Abstract
Platycodonis radix is extensively used for treating cough, excessive phlegm, sore throat, bronchitis and asthma in the clinic. Meanwhile, the stems, leaves and seeds of Platycodon grandiflorum (PG) have some pharmaceutical activities such as anti-inflammation and anti-oxidation effects, etc. These effects must be caused by the different metabolites in various parts of herb. In order to profile the different parts of PG, the ultra-high performance liquid chromatography combined with quadrupole time-of- flight mass spectrometry (UPLC-QTOF-MSE) coupled with UNIFI platform and multivariate statistical analyses was used in this study. Consequently, for the constituent screening, 73, 42, 35, 44 compounds were characterized from the root, stem, leaf and seed, respectively. The stem, leaf and seed contain more flavonoids but few saponins that can be easily discriminated in the root. For the metabolomic analysis, 15, 5, 7, 11 robust biomarkers enabling the differentiation among root, stem, leaf and seed, were discovered. These biomarkers can be used for rapid identification of four different parts of PG grown in northeast China.
Keywords: Platycodon grandiflorum, nontargeted metabolomic analysis, different part, UPLC-QTOF-MSE
1. Introduction
It is well-known that there are both chemical and pharmacological differences in different parts of herbs. Taking Aristolochia mollissima Hance as an example, the fruits are used to treat cough and asthma, the roots have obvious antihypertensive effects, while the stems and leaves are rheumatoid medicines. This phenomenon also exists in other herbs, such as Lycium barbarum, Polygonum Multiflorum Thunb., Trichosanthes kirilowii Maxim, Ephedra sinice Stapf, etc. [1].
As both food and medicine, Platycodon grandiflorum (Jacq.) A. DC. (PG) is known as “Jiegeng” in China, “Huridunzhaga” in Mongolia, “Kikyo” in Japan and “Doraji” in North Korea [2]. In clinical, the root of PG which has various biological activities, such as apophlegmatic and antitussive [3], anti-inflammation [4], immunoregulation [5], anti-oxidant [6], etc., has been widely used for the treatment of cough, excessive phlegm, and sore throat. In addition, the stem and leaf of PG also have anti-inflammatory [7] and anti-oxidant [8,9] activities, while research on the pharmacological effects of PG seed is currently non-existent.
PG is a rich source of different natural products with various structural patterns. Around 100 compounds have been isolated from the roots of PG, including steroidal saponins, flavonoids, phenolic acids, polyacetylenes, sterols, etc. [2]. Triterpenoid saponins, mainly of the oleanane family pentacyclic type, are the active components of the root of PG [10]. Several flavonoids and phenolic acids were isolated from the aerial parts of PG [11]. Two glycosides and four flavonoids were isolated from the seeds of PG [12]. Recently, instead of traditional separation and identification method, a combination of ultra-high performance liquid chromatography (UHPLC) separation, quadrupole time-of-flight tandem mass spectrometry (QTOF-MS/MS) detection and automated data processing software UNIFI with scientific library was innovatively used for screening and identifying chemical components in herbal medicines [13,14] and traditional Chinese medicine formulas [15]. In 2015, Lee et al. reported the global profiling of various metabolites in PG by UPLC-QTOF/MS [16]. In that paper, a total of 20 metabolites were characterized from the roots, and 56 compounds from stems and leaves of PG grown in Korea. Herbs collected from different regions will show certain differences both in chemical constituents and in pharmacological activities [17]. For example, saponins in the root of PG from different sites in Gyeongnam Province, Korea showed different contents [18]. The 1H-NMR-based metabolomics with OPLS-DA statistical models was used to cluster the ginseng samples from Korea and China, and the result suggested that the chemical profiles from two countries are quite different due to their different geographical origins [19]. Hence, in order to illustrate different chemical constituents from the different regions and from the different parts of the plants, and to better clarify the pharmacological fundamental substances of PG, the root, stem, leaf and seed of PG produced in Jilin Province, China were taken as samples in this paper.
Metabolomics, including targeted and untargeted complementary approaches, is primarily concerned with identification and quantitation of small-molecule metabolites (<1500 Da) [20]. Recently, because of its ability to profile diverse classes of metabolites, untargeted metabolomics has been widely used to compare the overall metabolic composition of different samples [21]. An untargeted analysis approach is mainly applied in metabolite identification through mass-based search followed by manual verification [20] Being a sensitive, efficient, reliable, accurate and nondestructive method, UPLC-QTOF-MS has been widely used recently in this kind of analysis, such as exploring the early detection of mycotoxins in wheat [22], estimating compliance to a dietary pattern [23], exploring the bioavailability of the secoiridoids from a seed/fruit extract in human healthy volunteers [24], evaluating the enantioselective metabolic perturbations in MCF-7 cells after treatment with R-metalaxyl and S-metalaxyl [25].
In this study we focus on both the quickly chemical components’ screening and the non-targeted metabolomic analysis of the root, stem, leaf and seed of PG. UPLC-QTOF-MSE, UNIFI platform and multivariate statistical analyses, such as principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were used to profile the four different plant parts and to find the biomarkers among these four parts of PG grown in northeast China.
