Abstract
An external quality assessment of yellow fever virus (YFV) molecular detection in European laboratories was organised in rapid response to an increase in human cases in Brazil in 2018 with risk of import to Europe. Detection of YFV was assessed among 32 laboratories in 23/31 European Union (EU) and European Economic Area (EEA) countries and two laboratories in one non-EU/EEA country. Adequate capabilities were lacking in 10/23 countries; five did not participate as they lacked implemented assays.
Keywords: yellow fever virus, laboratory surveillance, emerging or re-emerging diseases, imported viral diseases, vector-borne infections
In March 2018, the Emerging Viral Diseases-Expert Laboratory Network (EVD-LabNet), funded by the European Centre for Disease Prevention and Control (ECDC) in Stockholm, Sweden, organised an external quality assessment (EQA) of molecular detection of yellow fever virus (YFV). The EQA was a rapid response to the recent outbreaks with YFV in South America and the increasing number of unvaccinated citizens from European Union /European Economic Area (EU/EEA) countries that acquired infection while travelling to outbreak regions [1-3].
Participation of European Expert Laboratories
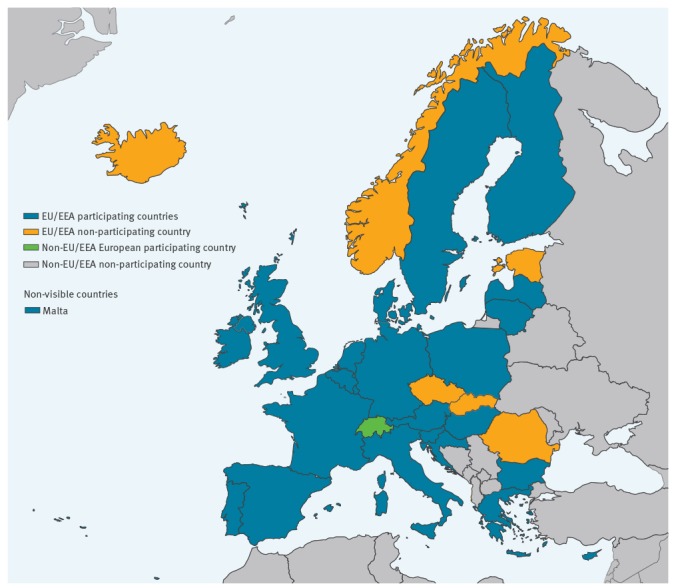
In total, 71 laboratories were invited to participate in the EQA: 60 laboratories in 30 EU/EEA countries (excluding Liechtenstein, which has no EVD-LabNet member laboratory), seven laboratories in seven EU pre-accession countries and four laboratories in two other European countries. Thirty-two laboratories in 23 EU/EEA countries and two laboratories in one other European country participated in the EQA (Figure). Of the seven EU/EEA countries that did not participate, five laboratories indicated that they did not have a test available at the time of the EQA, one laboratory had no funds and one laboratory had no permission to participate. Of the seven invited laboratories in the EU pre-accession countries, five had no test available and two did not indicate a reason for not participating.
Figure.
Overview of the number of Emerging Viral Diseases-Expert Laboratory Network laboratories per country that participated in the yellow fever virus molecular external quality assessment, March 2018 (n = 34)
EU/EEA participating countries: Italy 6 laboratories; Germany 4 laboratories; Spain 2 laboratories; all other countries 1 laboratory. Non-EU/EEA participating countries: Switzerland 2 laboratories.
