bases before or 3 bases after the tsRNA-annotated end site. Last, salivary tsRNAs were mapped to bacterial tRNA (http://gtrnadb.ucsc.edu/) to determine homology. Homology mapping was performed with a primary round of Bowtie with the previously described parameters and then a secondary round of Bowtie2 (version 2.2.9) on the unmapped reads with the local command to allow soft clipping (Langmead and Salzberg 2012).
Growth Inhibition Assay
Fusobacterium nucleatum subsp. nucleatum 23726 and Streptococcus mitis ATCC 6249 were used in this study (henceforth, F. nucleatum and S. mitis). F. nucleatum and S. mitis were cultured in Columbia and brain-heart infusion broth, respectively, and incubated at 37 ºC under anaerobic condition (10% H2, 10% CO2, 80% N2) until exponential phase. Cells were collected by centrifugation and resuspended in fresh medium to obtain 106 CFU (colony-forming units) / mL. Meanwhile, 6 commercially synthesized (IDT, Inc.) sRNAs were serially diluted in medium with concentrations between 0 and 100 μM; 100 μL of bacterial suspension and sRNA solutions were mixed into individual wells of a 96-well plate. As control, sRNAs were pretreated with RNase A for 30 min before being added to the bacterial cultures. Plates were incubated at 37 °C under anaerobic conditions overnight. Bacterial growth was determined by measuring the optical density at 600 nm with a microplate reader.
To test the impact of tsRNAs on bacterial growth dynamics, F. nucleatum and S. mitis cells were cultured and collected as described earlier. Bacterial cells were seeded into their respective growth medium in 96-well plates containing 100μM tsRNA-000794, tsRNA-020498, or control RNAs, including piRNA-0016792 and scrambled RNA (sequence provided in the Appendix Table), with a final 106 CFU/mL. Plates were incubated under anaerobic condition at 37 ºC, and OD600 was measured with a microplate reader at time intervals indicated in the results.
Measurement of Protein Synthesis
The Click-iT AHA Alexa Fluor 488 Protein Synthesis HCS Assay (Invitrogen) was used to measure the protein synthesis in bacteria. This assay labels newly synthesized proteins with an analog of methionine that reacts to Alexa Fluor 488 alkyne by click chemistry.
F. nucleatum and S. mitis cells from the mid-exponential growth phase were used to inoculate fresh media containing RNase inhibitor (1 U/µL) and different concentrations of tsRNA-000794 or tsRNA-020498, with a final bacterial OD600 of 0.05. Cultures were incubated for 24 h at 37 ºC under anaerobic condition and transferred to chemically defined media containing 10µM glucose with different concentrations of tsRNA and diluted (1:1,000) Click-iT AHA reagent. Bacteria were incubated in the anaerobic chamber at 37 °C for 30 min before being fixed with 75% ethanol for 25 min at room temperature. Fixed bacteria were labeled with Alexa Flour 488 for 30 min according to the manufacturer’s guidelines. Labeled cells were visualized with an inverted confocal microscope (Leica SPE I) equipped with a Leica ACS APO 100X Oil CS objective (NA:1.15).
All images were acquired with the same settings on the microscope and processed with standard FIJI analysis (Schindelin et al. 2012) to quantify the fluorescence intensity per unit area covered by bacterial cells. The regions of interest were selected arbitrarily. However, to ensure that the selected cells were representative of the overall population, we chose 3 to 5 representative images from each experiment (3 independent experiments in total). For each image, we picked >10 cells representing the overall cell population for further quantification.