2. Results
2.1. Identification of Components from Different Parts of PG
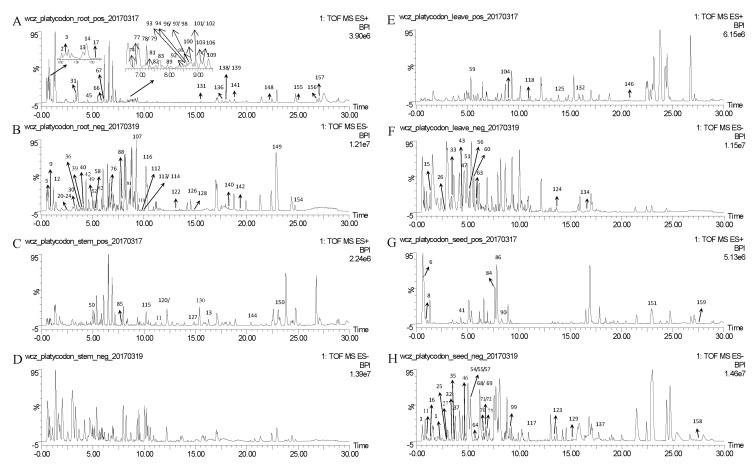
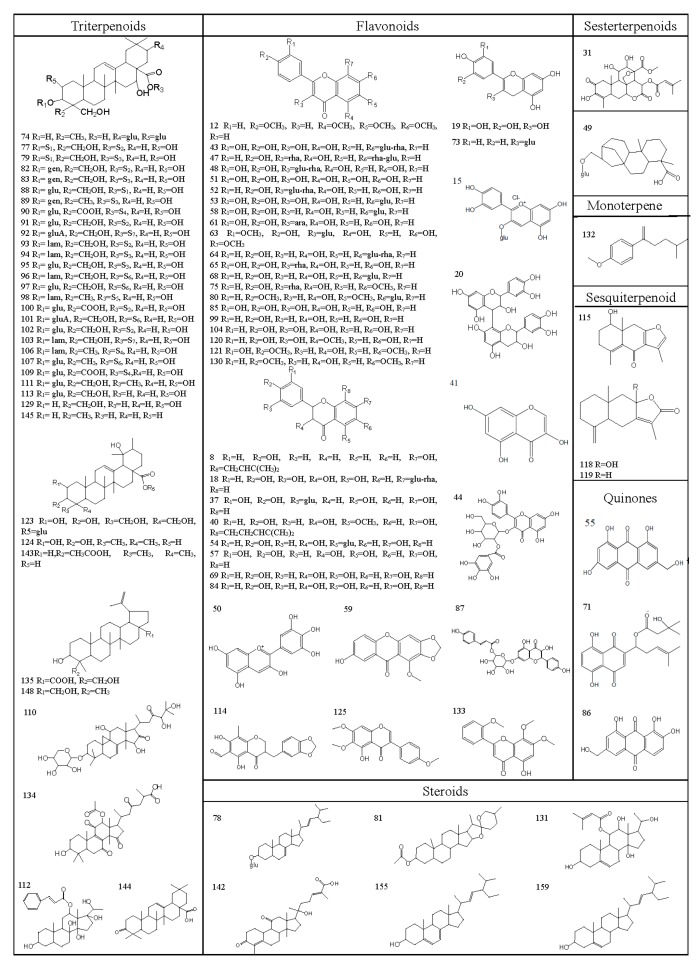
As a result, a total of 159 compounds were identified or tentatively characterized in both positive and negative mode from the four parts of PG, the base peak intensity (BPI) chromatograms are shown in Figure 1, and their chemical structures are shown in Figure 2. More specifically, 73, 42, 35, 44 compounds were characterized from the root, stem, leaf and seed respectively (Table 1), including triterpenoid saponins, organic acids, steroids, phenols, flavonoids, alcohols, amino acids, coumarins, terpenoids, alkaloids and amides and so on.
Figure 1.
The representative base peak intensity (BPI) chromatograms of root in positive (A) and negative (B) modes; of stem in positive (C) and negative (D) modes; of leaf in positive (E) and negative (F) modes; of seed in positive (G) and negative (H) modes.
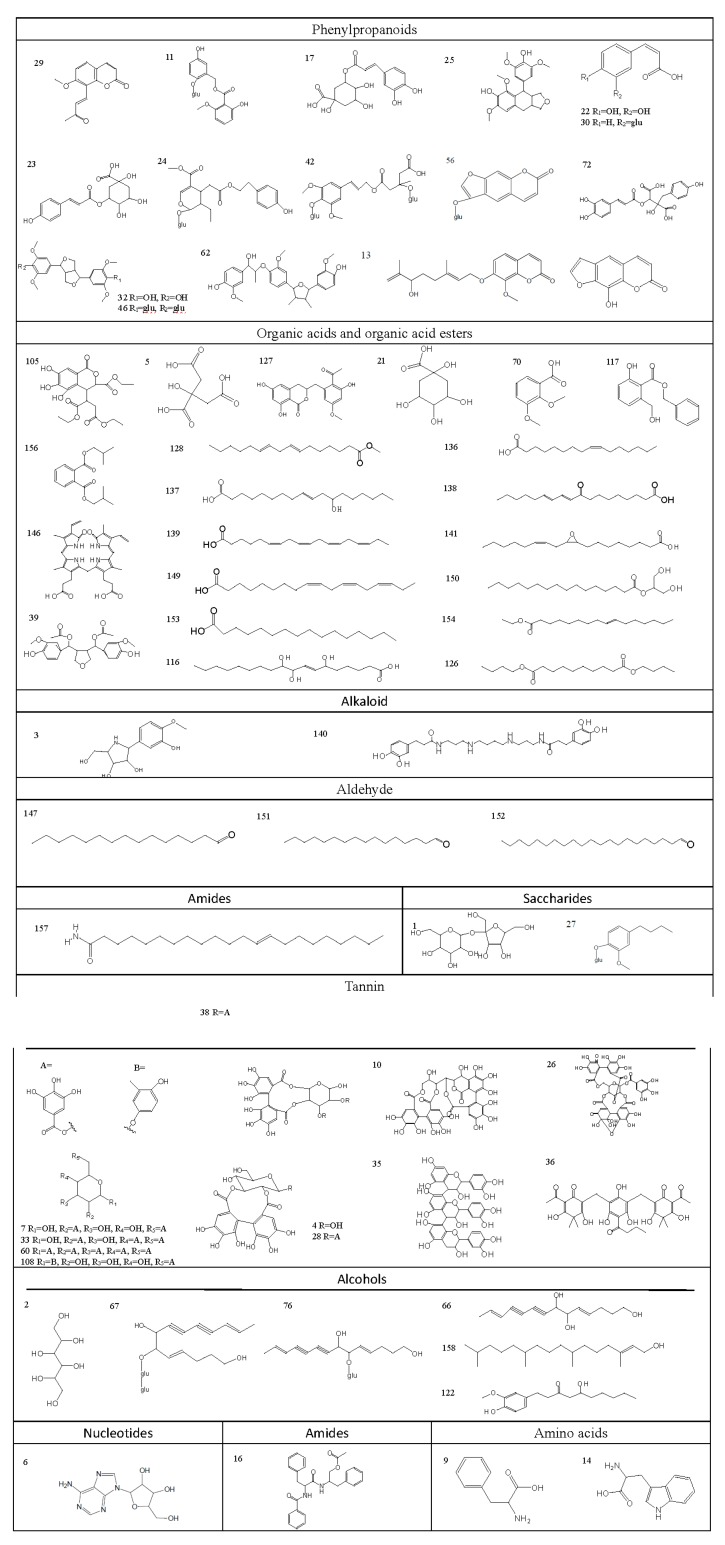
Figure 2.