External quality assessment set-up
For the preparation of the EQA panel, Vero E6 cell culture supernatants were infected with different YFV strains (vaccine strain YFV-17D, South American strain genotype 1E Brazil 2008, West African strain Ivory Coast 1999, GenBank AY603338), inactivated by heat (56 °C, 1 h) and gamma irradiation (25 kilogray) and tested for non-infectivity. Plasma or urine was spiked to prepare a set of 12 representative samples (Table 1). As a specificity control, plasma samples containing inactivated lineage 1 West Nile virus (WNV, strain New York), the four dengue virus serotypes (DENV-1, strain Thai 1958; DENV-2, TH-36 strain; DENV-3, H87 strain; DENV-4, H241 strain), and Zika virus (ZIKV, strain MR766) and a negative control (plasma and urine) were included (Table 1). The samples were number coded, freeze-dried (Christ, AlphaI-5, Hanau, Germany) and stored at 4 °C until dispatch. Sample quality and YFV load were estimated in two different panels using a reference real-time quantitative polymerase chain reaction (RT-qPCR) [4], and by a pan-flavirus RT-qPCR [5]. The non-YFV viral loads were determined using RT-qPCR (WNV) [6] and in house RT-qPCRs (DENV and ZIKV). The panel was further validated by two independent reference laboratories using a YFV specific RT-qPCR [4,7]. On 26 February, the freeze-dried panels were shipped at room temperature to the participants and all reported the panel to be in good condition on arrival. EQA results could be submitted until 31 March 2018 via an online system.
Table 1. Composition and panel results of the yellow fever virus molecular detection external quality assessment, March 2018 (n = 46).
| Parameter | Sample number | Virus (Strain) | Viral RNA copies/sample | Matrix | Correct result | Inconclusive result | False negative result | False positive result | Flavivirus not-specified | Total % of test with correct result |
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | 8 | YFV American (Brazil) | 2.8E + 06 | Plasma | 35 | 1 | 7 | NA | 3 | 76 |
| 9 | YFV American (Brazil) | 2.6E + 05 | Plasma | 35 | 1 | 8 | NA | 3 | 76 | |
| 2 | YFV American (Brazil) | 3.5E + 04 | Plasma | 33 | 1 | 9 | NA | 3 | 71.7 | |
| 3 | YFV American (Brazil) | 3.4E + 03 | Plasma | 28 | 2 | 13 | NA | 3 | 60.8 | |
| 5 | YFV American (Brazil) | 3.5E + 05 | Urine | 34 | 1 | 8 | NA | 3 | 73.9 | |
| 11 | YFV American (Brazil) | 2.3E + 03 | Urine | 25 | 1 | 17 | NA | 3 | 54.3 | |
| 6 | YFV Africa (Ivory Coast 1999) | 4.7E + 06 | Plasma | 35 | 1 | 7 | NA | 3 | 76 | |
| 12 | YFV Africa (Ivory Coast 1999) | 4.7E + 05 | Plasma | 34 | 1 | 8 | NA | 3 | 73.9 | |
| 15 | YFV Africa (Ivory Coast 1999) | 4.9E + 04 | Plasma | 32 | 1 | 10 | NA | 3 | 69.5 | |
| 13 | YFV Africa (Ivory Coast 1999) | 3.7E + 03 | Plasma | 33 | 1 | 9 | NA | 3 | 71.7 | |
| 14 | YFV-vaccine (17D) | 6.5E + 04 | Plasma | 38 | 0 | 5 | NA | 3 | 82.6 | |
| 1 | YFV-vaccine (17D) | 5.4E + 03 | Plasma | 32 | 2 | 10 | NA | 2 | 69.5 | |
| Specificity | ZIKV (MR766) DENV-1 (Thailand 1958) |
4.2E + 05 3.5E + 06 |
||||||||
| DENV-2 (TH-36) | 2.2E + 06 | |||||||||
| 4 | DENV-3 (H-87) | 3.9E + 05 | Plasma | 40 | 2 | NA | 2 | 3 | 86.9 | |
| DENV-4 (H241) | 6.6E + 04 | |||||||||
| WNV (New York) | 1.27 + 04 | |||||||||
| Contamination control | 7 | Negative | NA | Plasma | 43 | 0 | NA | 2 | 1 | 93.4 |
| 10 | Negative | NA | Urine | 46 | 0 | NA | 0 | 0 | 100 |
DENV: dengue virus; NA: not applicable; WNV: West Nile virus; YFV: yellow fever virus; ZIKV: Zika virus.