Monitoring the Abundance of Extracellular Exosome-Bound sRNAs in Human Normal Oral Keratinocyte Cells
The normal oral keratinocyte (NOKSI) cell line was a gift from Dr. Silvio Gutkind and tested negative for mycoplasma contamination. Cells were cultured with defined keratinocyte serum-free medium (Thermo Fisher Scientific). After cells reached >90% confluence, bacteria resuspended in keratinocyte serum-free medium were added to the NOKSI cell monolayer at a ratio of 100:1. Experiments were stopped at 2 and 4 h. The medium was separated from cells and exosome isolation performed with a differential centrifugation method previously described (Bonifacino et al. 2006) with modifications. Briefly, cells were removed by centrifugation at 300 × g for 10 min. To remove bacteria and dead NOKSI debris, the supernatant was centrifuged at 3,000 × g for 20 min, followed by centrifugation at 17,000 × g for 15 min. The supernatant was filtered through a 0.2-μm filter, followed by ultracentrifugation at 120,000 × g for 90 min to pellet the exosomes. Isolated exosomes were lysed with QIAzol reagent (Qiagen) and store at −80 °C. The total exosomal RNA was isolated with the miRNeasy Micro kit (Qiagen).
The abundance of 6 tested sRNAs in the exosomes was measured with droplet digital polymerase chain reaction (ddPCR) with custom TaqMan small RNA assays ordered from Applied Biosystems. Total exosomal RNA was converted to cDNA as described previously. The TaqMan PCR reaction mixtures were assembled with 10 μL of 2 × ddPCR Supermix (Bio-Rad), 4 μL of cDNA template, 5 μL of water and 1 μL of custom 20 × TaqMan probes/primers specific for each assay. Droplet formation was carried out with a QX100 droplet generator. The droplets were transferred to a 96-well polymerase chain reaction plate and amplified to the endpoint with the following conditions: 95 °C for 10 min, 40 cycles of 94 °C for 30 s, 60 °C for 1 min, and 98 °C for 10 min at a ramp rate of 2.5 °C/s for all steps. The samples were measured with a BioRad QX100 droplet reader. The fold changes were calculated per comparison with nonchallenged keratinocytes.
Statistical Analysis
For all analysis, we used 1- or 2-way analysis of variance statistical tests for multiple group comparisons.
Results
Detection of Diverse Host-Derived tsRNAs in Cell-Free Saliva
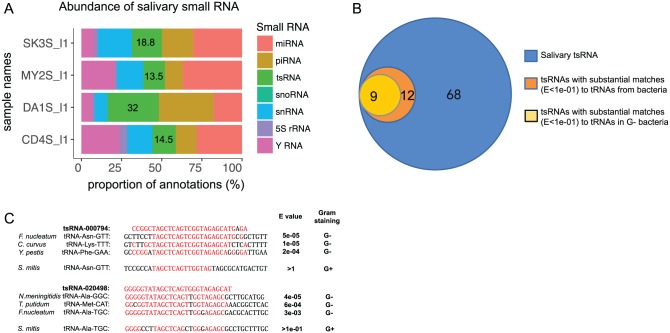
Previously, we used RNA-seq to investigate cell-free human saliva and found that it contains abundant host-derived sRNAs (Bahn et al. 2015). In this study, we performed a more comprehensive bioinformatics analysis of this data set. From the 4 human subjects we previously studied, we identified a total of 68 distinct host-derived tsRNAs, which included 47 fragments derived from 5′ ends of tRNA (69%) and 21 derived from 3′ ends (31%), accounting for 13.5% to 32% of the total host-derived salivary sRNA (Appendix Table; Fig. 1A, B). Interestingly, when aligned against the Human Oral Microbiome Database (HOMD; http://www.homd.org), 12 of 68 (18%) host-derived tsRNAs substantially matched (E value <1e-01) specific microbial tRNA partial sequences (Appendix Table; Fig. 1B). Furthermore, the majority (9 of 12) matched the partial sequence of tRNAs of Gram-negative oral bacteria, including known opportunistic pathogens such as F. nucleatum, Neisseria meningitidis, and Treponema putidum (Fig. 1C).
Figure 1.