Chemical structures of compounds identified in PG.
Table 1.
Compounds identified from different parts of PG by UPLC-QTOF-MSE.
| No. | tR (min) | Formula | Experimental (Da) | Theoretical (Da) | Mass Error (ppm) | Adducts | MSE Fragmentation | Component Name | Source |
|---|---|---|---|---|---|---|---|---|---|
| 1 * | 0.59 | C12H22O11 | 342.1169 | 342.1162 | 2.04 | −H | 323.0984, 195.0510, 161.0465 | Sucrose | D |
| 2 * | 0.60 | C6H14O6 | 182.0797 | 182.0790 | 3.04 | +Na | 205.0689, 152.0713 | Mannitol | R |
| 3 | 0.67 | C12H17NO5 | 255.1114 | 255.1107 | 2.91 | +H | 256.1114, 226.1074, 122.0375 | Radicamine A | R |
| 4 | 0.68 | C20H18O14 | 482.0682 | 482.0697 | −2.95 | −H | 343.0676, 301.0007, 274.0119, 191.0554, 152.0124 | 2,3-(S)-Hexahydroxydiphenoyl-d-glucose a | S, L |
| 5 * | 0.71 | C6H8O7 | 192.0278 | 192.0270 | 3.93 | −H | 191.0205, 173.0077, 111.0089 | Citric acid | R |
| 6 * | 0.75 | C10H13N5O4 | 267.0974 | 267.0968 | 2.23 | +H | 218.1020, 136.0634 | Adenosine | D |
| 7 | 0.82 | C20H20O14 | 484.0857 | 484.0853 | 0.78 | −H | 313.0568, 183.0308, 169.0156, 152.0123 | 2,6-Di-O-Galloyl-β-d-glucose a | S, L |
| 8 | 0.85 | C20H20O4 | 324.1347 | 324.1362 | −4.38 | +H | 203.0708, 175.0758, 164.0463, 149.0602, 103.0556 | Isobavachin a | D |
| 9 * | 0.86 | C9H11NO2 | 165.0796 | 165.0790 | 3.98 | −H | 164.0724, 147.0456, 103.0549 | Phenylalanine | R |
| 10 | 0.95 | C34H24O22 | 784.0751 | 784.0759 | −1.05 | −H | 421.0417, 337.0214, 249.0416, 182.0223, 168.0074, 149.9967 | Casuariin a | S |
| 11 | 0.97 | C21H24O11 | 452.1341 | 452.1319 | 4.86 | −H | 299.0771, 289.0737, 271.0611, 165.0206, 137.0257 | Curculigoside B a | D |
| 12 | 1.02 | C19H18O6 | 342.1089 | 342.1103 | −4.14 | −H | 211.0628, 181.0506, 179.0349, 161.0240, 151.0404 | 5,6,7,4′-Tetramethoxyflavone a | R |
| 13 | 1.24 | C20H24O5 | 344.1609 | 344.1624 | −3.98 | +Na | 222.0916, 194.0973, 182.0611, 127.0394 | Schininallylol a | R |
| 14 * | 1.35 | C11H12N2O2 | 204.0903 | 204.0899 | 2.29 | +H | 188.0706, 144.0808, 132.0813, 118.0661 | Tryptophan | R |
| 15 | 1.36 | C21H21ClO11 | 484.0775 | 484.0772 | 0.45 | −H | 309.0630, 287.0594, 124.0163, 109.0291 | Cyanidin 3-glucoside a | L |
| 16 | 1.37 | C27H28N2O4 | 444.2034 | 444.2049 | −3.41 | −H | 235.1215, 175.0626, 173.0464, 131.0364, 105.0356 | Aurantiamide acetate a | D |
| 17 * | 1.73 | C16H18O9 | 354.0950 | 354.0951 | −0.22 | +H | 192.0663, 163.0396, 145.0294, 135.0452 | Chlorogenic acid | R |
| 18 | 2.15 | C27H32O16 | 612.1712 | 612.1690 | 3.51 | −H | 593.1511, 461.1313, 303.0532, 285.0428, 177.0209, 151.0052 | (2R,3R)-Taxifolin7-O-α-l-rhamnopyranosyl-(1→6)-β-d-glucopyranoside | D |
| 19 | 2.30 | C30H26O12 | 578.1430 | 578.1424 | 0.98 | −H | 449.0876, 425.0875, 407.0777, 289.0718, 125.0257 | Procyanidin B1 a | D |
| 20 * | 2.31 | C15H14O7 | 306.0738 | 306.0740 | −0.53 | +HCOO | 179.0349, 167.0343, 163.0406, 161.0241, 109.0315 | Gallocatechin | R |
| 21 * | 2.34 | C7H12O6 | 192.0637 | 192.0634 | 1.53 | −H | 173.0480, 127.0406, 116.0514, 111.0456 | Quinine acid | R |
| 22 * | 2.35 | C9H8O4 | 180.0425 | 180.0423 | 1.60 | −H | 161.0241, 135.0451, 133.0297, 109.0315, 108.0224 | Caffeic acid | R |
| 23 | 2.36 | C16H18O8 | 338.0993 | 338.1002 | −2.62 | −H | 191.0567, 177.0195, 161.0243, 119.0505, 105.0351 | 3-O-trans-Coumaroylquinic acid | R |
| 24 | 2.70 | C25H34O12 | 526.2045 | 526.2050 | −1.04 | −H | 363.1452, 315.1244, 179.0713, 167.0711, 149.0612 | LucidumosideA a | R |
| 25 | 2.78 | C22H26O7 | 402.1670 | 402.1679 | −1.88 | +HCOO | 327.0884, 303.0885, 297.0421, 209.0844, 137.0256 | Neociwujiaphenol a | D |
| 26 | 2.81 | C41H28O27 | 952.0809 | 952.0818 | −0.97 | −H | 605.0777, 479.0469, 481.0642, 453.0677, 246.0169 | Geraniin a | L |
| 27 | 2.98 | C17H26O7 | 342.1678 | 342.1679 | −0.01 | +HCOO | 281.0651, 163.1130, 121.0300 | Citrusin C | D |