Outcomes
Thirty-four laboratories submitted results for a total of 46 panel tests. There were 20 different in-house (including own design) and four commercial RT-PCRs represented in the EQA; 15 YFV-specific, three vaccine-strain specific and six pan-flavivirus RT-PCRs. In 10 of 13 laboratories submitting results based on a pan-flavivirus RT-PCR, this RT-PCR was combined with amplicon sequencing to determine the specific flavivirus involved (Table 2).
Table 2. Overview of RT-PCR systems for yellow fever virus detection used by 34 laboratories in the Emerging Viral Diseases-Expert Laboratory Network external quality assessment of molecular detection of yellow fever virus, March 2018 (n = 46).
| RT-PCR used and specificity | YFV genome target | number of laboratories |
|---|---|---|
| In-house YFV specific (wildtype/ vaccine strain) | ||
| Domingo et al. 2012 [4] | 5'-UTR | 4 |
| Drosten et al. 2002 [7] | 5'-UTR | 7 |
| Fischer et al. 2017 [13] | NS1 | 2 |
| Weidmann et al. 2010 [14] | 5'-UTR | 3 |
| Own design | not specified | 7 |
| Own design-adapted from [4] | 5'-UTR | 1 |
| Fast Track diagnostics Tropic fever Africa (commercial) | unknown | 1 |
| Genesig (commercial) | unknown | 1 |
| Real Star YFV RT-PCR kit 1.0 (commercial) | unknown | 1 |
| In-house YFV vaccine strain specific | ||
| Bae et al., 2003 [8] | NS3 | 2 |
| Fernandes-Monteiro et al. 2015 [10] | NS5 | 1 |
| Mantel et al. 2008 [9] | NS5 | 2 |
| Pan-flavivirus | ||
| Ayers et al. 2006 + seq [15] | NS5 | 1 |
| Moureau et al. 2007 + seq [16] | NS5 | 1 |
| Patel et al. 2013 + seq [17] | NS5 | 3 |
| Scaramozzino et al. 2001 + seq [18] | NS5 | 4 |
| Own design (pan-flavivirus) + seq | unknown | 1 |
| Patel et al. 2013 [17] | NS5 | 1 |
| Scaramozzino et al. 2001 [18] | NS5 | 1 |
| Genekam (commercial pan-flavivirus) | unknown | 1 |
| Other, no YFV detection | ||
| Fast Track diagnostics Tropic fever Asia | NA | 1 |
| Total number of submitted tests | NA | 46 |
NA: not applicable; YFV: yellow fever virus.
Twenty-three laboratories used one method for detection, while 10 used a combination of two methods and one laboratory combined three methods (Supplement). For the laboratories that used more than one test, the submitted results could have represented a routine combinatorial diagnostic approach e.g. combining a YF-17D specific with a YFV wildtype-specific PCR assay. Although the individually reported assays could not give a perfect overall result, the logical combination of more than one assay might give a satisfactory end result at the laboratory level.
At the laboratory level 18 of 34 participating laboratories had at least one test that scored the presence/absence of YFV RNA in the panel 100% correctly (Category I Supplement). At the country level this corresponded to 13 of 23 EU/EEA countries and one non-EU/EEA European country. A within-laboratory combination of test results did not yield any additional laboratories with a result that was 100% correct. These results indicate that there is a definite need for improvement of current YFV molecular diagnostics in 16 of the 34 participating laboratories and in 10 of the 23 participating EU/EEA countries respectively. At the test level, 20 of 46 submitted test results were 100% correct (Supplement).
Samples containing YF wildtype strains were missed by the three vaccine-strain-specific RT-PCR tests (Table 2, Supplement) [8-10]. Of the five laboratories submitting results based on YFV 17D-strain-specific tests, two laboratories in one EU/EEA country did not have an alternative or complementary parallel test available; meaning that these laboratories would miss YF in travellers returning from regions with YFV activity.