Salivary host-derived tsRNAs. (A) Relative abundance of host-derived tsRNAs in total sRNA reads among 4 subjects (SK3S_I1, MY2S_I2, DA1S_I1, and CD4S_I1). (B) A subset of host-derived tsRNAs have substantial matches to bacteria, particularly Gram-negative oral bacterial tRNA sequences. (C) Alignment of tsRNA-000794 and tsRNA-020498 with partial sequences of specific tRNAs from oral Gram-negative bacterial species. Letters in red indicate the nucleotides conserved between or among tsRNA and partial tRNA sequences from selected oral bacterial species. sRNA, small regulatory noncoding RNA; tRNA, transfer RNA; tsRNA, tRNA-derived small sRNA.
tsRNA-000794 and tsRNA-020498 Inhibit F. nucleatum Growth
Evidence from both eukaryotic and prokaryotic organisms suggests that endogenous tsRNAs can modulate cellular growth (Yamasaki et al. 2009; Zhang et al. 2009). Furthermore, it was shown that Archaea-derived tsRNAs can inhibit bacterial and eukaryotic protein biosynthesis (Gebetsberger et al. 2016). We hypothesize that salivary host-derived tsRNAs, particularly those with high sequence similarity to bacterial tRNAs, may play an important role in mediating bacterial-host interactions.
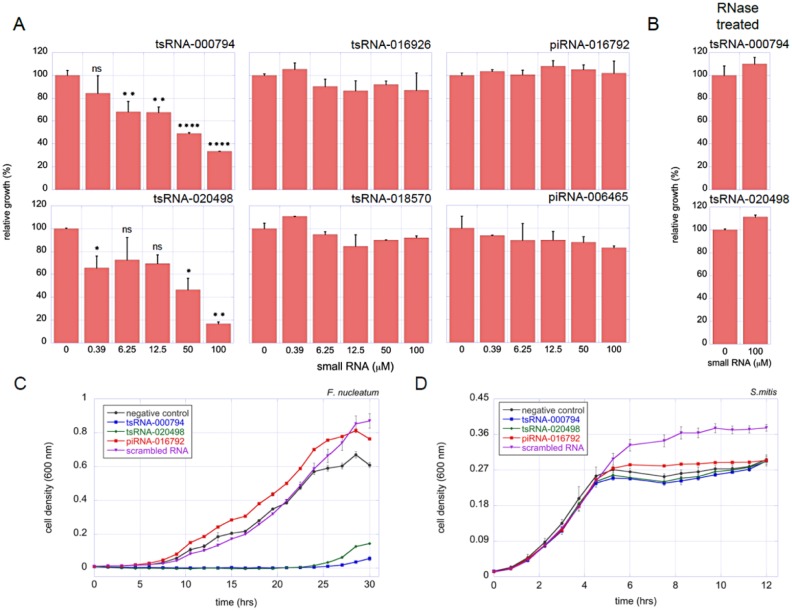
To test this hypothesis and provide proof-of-concept data, we selected 2 host-derived 5′ tRNA halves, tsRNA-000794 and tsRNA-020498, for further analysis based on their high sequence similarity to tRNAs from a group of Gram-negative oral bacteria, which includes F. nucleatum, a key oral opportunistic pathogen (Fig. 1C). As controls, we included 2 other tsRNAs that did not have significant sequence matches (E value >1) to the microbial tRNA database (http://gtrnadb.ucsc.edu) or HOMD: tsRNA-016926 and tsRNA-018570. In addition, 2 host-generated salivary piRNAs were included as controls—piRNA-016792 and piRNA-006465—which displayed no significant sequence similarity to bacterial genome sequences in HOMD. Different concentrations of these 6 commercially synthesized sRNAs (IDT, Inc.) were added to cultures of either F. nucleatum or the health-associated Gram-positive oral bacterium S. mitis, the tRNAs and genomes of which do not have sequences with high-similarity matches to any of the tested sRNA (E value >1e-01). The overnight growth of the differentially treated bacterial cultures was recorded. The results showed that while the 4 control sRNAs had no noticeable effect on the growth of F. nucleatum, tsRNA-000794 and tsRNA-020498 clearly inhibited the growth of F. nucleatum in a dose-dependent manner (Fig. 2A). At 50 µM, tsRNA-000794 and tsRNA-020498 displayed >50% growth inhibition. Furthermore, the inhibition was abolished when tsRNAs were pretreated with RNase A (Fig. 2B). In contrast, S. mitis was not sensitive to any of the sRNAs tested (Appendix Fig., Appendix Table). These results support our hypothesis that host-derived tsRNAs may be involved in modulating microbial-host interaction. The data also suggest that a certain level of sequence similarity is required between the tsRNAs and the target species.