| 28 | 2.99 | C27H22O18 | 634.0813 | 634.0806 | 1.16 | −H | 601.0460, 463.0518, 419.0617, 301.0007, 291.0156, 275.0208 | Sanguiin H-4 a | S |
| 29 | 3.05 | C14H12O4 | 244.0745 | 244.0736 | 3.14 | +HCOO | 203.0721, 187.0402, 161.0250, 123.0457, 109.0303 | cis-Osthenone | D |
| 30 | 3.24 | C15H18O8 | 326.1003 | 326.1002 | 0.33 | +HCOO | 162.0552, 129.0199, 121.0304 | 4-O-β-d-glucopyranosyl-trans-cinnamic acid a | R, D |
| 31 * | 3.28 | C26H32O11 | 520.1968 | 520.1945 | 4.46 | +H | 443.0984, 341.1392, 163.075 | Brusatol | R |
| 32 | 3.42 | C22H26O8 | 418.1631 | 418.1628 | 0.72 | −H | 359.1465, 179.0726, 164.0477, 149.0251, 125.0254 | (+)-Syringaresinol | D |
| 33 | 3.55 | C27H24O18 | 636.0969 | 636.0963 | 0.99 | −H | 483.0791, 465.0679, 331.0667, 313.0578, 169.0163 | 2,4,6-Tri-O-galloyl-β-d-glucose a | S,L |
| 34 * | 3.65 | C11H6O4 | 202.0260 | 202.0266 | −2.32 | +HCOO | 163.0419, 149.0244, 134.0373, 133.0304, | Xanthotoxol | S, L |
| 35 | 3.67 | C45H38O18 | 866.2079 | 866.2058 | 2.37 | −H | 575.1207, 407.0781, 289.0730, 179.0356 | Arecatannin A1 a | D |
| 36 | 3.76 | C32H36O12 | 612.2223 | 612.2207 | 2.59 | −H | 562.1866, 518.1583, 210.0880, 135.0462 | Filixic acid ABA a | R |
| 37 | 3.78 | C21H22O12 | 466.1128 | 466.1111 | 3.59 | −H | 285.0428, 177.0208, 165.0568, 151.0053, 137.0257, 124.0178 | Taxifolin-3-O-glucoside a | D |
| 38 | 3.80 | C34H26O22 | 786.0915 | 786.0916 | −0.08 | −H | 615.0646, 597.0511, 445.0416, 301.0021, 125.0258 | Collinin a | S |
| 39 | 3.82 | C24H28O9 | 460.1739 | 460.1733 | 1.24 | −H | 414.1699, 389.1244, 193.0528, 137.0261, 125.0258 | Sanjidin A a | R |
| 40 | 4.30 | C22H24O6 | 384.1560 | 384.1573 | −2.96 | +HCOO | 325.1065, 313.1078, 310.0838, 150.0322 | Sophoflavescenol a | R |
| 41 | 4.33 | C9H6O5 | 194.0211 | 194.0215 | −2.35 | +H | 177.0183, 153.0178, 138.0309, 127.0398 | 3,5,7-Trihydroxychromone | D |
| 42 | 4.38 | C29H42O18 | 678.2395 | 678.2371 | 3.54 | −H | 497.1692, 453.1789, 323.0997, 291.1258, 161.0471 | TangshenosideI | R |
| 43 | 4.48 | C27H30O16 | 610.1554 | 610.1534 | 3.24 | −H | 463.0844, 313.0580, 265.0370, 190.9983, 151.0043 | Quercetin-7-O-rutinoside | L |
| 44 | 4.50 | C28H24O16 | 616.1084 | 616.1064 | 3.27 | −H | 313.0580, 190.9983, 177.0206, 169.0158, 151.0043 | 2′′-O-Galloylhyperoside a | S, L |
| 45 | 4.53 | C11H12O3 | 192.0791 | 192.0786 | 2.21 | +H | 193.0863, 167.0703, 161.0603 | Myristicin | R |
| 46 | 4.66 | C34H46O18 | 742.2707 | 742.2684 | 2.90 | +HCOO | 579.2040, 417.1564, 181.0520, 149.0248 | Syringaresinol-di-O-β-d-glucoside a | D |
| 47 | 4.72 | C33H40O19 | 740.2178 | 740.2164 | 1.92 | −H | 593.1506, 575.1401, 429.0824, 335.0414, 284.0336 | Grosvenorine a | S, L |
| 48 * | 4.93 | C27H30O16 | 610.1550 | 610.1534 | 2.59 | −H | 401.0912, 301.0365, 299.0205, 247.0609 | Rutin | S, L, D |
| 49 | 4.94 | C26H42O8 | 482.2874 | 482.2880 | −1.13 | +HCOO | 261.1352,179.1074,149.0608, 125.0589 | 17-O-β-d-Glucopyra-nosyl-16β-H-ent-kauran-19-oicacid a | R |
| 50 * | 4.96 | C15H10O7 | 302.0427 | 302.0427 | 0.15 | +H | 161.0264, 123.0099, 109.0306, 107.0153 | Delphinidin | S, L |
| 51 | 4.97 | C15H10O8 | 318.0368 | 318.0376 | −2.25 | +HCOO | 300.0266, 264.0562, 176.0132, 148.0176 | Quercetagetin | L |
| 52 | 5.12 | C27H30O15 | 594.1609 | 594.1585 | 4.02 | −H | 285.0403, 161.0459, 151.0038, 135.0452 | Kaempferol-3-O-neohesperidoside | R |
| 53 | 5.14 | C21H20O12 | 464.0945 | 464.0955 | −2.16 | −H | 313.0549, 300.0266, 284.0330, 151.0041 | Quercimeritrin | S,L,D |
| 54 | 5.17 | C21H22O11 | 450.1178 | 450.1162 | 3.61 | −H | 193.0156, 179.0574, 175.0051, 151.0052, 135.0468 | Dihydrokaempferol-5-O-β-d-glucopyranoside | D |
| 55 | 5.24 | C15H10O6 | 286.0463 | 286.0477 | −4.95 | −H | 256.0372, 177.0180, 164.0487, 150.0300, 123.0439, 107.0134 | ω-Hydroxyemodin a | D |