As all the YFV positive samples in the panel were representative for the range of viremia observed in clinical samples of acute YFV cases [11,12] it highlights the need for improvement of sensitivity in all laboratories that missed the samples with lower RNA loads (Category II Supplement). Furthermore, combinational testing by laboratories showed different performances related to the sensitivity of the individual assays and/or the performance across the laboratories.
Differences in the detection of the samples as an effect of virus strain were not significant and were mostly related to the characteristics of the assay used (Table 2, Supplement).
Among the exceptions, was one laboratory that missed all wild-type YFV samples using RT-PCR systems based on Drosten et al. [8] and Patel et al. [12]. This could imply the issues were at a laboratory-specific level, as multiple laboratories analysed the complete panel correctly based on these RT-PCR tests (Supplement). Another laboratory missed all YFV RNA positive samples (both vaccine strain and wildtype YFV based) as they used a commercial kit that does not detect YFV. One laboratory had only capacity for pan-flavivirus testing without sequencing and could not distinguish between the YFV RNA positive samples and the sample containing a mixture of flavivirus RNA. Finally, four laboratories indicated to have detected YFV RNA in one or more YFV RNA negative samples which is suggestive for contamination issues.
Discussion
Since December 2017, Brazil has experienced an increase in YFV cases with 1,266 confirmed and 1,232 suspected human cases in the period 1 July 2017 to 16 May 2018 [19]. The majority of notifications were from January to April 2018. The appearance of epizootics in non-human primates and of the first human cases in close vicinity to the metropolitan areas of Sao Paulo and Rio de Janeiro in early 2018, raised concerns about higher risks of exposure for international travellers [1,3]. On 15 January 2018, the first imported case related to the Sao Paulo metropolitan area was reported from the Netherlands [2] in a returning traveller showing the potential risk of international spread [20]. It was estimated that 1.2 million travellers would return from Brazil to Europe during the peak of the YFV season (December to May) [3], therefore, on 25 January 2018, the ECDC asked EVD-LabNet to assess the quality of YFV molecular diagnostics in its member laboratories. During and after the EQA, there have been eight additional YFV cases imported to Europe; one to the Czech Republic, two to France, three to Germany (one of which reported by UK), one to Romania and one to Switzerland [21]. This warrants the need for a reliable diagnostic capability and adequate capacity to support individual patient care, surveillance and response activities in Europe.
Of the 30 EU/EEA countries, five non-participating countries indicated that they have no test, two non-participating countries had a test available and 10 of the 23 participating countries showed insufficient capability for molecular testing for YFV. This indicates room for improvement in at least half of the countries. As this EQA assessed the diagnostic workflow as a whole from sample receipt to result reporting, it is not possible to recommend specific RT-PCR tests over others, as the influence of e.g. nucleic acid extraction methodology cannot be excluded.
Two laboratories only applied a vaccine strain-specific RT-PCR, meaning that they would miss YF cases in returning travellers. This issue was already noted in a previous YF EQA [4] and could be solved by the laboratories through use of another primary assay or using a complementary one. These laboratories were located in a country with endemic presence of Ae. albopictus that is a potential vector for YFV transmission [22-24]. One laboratory only had capacity for pan-flavivirus testing without sequencing, which without specific follow-up could potentially create interpretation issues in returning travellers presenting with other flavivius diseases or in areas where flaviviruses are endemic; DENV and YFV have an overlapping clinical manifestation and geographic distribution [25]. WNV, tick-borne encephalitis virus (TBEV) and Usutu virus are endemic in partially overlapping areas in Europe [26]. This laboratory would benefit from pre-arranged access to confirmatory testing.
The variety in RT-PCR systems used in the EQA reflects the absence of standardisation in wildtype YFV diagnostics across European laboratories potentially hampering consistent case finding and reporting.