Figure 2.
tsRNA-000794 and tsRNA-020498 induced growth inhibition in Fusobacterium nucleatum. (A) Different concentrations of tsRNA-000794, tsRNA-020498, tsRNA-016926, tsRNA-018570, piRNA-016792, and piRNA-006465 were added to F. nucleatum cultures. Cultures were incubated overnight at 37 ºC under anaerobic condition before OD600 was measured. The growth inhibition is expressed as percentage of absorbance to that of the negative control (no addition of tRF). (B) tsRNA-000794 and tsRNA-020498 were pretreated with RNase A before being added to the F. nucleatum culture, and impact on bacterial growth was similarly monitored. tsRNA-000794 (100 µM), tsRNA-020498 (100 µM), piRNA-016792 (100 μM), or scrambled control RNA (100 μM) was added to F. nucleatum (C) and Streptococcus mitis (D), and the growth curves were measured. Assays were performed in triplicates, and mean ± SEM values are shown. ns, P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001. All the columns without “ns” or stars are nonsignificant according to statistical tests. piRNA, PIWI-interacting RNA; tsRNA, transfer RNA–derived small sRNA; tRF, tRNA-derived fragment.
We further tested the effect of tsRNA on the growth dynamics of bacteria. As shown in Figure 2C, the growth dynamic of F. nucleatum was severely negatively affected by the addition of 100µM tsRNA-000794 or tsRNA-020498, while addition of piRNA-0016792 or scrambled control RNA not only failed to inhibit the growth of F. nucleatum but resulted in higher cell density at a late log phase as compared with the negative control. Meanwhile, the addition of all the tested RNAs did not exert a negative impact on the growth dynamic of S. mitis (Fig. 2D). Interestingly, in the presence of scrambled RNA, S. mitis culture reached even higher optical density when cells entered the stationary phase as compared with the nonaddition negative control, a phenomenon worth further investigation in the future.
tsRNA-000794 and tsRNA-020498 Inhibit F. nucleatum Protein Biosynthesis
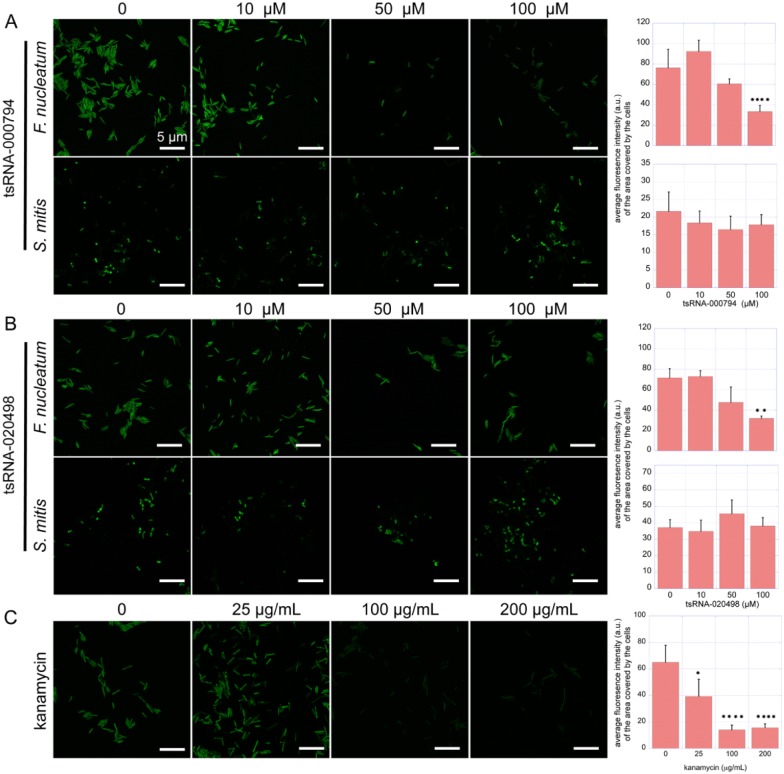
Endogenous tsRNAs are capable of globally downregulating protein synthesis in eukaryotic and prokaryotic organisms (Yamasaki et al. 2009; Zhang et al. 2009; Gebetsberger et al. 2012). To test if the observed tsRNA-induced growth inhibition was a result of reduced protein biosynthesis, we monitored the amount of global protein synthesis in F. nucleatum with the modified Click-iT AHA Alexa Fluor 488 assay. The results showed that the addition of tsRNA-000794 and tsRNA-020498 at 100 µM resulted in a drastic reduction in the incorporation of the methionine analog, as reflected by significantly reduced fluorescence