| 56 | 5.25 | C17H16O9 | 364.0780 | 364.0794 | −3.53 | +HCOO | 337.0566, 278.0432, 202.0248, 185.0254, 149.0251 | Bergaptol-O-β-d-glucopyranoside | L |
| 57 | 5.26 | C15H12O7 | 304.0568 | 304.0583 | −4.92 | −H | 285.0366, 243.0329, 152.0099, 150.0300, 125.0238 | Dihydroquercetin | D |
| 58 | 5.27 | C21H20O11 | 448.1005 | 448.1006 | −0.19 | −H | 285.0406, 283.0256, 179.0569 | Luteolin-7-O-glucopyranoside | R,D |
| 59 | 5.40 | C15H10O6 | 286.0479 | 286.0477 | 0.70 | +H | 149.0216, 139.0371, 123.0433, 111.0439 | 7-Hydroxy-1-methoxy-2-methoxyxanthone a | S, L |
| 60 | 5.57 | C41H32O26 | 940.1163 | 940.1182 | −1.96 | −H | 769.0887, 617.0782, 313.0565, 291.0150, 169.0158 | 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside a | S, L |
| 61 | 5.72 | C20H18O11 | 434.0853 | 434.0849 | 0.80 | −H | 300.0301, 195.0321, 151.0050, 109.0305 | Quercetin-3-O-arabinoside | S |
| 62 | 5.76 | C30H36O8 | 524.2409 | 524.2410 | −0.19 | +HCOO | 453.1908, 339.1256, 195.0667, 165.0570 | Saucerneol C a | R |
| 63 | 5.79 | C23H24O13 | 508.1224 | 508.1217 | 1.36 | −H | 315.0519, 207.0291, 193.0506, 151.0044, 137.0246 | Limocitrin-3-O-β-d-glucopyranoside a | L |
| 64 | 5.83 | C27H30O14 | 578.1637 | 578.1636 | 0.24 | −H | 269.0475, 227.0364, 177.0203, 151.0050, 119.0513 | Apigenin-7-O-β-d-rutinoside | D |
| 65 | 5.84 | C21H20O11 | 448.1016 | 448.1006 | 2.36 | −H | 295.0843, 284.0340, 179.0362, 151.0411, 123.0102 | Quercetin-3-O-α-l-rhamnoside | S |
| 66 | 5.86 | C14H18O3 | 234.1243 | 234.1256 | −4.58 | +H | 175.0746, 163.0746, 133.0647, 119.0860, 111.0811 | Lobetyol | R |
| 67 | 6.02 | C26H38O13 | 558.2326 | 558.2312 | 2.37 | +Na | 217.1197, 199.1096, 145.0642, 128.0613, 115.0541 | Lobetyolinin | R |
| 68 | 6.12 | C21H20O10 | 432.1040 | 432.1056 | −3.82 | −H | 268.0367, 227.0341, 177.0181, 151.0037, 124.0168 | Cosmosiin | D |
| 69 | 6.17 | C15H12O6 | 288.0643 | 288.0634 | 3.23 | −H | 271.0623, 177.0181, 151.0037, 133.0297, 125.0254, 107.0143 | Dihydrokaempferol | D |
| 70 | 6.32 | C9H10O4 | 182.0584 | 182.0579 | 2.65 | −H | 166.0263, 151.0040, 135.0452, 108.0226 | 2,6-Dimethoxy benzoic acid | D |
| 71 | 6.59 | C21H24O7 | 388.1509 | 388.1522 | −2.96 | +HCOO | 358.1066, 301.0369, 243.0306, 231.0308, 151.0047 | β-Hydroxyisovalerylshikonin a | D |
| 72 | 6.61 | C20H18O10 | 418.0892 | 418.0900 | −1.74 | +HCOO | 358.1066, 243.0306, 231.0308, 178.9997, 151.0047, 121.0304 | Cimicifugic acid D a | D |
| 73 | 6.70 | C21H24O10 | 436.1373 | 436.1369 | 0.76 | −H | 273.0781, 255.0666, 179.0358, 149.0248, 123.0457 | Epiafzelechin-3-O-β-d-allopyranoside a | D |
| 74 | 6.75 | C42H68O16 | 828.4491 | 828.4507 | −1.99 | +H | 667.4052, 651.4104, 505.3529, 487.3428, 469.3321, 421.3113 | Platycosaponin A | R |
| 75 | 6.79 | C22H22O10 | 446.1231 | 446.1213 | 3.66 | +HCOO | 285.0424, 187.0053, 163.0414, 124.0179 | Rhamnocitrin-3-O-rhamnoside a | S |
| 76 | 6.81 | C20H28O8 | 396.1793 | 396.1784 | 2.03 | +HCOO | 215.1094, 185.0984, 159.0826, 143.0724, 125.0616 | Lobetyolin | R |
| 77 * | 6.85 | C64H104O34 | 1416.6388 | 1416.6409 | −1.49 | +H | 811.4487, 763.42581, 647.37911, 485.3261 | Deapio platycoside E | R |
| 78 | 6.93 | C35H58O6 | 574.4227 | 574.4233 | −1.03 | +H | 472.3166, 463.3096, 378.2044, 302.1716 | α-Spinasterol glucoside | R |
| 79 * | 6.98 | C69H112O38 | 1548.6799 | 1548.6832 | −2.13 | +H | 1007.5104, 845.4571, 683.4034, 521.3493, 485.3282 | Platycoside E | R |
| 80 | 6.99 | C23H24O11 | 476.1314 | 476.1319 | −0.84 | +HCOO | 433.1097, 345.0819, 313.0554, 183.0309, 151.0041 | 5-Hydroxy-6,4′-dimethoxy-flavone-7-O-β-d-gluco-pyranoside | S |
| 81 | 7.35 | C29H46O4 | 458.3396 | 458.3396 | −0.05 | +H | 341.2455, 217.1953, 149.1333, 121.1027 | Neotigogenin acetate a | R |