The results of the EQA were reported back to the individual laboratories in April 2018. Within one month, three laboratories in three EU/EEA countries informed the EVD-LabNet that they have taken successful actions to improve their capability. New protocols have been shared with two laboratories underlining the value of EQA exercises in laboratory preparedness and response activities.
Acknowledgements
This work was supported by the European Centre for Disease Prevention and Control (ECDC) under specific contract no. 3-ECD.8456 for implementation of the framework contract ECDC/2016/002. We thank all EQA participants: Medical University of Vienna, Department of Virology, Vienna, Austria; Clinical Sciences, Institute of Tropical Medicine, Antwerp, Belgium; Microbiology Department, National Center of Infectious and Parasitic Diseases, Sofia, Bulgaria; Research Unit, University Hospital for Infectious Diseases Dr. Fran Mihaljevic´, Zagreb, Croatia; Molecular Virology, Cyprus Institute of Neurology and Genetics, Nicosia, Cyprus; Microbiological Diagnostics and Virology, Statens Serum Institut, Copenhagen, Denmark; Virology and Immunology, HUSLAB, Helsinki University Hospital, Helsinki, Finland; Unite des Virus Emergents UVE, Aix Marseille Univ, Marseille, France; Institute of Novel and Emerging Infectious Diseases at the Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany; Institut für Virologie, Marburg, Germany; Institute of Virology, Charite - Universitätsmedizin Berlin, Berlin, Germany; Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany; Department of Microbiology, Aristotle University of Thessaloniki, Thessaloniki, Greece; Division for Virology, National Center for Epidemiology, Budapest, Hungary; National Virus Reference Laboratory, U.C.D., Dublin, Ireland; National Reference Laboratory for Arboviruses-Department of Infectious, Parasitic and Immune-Mediated Diseases, Istituto Superiore di Sanità, Rome, Italy; U.O. Microbiology (CRREM), Az. Ospedaliero-Universitaria di Bologna, Policlinico S.Orsola-Malpighi, Bologna, Italy; Microbiology and Virology Unit, Padua University Hospital, Padua, Italy; Laboratory of Virology, National Institute for Infectious Diseases Lazzaro Spallanzani IRCCS, Rome, Italy; Scientific Department, Army Medical Center, Rome, Italy; Microbiology and Virology, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy; Infectology Centre of Latvia, National Microbiology Reference Laboratory, Riga, Latvia; Clinical Research Unit, National Public Health Surveillance Laboratory, Vilnius, Lithuania; Laboratoire National de Santé, Dudelange, Luxembourg; Virology Laboratory, BTICC of Military Insitute of Hygiene and Epidmeiology, Pulawy, Poland; Center for Vectors and Infectious Diseases Research, National Institute of Health, Lisbon, Portugal; Institute of Microbiology and Immunology, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia; Laboratorio de Arbovirus y Enfermedades Viricas Importadas, Instituto de Salud Carlos III, Madrid, Spain; Hospital Clinic, Barcelona, Spain; Public Health Agency of Sweden, Unit for Diagnostics Preparedness of Notifiable and High Consequence Pathogens, Solna, Sweden; Spiez Laboratory, Spiez, Switzerland; Laboratory of Virology, University hospitals of Geneva, Switzerland; Department of Viroscience, Erasmus MC, Rotterdam, The Netherlands; Rare and Imported Pathogens Laboratory, National Infection Services, Public Health England, Porton Down, Salisbury, United Kingdom. In addition we thank the technicians providing laboratory support at RKI, Erasmus MC and Aix University Marseille.
Conflict of interest: None declared.