signal intensity (Fig. 3A, B). At 100 µM, treatment with either of the tsRNAs led to a similar level of reduction in fluorescence intensity per area unit covered by bacterial cells as compared with the samples treated with 25 µg/mL of kanamycin (Fig. 3C), while more drastic fluorescence reduction was observed for cultures treated with higher concentrations of kanamycin (50 and 100 µg/mL). Conversely, treatment of S. mitis with tsRNA-000794 or tsRNA-020498 did not affect the fluorescence intensity as compared with the negative control (Fig. 3A, B), which is consistent with the lack of an observed effect on growth (Appendix Fig.).
Figure 3.
tsRNA-000794 and tsRNA-020498 affect protein synthesis in Fusobacterium nucleatum. The effect of the addition of tsRNA-000794 and tsRNA-020498 on protein synthesis in F. nucleatum and Streptococcus mitis (A and B) and the effect of kanamycin on protein synthesis in F. nucleatum (C). The left panels show representative fluorescence images of differentially treated cultures. The right panel shows the quantification of fluorescence intensity per area unit covered by bacterial cells. au, arbitrary units. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001. All the columns with no “ns” or stars are nonsignificant according to statistical tests. tsRNA, transfer RNA–derived small sRNA.
F. nucleatum Induces Release of Exosome-Associated tsRNA-000794 and tsRNA-020498 by Human NOKSI Cells
We next investigated if tsRNA-000794 and tsRNA-020498 secretion can be induced by F. nucleatum and if the tsRNAs are secreted via exosomes. We designed a coculture system in which NOKSI cells were challenged with F. nucleatum. NOKSI cells were cocultured with bacteria at a multiplicity of infection of 100 and harvested after 2 and 4 h. The secreted extracellular exosomes were extracted, and the absolute copy number of the sRNAs within the exosomes was measured with ddPCR. Results showed that a significant increase (P < 0.05) in tsRNA-000794 and tsRNA-020498 was detected within exosomes when cells were challenged with F. nucleatum, especially at the 4-h time point (Fig. 4A). No significant change was observed for control tsRNAs (Fig. 4A). Furthermore, when NOKSI cells were challenged with S. mitis, there was no increase detected for any of the tested sRNAs in the secreted exosome (Fig. 4B). These data suggest that oral keratinocytes can respond to the presence of specific oral bacteria by releasing specific tsRNAs. Interestingly, exosome-associated tsRNA-020498 level was decreased at the 4-h time point when cells were challenged with S. mitis as compared with nonchallenged control, a phenomenon worth further study.
Figure 4.