| 82 | 7.57 | C58H94O29 | 1254.5905 | 1254.5881 | 1.95 | +H | 931.4894, 845.4518, 799.4485, 295.1007 | Deapioplatycodin D3 | R |
| 83 * | 7.68 | C63H102O33 | 1386.6326 | 1386.6303 | 1.65 | +H | 1255.5937, 931.4894, 845.4518, 799.4484, 665.3879, 441.1585 | Platycodin D3 | R |
| 84 | 7.69 | C15H12O6 | 288.0629 | 288.0634 | −1.59 | +H | 255.0652, 179.0353, 163.0400, 153.0196, 145.0295 | 3-Hydroxynaringenin a | D |
| 85 * | 7.77 | C15H10O7 | 302.0422 | 302.0427 | −1.40 | +H | 243.0319, 151.0055, 125.0260, 107.0157 | Quercetin | S, L |
| 86 | 7.86 | C15H10O6 | 286.0488 | 286.0477 | 3.61 | +H | 269.0460, 257.0450, 241.0490, 161.0239, 135.0453 | 6-Hydroxyaloeemodin a | D |
| 87 | 7.91 | C30H26O13 | 594.1373 | 594.1373 | −0.14 | −H | 447.0966, 429.0832, 285.0440, 145.0316, 119.0513 | Buddlenoid A a | S, L |
| 88 | 7.92 | C47H76O20 | 960.4934 | 960.4930 | 0.39 | +HCOO | 869.4537, 715.3371, 529.2698, 295.2034 | Platycoside F | R |
| 89 * | 7.94 | C63H102O32 | 1370.6373 | 1370.6354 | 1.40 | +H | 827.4398, 783.4476, 637.3944, 459.3430, 409.3090, 325.1130 | Platycoside G3 | R |
| 90 | 8.33 | C57H90O29 | 1238.5577 | 1238.5568 | 0.71 | +H | 1107.5237, 957.4692, 895.4676, 811.4125, 697.3760, 661.3582, 485.3245, 409.3094 | Platyconic acid A | D |
| 91 * | 8.46 | C52H84O24 | 1092.5397 | 1092.5353 | 4.07 | −H | 959.4846, 941.4753, 681.3871, 663.3768, 649.3607, 503.3364, 485.3366, 295.1038, 277.0942 | Deapioplatycodin D | R |
| 92 | 8.48 | C59H92O30 | 1280.5649 | 1280.5673 | −1.90 | +H | 1017.4875, 999.4760, 931.4860, 829.4192, 697.3796, 679.3651, 651.3761, 519.3316, 503.3334, 487.3377 | Platycodin L | R |
| 93 * | 8.51 | C58H94O29 | 1254.5847 | 1254.5881 | −2.65 | +H | 931.4894, 845.4518, 799.4485, 483.3065, 457.1533, 427.1433, 325.1116, 295.1007 | Deapioplatycodin D2 | R |
| 94 * | 8.62 | C63H102O33 | 1386.6300 | 1386.6303 | −0.26 | +H | 977.4981, 845.4558, 829.4604, 683.4031, 667.4073, 653.3919, 521.3488, 485.3273 | Platycodin D2 | R |
| 95 * | 8.68 | C57H92O28 | 1224.5778 | 1224.5775 | 0.23 | +H | 799.4485, 683.3961, 667.4052, 521.3444, 503.3364, 485.3257 | Platycodin D | R, D |
| 96 * | 8.73 | C65H104O34 | 1428.6407 | 1428.6409 | −0.15 | +H | 1297.6065, 1165.5621, 845.4520, 841.4580, 681.3837, 665.3903, 653.3884, 617.3663, 519.3298, 485.3243 | 2′-O-Acetylplatycodin D2 | R, D |
| 97 | 8.78 | C59H94O29 | 1266.5869 | 1266.5881 | −0.93 | +H | 1003.5108, 841.4569, 823.4458, 683.3979, 189.0749, 171.0641 | Platycodin A | R, D |
| 98 | 8.80 | C65H104O33 | 1412.6458 | 1412.6460 | −0.16 | +H | 985.4990, 823.4461, 635.3794, 617.3695, 503.3369, 453.1605, 321.1182, 303.1076, 189.5707 | 3′′-O-Acetylpolygalacin D2 | R |
| 99 | 8.86 | C15H10O5 | 270.0539 | 270.0528 | 4.15 | −H | 151.0043, 123.0099, 117.0359, 107.0154 | Apigenol | D |
| 100 | 8.87 | C52H82O25 | 1106.5163 | 1106.5145 | 1.57 | +H | 975.4806, 931.4908, 829.4243, 811.4113, 697.3814, 679.3695, 517.3151, 503.3373, 455.3161 | Platyconic acid C | R |
| 101 | 8.94 | C59H92O30 | 1280.5705 | 1280.5673 | 2.47 | +H | 1017.4875, 829.4192, 697.3796, 637.3939, 519.3316, 321.1178 | Platycodin K | R, D |
| 102 | 9.04 | C54H86O25 | 1134.5444 | 1134.5458 | −1.23 | +H | 1003.5108, 841.4569, 823.4458, 683.3979, 321.1160, 189.0749 | Platycoside B | R |
| 103 * | 9.10 | C65H104O34 | 1428.6370 | 1428.6409 | −2.71 | +H | 1297.6065, 955.4894, 841.4580, 813.4279, 797.4332, 681.3837, 665.3903, 653.3884, 635.3780 | 3′-O-acetyl-platycodin D2 | R |
| 104 | 9.11 | C15H10O6 | 286.0483 | 286.0477 | 1.85 | +H | 231.0662, 229.0504, 195.0289, 153.0187, | Kaempferol | L |
| 105 | 9.14 | C20H24O11 | 440.1314 | 440.1319 | −1.01 | −H | 393.0860, 303.0523, 257.0104, 231.0303, 177.0204 | (-)-Chebulic acid triethyl ester a | S, L |
| 106 | 9.18 | C65H104O33 | 1412.6430 | 1412.6460 | −2.14 | +H | 823.4461, 503.3369, 485.3255, 455.3156, 321.1182, 189.0757 | 2′′-O-acetylpolygalacin D2 | R, D |