Authors’ contributions: CD, HE, MK, AN, KL, RC, CR were involved with the study design and study preparation. CR, CD,HE were involved with the data collection and analysis. CR, CDC drafted the manuscript. RC, AN, MK, KL co-wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.European Centre for Disease Prevention and Control (ECDC). Rapid Risk Assessment: Yellow fever among travellers returning from South America. Stockholm: ECDC; 2017. Available from: http://ecdc.europa.eu/en/publications/Publications/14-03-2017-RRA-Yellow%20fever,%20Flaviviridae-Suriname,%20Southern%20America.pdf
- 2.ProMED-mail YELLOW FEVER - NETHERLANDS: ex BRAZIL (SAO PAULO). Archive number. 20180115. 5561671 15 Jan 2018. Available from: http://www.promedmail.org [Google Scholar]
- 3.Gossner CM, Haussig JM, de Bellegarde de Saint Lary C, Kaasik Aaslav K, Schlagenhauf P, Sudre B. Increased risk of yellow fever infections among unvaccinated European travellers due to ongoing outbreak in Brazil, July 2017 to March 2018. Euro Surveill. 2018;23(11):1800106. 10.2807/1560-7917.ES.2018.23.11.18-00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domingo C, Patel P, Yillah J, Weidmann M, Méndez JA, Nakouné ER, et al. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. J Clin Microbiol. 2012;50(12):4054-60. 10.1128/JCM.01799-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel P, Landt O, Kaiser M, Faye O, Koppe T, Lass U, et al. Development of one-step quantitative reverse transcription PCR for the rapid detection of flaviviruses. Virol J. 2013;10(1):58. 10.1186/1743-422X-10-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linke S, Ellerbrok H, Niedrig M, Nitsche A, Pauli G. Detection of West Nile virus lineages 1 and 2 by real-time PCR. J Virol Methods. 2007;146(1-2):355-8. 10.1016/j.jviromet.2007.05.021 [DOI] [PubMed] [Google Scholar]
- 7.Drosten C, Göttig S, Schilling S, Asper M, Panning M, Schmitz H, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40(7):2323-30. 10.1128/JCM.40.7.2323-2330.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae HG, Nitsche A, Teichmann A, Biel SS, Niedrig M. Detection of yellow fever virus: a comparison of quantitative real-time PCR and plaque assay. J Virol Methods. 2003;110(2):185-91. 10.1016/S0166-0934(03)00129-0 [DOI] [PubMed] [Google Scholar]
- 9.Mantel N, Aguirre M, Gulia S, Girerd-Chambaz Y, Colombani S, Moste C, et al. Standardized quantitative RT-PCR assays for quantitation of yellow fever and chimeric yellow fever-dengue vaccines. J Virol Methods. 2008;151(1):40-6. 10.1016/j.jviromet.2008.03.026 [DOI] [PubMed] [Google Scholar]
- 10.Fernandes-Monteiro AG, Trindade GF, Yamamura AM, Moreira OC, de Paula VS, Duarte AC, et al. New approaches for the standardization and validation of a real-time qPCR assay using TaqMan probes for quantification of yellow fever virus on clinical samples with high quality parameters. Hum Vaccin Immunother. 2015;11(7):1865-71. 10.4161/21645515.2014.990854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbosa CM, Di Paola N, Cunha MP, Rodrigues-Jesus MJ, Araujo DB, Silveira VB, et al. Yellow Fever Virus RNA in Urine and Semen of Convalescent Patient, Brazil. Emerg Infect Dis. 2018;24(1):176-8. 10.3201/eid2401.171310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wouthuyzen-Bakker M, Knoester M, van den Berg AP. GeurtsvanKessel CH, Koopmans MP, Van Leer-Buter C, et al. Yellow fever in a traveller returning from Suriname to the Netherlands, March 2017. Euro Surveill. 2017;22(11):30488. 10.2807/1560-7917.ES.2017.22.11.30488 [DOI] [PMC free article] [PubMed]