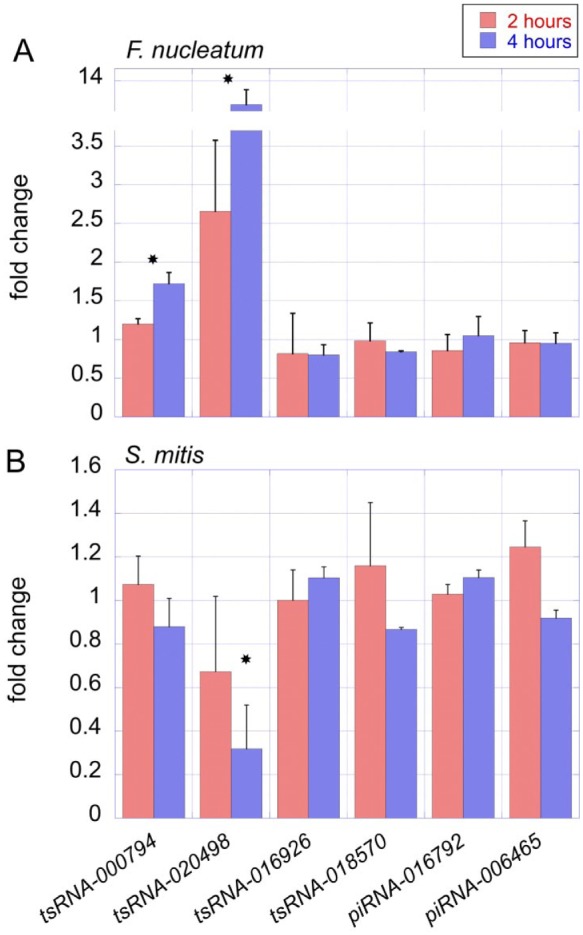
Fusobacterium nucleatum specifically induces the secretion of tsRNA-000794 and tsRNA-020498 from human oral keratinocytes. Human oral keratinocyte NOKSI cells were cocultured with F. nucleatum or Streptococcus mitis at a multiplicity of infection of 100. (A) The level of exosome-associated sRNAs in NOKSI medium when cells were challenged with F. nucleatum was measured by ddPCR. The fold changes were calculated per the comparison with nonchallenged keratinocytes. (B) The levels of exosome-associated sRNAs in NOKSI medium when cells were challenged with S. mitis were measured by ddPCR. The fold changes were calculated per the comparison with nonchallenged keratinocytes. Assays were performed in triplicates, and mean ± SEM values are shown. *P < 0.05. All the columns with no “ns” or stars are nonsignificant according to statistical tests. ddPCR, droplet digital polymerase chain reaction; NOKSI, normal oral keratinocyte; sRNA, small regulatory noncoding RNA; tsRNA, transfer RNA–derived small sRNA.