| 107 | 9.23 | C59H94O28 | 1250.5904 | 1250.5932 | −2.23 | −H | 1208.5857, 1159.5571, 635.3812, 499.3046, 131.0337 | 2′-O-acetyl Polygalacin D | R |
| 108 | 9.32 | C20H22O11 | 438.1170 | 438.1162 | 1.78 | −H | 419.0956, 235.0654, 163.0050 | 6′-O-Galloyl-homoarbutin a | S, L |
| 109 | 9.37 | C54H84O26 | 1148.5293 | 1148.5251 | 3.63 | +H | 1017.4908, 999.4786, 535.3279, 631.3477, 517.3170, 499.3050, 453.3001, 321.1190, 189.0764 | Platyconic acid D | R |
| 110 | 9.45 | C35H54O11 | 650.3666 | 650.3666 | 0.04 | +HCOO | 451.2830, 441.2997, 197.1183, 149.0465, 131.0354 | 15α-Hydroxy-ximicifugoside H2 a | R |
| 111 | 9.59 | C37H60O12 | 696.4087 | 696.4085 | 0.28 | −H | 487.3424, 469.3302, 425.3438 | 3-O-d-glucopyranosyl platycodigenin methyl ester | S |
| 112 | 9.80 | C30H42O7 | 514.2938 | 514.2931 | 1.41 | −H | 436.2610, 319.1910, 301.1814, 265.1468 | Marstenacigenin A | R |
| 113 | 9.91 | C36H58O12 | 682.3893 | 682.3928 | −4.81 | +HCOO | 635.3797, 449.3263, 407.2948, 179.0565 | 3-O-d-glucopyranosyl platycodigenin | R |
| 114 | 9.94 | C19H16O7 | 356.0886 | 356.0896 | −2.55 | +HCOO | 401.0868, 313.0718, 121.0297 | 6-Formyl-isoophiopogonanone A a | R |
| 115 | 10.17 | C15H18O3 | 246.1258 | 246.1256 | 0.84 | +H | 229.1220, 163.0756, 149.0598, 119.0865, 105.0713 | Curcolone a | S, L |
| 116 | 10.25 | C18H34O5 | 330.2418 | 330.2406 | 3.57 | −H | 311.2224, 293.2140, 211.1348, 185.1189, 129.0928 | Sanleng acid a | R, S, D |
| 117 | 10.91 | C15H14O4 | 258.0901 | 258.0892 | 3.45 | −H | 239.0705, 163.0397, 151.0421, 133.0313, 121.0296 | Benzyl-2-hydroxy-6-methoxybenzoate | D |
| 118 * | 10.95 | C15H20O3 | 248.1413 | 248.1412 | 0.27 | +H | 231.1379, 219.1381, 203.1425, 119.0864, 107.0867 | Atractylenolide ІІІ | L |
| 119 | 11.13 | C15H20O2 | 232.1464 | 232.1463 | 0.24 | +H | 215.1424, 187.1486, 159.1172, 135.1174, 107.0867 | Atractylenolide ІІ | S, L |
| 120 | 12.19 | C16H12O6 | 300.0637 | 300.0634 | 1.18 | +H | 285.0761, 242.0571, 167.0340, 136.0162, 108.0215 | 5-Methyl kaempferol | S, L |
| 121 | 12.26 | C17H14O6 | 314.0794 | 314.0790 | 1.05 | +H | 299.0552, 275.0673, 257.0445, 161.0597, 139.0397 | 3′,5-Dihydroxy-7,4′-dimethoxy flavone | S |
| 122 | 12.94 | C17H26O4 | 294.1833 | 294.1831 | 0.56 | −H | 235.1341, 141.0919, 129.0924 | 6-Gingerol a | R |
| 123 | 13.46 | C36H58O12 | 682.3905 | 682.3928 | −3.36 | −H | 635.3787, 473.3258, 443.3119, 425.3020, 179.0553 | Trachelosperoside B-1 a | D |
| 124 | 13.68 | C30H48O5 | 488.3514 | 488.3502 | 2.47 | −H | 455.3548, 439.3599, 281.2503, 293.2127, 171.1035 | 2α,19α-Dihydroxyursolic acid | L |
| 125 | 13.91 | C18H16O6 | 328.0949 | 328.0947 | 0.72 | +H | 314.0777, 296.0677, 184.0737, 136.0166 | 4′,7-Dimethyltectorigenin a | S, L |
| 126 * | 14.58 | C18H34O4 | 314.2466 | 314.2457 | 2.86 | −H | 201.1140, 199.0980, 155.1082, 127.1135 | Dibutyl sebacate | R |
| 127 | 14.85 | C19H18O7 | 358.1051 | 358.1053 | −0.47 | +H | 343.0809, 326.0778, 301.0705, 283.0599 | 3,4-Dihydro-6,8-dihydroxyl-3-(2′-acetyl-3′-hydroxyl-5′-methoxyphenyl)methyl-1H-[2] benzoplyran-1-one a | S, L |
| 128 | 14.86 | C17H30O2 | 266.2258 | 266.2246 | 3.76 | +HCOO | 311.2240, 155.1083, 139.1137 | Methyl 7, 10-hexadecadienoate | R |
| 129 | 15.36 | C30H48O7 | 520.3385 | 520.3400 | −2.93 | −H | 476.2774, 473.3256, 443.3168, 425.3093, 407.2940, 395.2941 | Platycodigenin | D |
| 130 | 15.39 | C17H14O5 | 298.0843 | 298.0841 | 0.51 | +H | 284.0679, 256.0730, 241.0495, 167.0339, 133.0648 | 5-Hydroxy-7, 4′-dimethoxyflavanone | S, L |
| 131 | 15.57 | C26H40O6 | 448.2818 | 448.2825 | −1.59 | +H | 393.2636, 350.1875, 242.1877 | Tenasogenin a | R |
| 132 | 15.89 | C14H20O | 204.1513 | 204.1514 | −0.51 | +H | 163.1118, 159.1169, 149.0956, 119.0863, 107.0502 | 2-(p-Anisyl)-5-methyl-1-hexen | L |