- 13.Fischer C, Torres MC, Patel P, Moreira-Soto A, Gould EA, Charrel RN, et al. Lineage-Specific Real-Time RT-PCR for Yellow Fever Virus Outbreak Surveillance, Brazil. Emerg Infect Dis. 2017;23(11):1867-71. 10.3201/eid2311.171131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidmann M, Faye O, Faye O, Kranaster R, Marx A, Nunes MR, et al. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J Clin Virol. 2010;48(3):187-92. 10.1016/j.jcv.2010.04.013 [DOI] [PubMed] [Google Scholar]
- 15.Ayers M, Adachi D, Johnson G, Andonova M, Drebot M, Tellier R. A single tube RT-PCR assay for the detection of mosquito-borne flaviviruses. J Virol Methods. 2006;135(2):235-9. 10.1016/j.jviromet.2006.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moureau G, Temmam S, Gonzalez JP, Charrel RN, Grard G, de Lamballerie X. A real-time RT-PCR method for the universal detection and identification of flaviviruses. Vector Borne Zoonotic Dis. 2007;7(4):467-77. 10.1089/vbz.2007.0206 [DOI] [PubMed] [Google Scholar]
- 17.Patel P, Landt O, Kaiser M, Faye O, Koppe T, Lass U, et al. Development of one-step quantitative reverse transcription PCR for the rapid detection of flaviviruses. Virol J. 2013;10(1):58. 10.1186/1743-422X-10-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scaramozzino N, Crance JM, Jouan A, DeBriel DA, Stoll F, Garin D. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol. 2001;39(5):1922-7. 10.1128/JCM.39.5.1922-1927.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brazilian Ministry of Health (IPFS). Monitoramento do Período Sazonal da Febre Amarela Brasil – 2017/2018; informe n° 26 | 2017/2018. [Monitoring of the Seasonal Period of Yellow Fever Brazil - 2017/2018; report n ° 26 | 2017/2018]. Brasilia: IPFS; 2018. (Brazilian). Available from: http://portalarquivos2.saude.gov.br/images/pdf/2018/maio/18/Informe-FA-26.pdf
- 20.Brent SE, Watts A, Cetron M, German M, Kraemer MU, Bogoch II, et al. International travel between global urban centres vulnerable to yellow fever transmission. Bull World Health Organ. 2018;96(5):343-354B. 10.2471/BLT.17.205658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Centre for Disease Prevention and Control (ECDC). Communicable disease threats report, week 22, 27 May-2 June 2018. Stockholm: ECDC; 2018 Available from: https://ecdc.europa.eu/sites/portal/files/documents/Communicable-disease-threats-report-2-june-2018.pdf
- 22.European Centre for Disease Prevention and Control (ECDC). Exotic vectors: mosquito maps. Stockholm: ECDC; 2016. Available from: http://ecdc.europa.eu/en/healthtopics/vectors/vector-maps/Pages/VBORNET_maps.aspx
- 23.Couto-Lima D, Madec Y, Bersot MI, Campos SS, Motta MA, Santos FBD, et al. Potential risk of re-emergence of urban transmission of Yellow Fever virus in Brazil facilitated by competent Aedes populations. Sci Rep. 2017;7(1):4848. 10.1038/s41598-017-05186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization (WHO). Yellow fever – Brazil; Disease outbreak news 27 February 2018. Geneva: WHO; 2018.Available from: http://www.who.int/csr/don/27-february-2018-yellow-fever-brazil/en/
- 25.Cleton N, Koopmans M, Reimerink J, Godeke GJ, Reusken C. Come fly with me: review of clinically important arboviruses for global travelers. J Clin Virol. 2012;55(3):191-203. 10.1016/j.jcv.2012.07.004 [DOI] [PubMed] [Google Scholar]
- 26.Reusken CBEM, Ieven M, Sigfrid L, Eckerle I, Koopmans M. Laboratory preparedness and response with a focus on arboviruses in Europe. Clin Microbiol Infect. 2018;24(3):221-8. 10.1016/j.cmi.2017.12.010 [DOI] [PubMed] [Google Scholar]