Discussion
tsRNAs have been reported as endogenous modulators of gene expression in prokaryotic and eukaryotic organisms (Gebetsberger et al. 2012; Goodarzi et al. 2015). A recent study also revealed the potential role of bacteria-generated tsRNAs in modulating the host immune response (Koeppen et al. 2016). Our data strongly suggested that host-derived tsRNAs, particularly those displaying strong sequence similarity to bacterial tRNAs, might serve as interdomain mediators to regulate the growth of host-associated bacteria. Various mechanistic models have been proposed to explain the role of endogenous tsRNAs in modulating gene expression, such as interacting with Argonaute proteins and silencing expression of target mRNAs (Shigematsu and Kirino 2015) or negatively affecting global protein biosynthesis via initiation (Ivanov et al. 2011; Gebetsberger et al. 2016) and elongation (Gebetsberger et al. 2012). Sobala and Hutvagner (2013) recently demonstrated that selected 5′-tRNA-derived sRNAs inhibit translation of an mRNA reporter in vitro, possibly by affecting translation elongation, which differs from the inhibition of translation initiation caused by 5′-tiRNAs. Similarly, in Haloferax volcanii, an archaeal species, a stress-induced 26-nt 5′-tRNA fragment derived from tRNAval inhibits translation by directly binding to the 30S small ribosomal subunit and inhibiting peptidyl transferase activity (Gebetsberger et al. 2012). Furthermore, these Archaea-derived tsRNAs can also inhibit eukaryal and bacterial protein biosynthesis, suggesting a role in mediating cross-domain interaction, as well as a functionally conserved mode of action (Gebetsberger et al. 2016). Our data are in line with these findings, as we observed tsRNA-induced inhibition of F. nucleatum growth and showed that it may directly result from tsRNA-mediated protein biosynthesis inhibition. However, we could not completely rule out the possibility that the observed protein biosynthesis reduction may be a consequence of tsRNA-induced decreased bacterial growth. Further studies are required to investigate the detailed mechanism. In our in vitro system, the amount of exogenously added tsRNAs is likely higher than physiologic concentrations. However, our data suggest that in the oral cavity these host-derived tsRNAs are likely packaged in exosomes, which would allow them to be delivered to the targeted bacteria in small but concentrated doses. Additional studies with better in vitro systems are needed to better understand the details of the molecular mechanism.
An intriguing finding is that the majority of the bacterial tRNA sequences that show high sequence similarity with the host-derived salivary tsRNAs are from Gram-negative bacteria (Fig. 1B). Within the oral cavity, the gingival sulcus is one of the sites of the most intense microbial-host interactions; a dysbiosis at this site can eventually lead to periodontitis (Teles et al. 2013; Costalonga and Herzberg 2014). Therefore, homeostatic host-bacterial interactions at the gingival sulcus are crucial for health. One of the hallmarks of periodontitis is a shift in the community from mostly Gram-positive to mostly Gram-negative species, which contain highly toxic and immunogenic lipopolysaccharide and other virulence factors (Darveau 2010). tsRNAs may represent a mechanism evolved in the host to suppress the growth of Gram-negative bacteria, either commensal opportunistic pathogens (e.g., F. nucleatum) or possibly exogenous pathogens (e.g., Yersinia pestis; Appendix Table) to maintain health, an intriguing hypothesis that warrants further testing.
More investigation is required to fully understand the mechanisms underlying the observed microbial-host interactions mediated by tsRNAs. Unanswered questions include how specific tsRNAs are induced and selected for transport in the presence of bacteria, how tsRNAs are transported outside of the cells, and the mechanism of target cell recognition. Understanding the mechanism by which tsRNAs affect the physiology of bacterial cells, either triggering the event by entering the target cells or remaining extracellularly, and determining if this is unique to the oral cavity or is more universal among microbiome-host interactions are important fundamental questions requiring further exploration. Nevertheless, our study expands the repertoire of known molecules that can act endogenously to modulate gene expression as well as exogenously, in this case as mediators of microbial-host homeostasis.
Author Contributions
X. He, contributed to design, data analysis, and interpretation, drafted and critically revised the manuscript; F. Li, contributed to design, data acquisition, analysis, and interpretation, critically revised the manuscript; B. Bor, K. Koyano, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript; L. Cen, contributed to data acquisition and analysis, critically revised the manuscript; X. Xiao, contributed to design, data analysis, and interpretation, critically revised the manuscript; W. Shi, D.T.W. Wong, contributed to conception, design, data analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034518770605 for Human tRNA-Derived Small RNAs Modulate Host–Oral Microbial Interactions by X. He, F. Li, B. Bor, K. Koyano, L. Cen, X. Xiao, W. Shi and D.T.W. Wong in Journal of Dental Research
Acknowledgments
We thank Dr. Melissa Agnello for her critical editing of the manuscript.
Footnotes
This work was supported by grants from the National Institute of Dental and Craniofacial Research to W. Shi and X. He (1R01DE023810, 1R01DE020102, 1R01DE026186) and from the National Institutes of Health to D. Wong (UH3 TR000923), X. Xiao (R01HG006264), and B. Bor (F32DE025548-1).
D. Wong is cofounder of RNAmeTRIX Inc., a molecular diagnostic company; he holds equity in RNAmeTRIX. Intellectual property that D. Wong invented and that University of California patented has been licensed to RNAmeTRIX. W. Shi is an employee of C3 Jian, Inc., which has licensed technologies from UC Regents that could be indirectly related to this research project. The authors declare no other potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is available online.
ORCID iD: F. Li  https://orcid.org/0000-0002-5819-734X
https://orcid.org/0000-0002-5819-734X
References
- Artis D. 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 8(6):411–420. [DOI] [PubMed] [Google Scholar]
- Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. 2015. The landscape of microRNA, PIWI-interacting RNA, and circular RNA in human saliva. Clin Chem. 61(1):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. 2006. Current protocols in cell biology. Wiley Online Library. doi: 10.1002/0471143030. [DOI] [Google Scholar]
- Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell. 136(4):642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Mazmanian SK. 2013. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 14(7):668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costalonga M, Herzberg MC. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 162(2, Pt A):22–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 8(7):481–490. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM. 2015. 5′ tRNA halves: the next generation of immune signaling molecules. Front Immunol. 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Pinedo AE, Yost S, Frias-Lopez J. 2015. Small RNA transcriptome of the oral microbiome during periodontitis progression. Appl Environ Microbiol. 81(19):6688–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RL, Hooper LV. 2012. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 12(7):503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 3(9):710–720. [DOI] [PubMed] [Google Scholar]
- Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N. 2016. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 14(10):1364–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, Zywicki M, Kunzi A, Polacek N. 2012. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012:260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. 2015. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 161(4):790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 43(4):613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knip M, Constantin ME, Thordal-Christensen H. 2014. Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 10(9):e1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, Demers EG, Dolben EL, Hammond JH, Hogan DA, et al. 2016. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 12(6):e1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, Bossios A, Sjostrand M, Gabrielsson S, Lotvall J, Valadi H. 2011. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23(22):2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss-Morais G, Waterhouse PM, Margis R. 2013. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol Direct. 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majem B, Rigau M, Reventos J, Wong DT. 2015. Non-coding RNAs in saliva: emerging biomarkers for molecular diagnostics. Int J Mol Sci. 16(4):8676–8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewski MM, Lambris JD. 2007. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol. 171(3):715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. 2013. TRNA-derived microrna modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 110(4):1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, et al. 2012. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 22(11):1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 14(3):141–153. [DOI] [PubMed] [Google Scholar]
- Rather MI, Nagashri MN, Swamy SS, Gopinath KS, Kumar A. 2013. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: implications for cancer therapeutics. J Biol Chem. 288(1):608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Miska EA. 2013. Molecular biology. Is there social RNA? Science. 341(6145):467–468. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M, Kirino Y. 2015. TRNA-derived short non-coding RNA as interacting partners of argonaute proteins. Gene Regul Syst Bio. 9:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H. 2007. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol. 3:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobala A, Hutvagner G. 2013. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 10(4):553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. 2013. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000. 62(1):95–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Croce CM. 2013. Micrornas play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 253(1):167–184. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 103(7):2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell. 136(4):615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A, Bellinger M, Jin H. 2015. Conversations between kingdoms: small RNAs. Curr Opin Biotechnol. 32:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. 2013. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci U S A. 110(11):4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sun X, Song B, Qiu X, Zhao J. 2017. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. 6(7):1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 185(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sun L, Kragler F. 2009. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 150(1):378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034518770605 for Human tRNA-Derived Small RNAs Modulate Host–Oral Microbial Interactions by X. He, F. Li, B. Bor, K. Koyano, L. Cen, X. Xiao, W. Shi and D.T.W. Wong in Journal of Dental Research