| 133 | 16.28 | C18H16O6 | 328.0957 | 328.0947 | 2.95 | +H | 314.0790, 299.0550, 286.0830, 271.0604, 150.0314 | 5-Hydro-7, 8, 2′-trimethoxyflavanone | S, L |
| 134 | 16.57 | C32H44O9 | 572.2965 | 572.2985 | −3.51 | −H | 481.2572, 429.2997, 227.0350, 183.1043 | Ganoderic acid H a | L |
| 135 | 17.23 | C30H48O4 | 472.3550 | 472.3553 | −0.49 | −H | 471.3448, 437.3061, 419.2937, 339.2705, 253.2187 | 2α-Hydroxybetulinic acid | S, L |
| 136 | 17.62 | C16H30O2 | 254.2252 | 254.2246 | 2.21 | +Na | 207.1743, 165.1274, 143.1067, 125.0961 | Palmitoleic acid | R |
| 137 | 17.78 | C18H34O3 | 298.2505 | 298.2508 | −1.05 | −H | 217.1615, 195.1391, 183.1401, 113.0984 | Ricinoleic acid | D |
| 138 | 18.00 | C18H30O3 | 294.2203 | 294.2195 | 2.51 | +Na | 277.2177, 165.1284, 151.1127, 109.1035 | (E,E)-9-Oxooctadeca-10,12-dienoic acid a | R |
| 139 | 18.01 | C18H28O2 | 276.2100 | 276.2089 | 3.85 | +H | 179.1424, 135.1180, 119.0862 | Stearidonic acid | R |
| 140 | 18.26 | C28H42N4O6 | 530.3100 | 530.3104 | −0.77 | −H | 529.3027, 511.2928, 293.2163 | Kukoamine A a | R |
| 141 | 19.02 | C18H32O3 | 296.2358 | 296.2351 | 2.19 | +Na | 279.2312, 161.1323, 147.1165, 133.1018, 121.1023 | Coronaric acid | R |
| 142 | 19.23 | C28H40O5 | 456.2878 | 456.2876 | 0.46 | −H | 409.2359, 343.1925, 339.2004, 275.2022 | Siraitic acid D a | R |
| 143 | 20.35 | C32H50O5 | 514.3662 | 514.3658 | 0.81 | −H | 495.3495, 469.3702, 451.3596, 449.3449 | 19α-Hydroxy-3-acetyl-ursolic acid | S |
| 144 | 20.39 | C30H46O3 | 454.3452 | 454.3447 | 1.03 | +H | 437.3422, 409.3470, 247.1695, 203.1796, 189.1642 | Oleanonic acid | S |
| 145 | 20.77 | C30H48O3 | 456.3604 | 456.3603 | 0.13 | −H | 455.3531, 443.3528, 233.1561 | 3-Epioleanolic acid | S |
| 146 | 20.78 | C33H36N4O6 | 584.2660 | 584.2635 | 4.08 | +Na | 567.2589, 535.2340, 501.2257, 467.20432, 417.1830 | Bilirubin a | L |
| 147 | 21.49 | C15H30O | 226.2309 | 226.2297 | 4.48 | +HCOO | 271.2302, 197.1911, 195.1754 | n-Pentadecanal | S |
| 148 | 22.20 | C30H50O2 | 442.3803 | 442.3811 | −1.76 | +H | 425.3776, 407.3666, 217.1950, 203.1791, 189.1641 | Betulin | R |
| 149 * | 22.93 | C18H30O2 | 280.2402 | 280.2400 | −0.25 | −H | 149.0972 | Linolenic acid | R |
| 150 * | 22.95 | C19H38O4 | 330.2774 | 330.2770 | 1.00 | +Na | 313.2738, 239.2368 | 1-Monopalmitin | S |
| 151 | 22.98 | C16H32O | 240.2452 | 240.2453 | −0.47 | +Na | 263.2344, 125.1317, 111.1175 | n-Hexadecanal | D |
| 152 | 24.06 | C21H42O | 310.3240 | 310.3236 | 1.28 | +HCOO | 355.3214, 125.0972 | n-Henicosanal | S |
| 153 | 24.40 | C16H32O2 | 256.2401 | 256.2402 | −0.49 | −H | 241.2176, 237.226, 227.2019, 125.0976 | Palmitic acid | S |
| 154 | 24.74 | C18H34O2 | 282.2569 | 282.2559 | 3.70 | −H | 253.2185, 163.1132, 125.0982, 111.0825 | Ethyl palmitate | R |
| 155 | 25.73 | C29H46O | 410.3565 | 410.3549 | 4.03 | +H | 395.3680, 203.1799, 145.1021, 133.1019 | Δ7-stigmasterol | R |
| 156 | 26.87 | C24H38O4 | 390.2771 | 390.2770 | 0.21 | +H | 301.1413, 189.0156, 165.0905, 149.0235 | Bis(2-ethylhexyl)phthalate | R |
| 157 | 27.09 | C22H43NO | 337.3356 | 337.3345 | 3.47 | +H | 321.3149, 212.2014, 198.1857, 153.1275 | Erucic amide a | R |
| 158 | 27.63 | C20H40O | 296.3093 | 296.3079 | 4.10 | +HCOO | 251.2393, 179.1459, 113.0987 | Phytol | S |
| 159 * | 28.49 | C29H48O | 412.3695 | 412.3705 | −2.48 | +H | 135.1178, 109.1025 | Stigmasterol | R |
* Identified with a reference standard. a Tentatively new identifications in Campanulaceae. The fragment ion mass highlighted as bold font is the characteristic MS fragmentation for each compound.
For the compounds which have isomers, they may be distinguished by their characteristic MS fragmentation patterns reported in literature, or may be compared with the retention times of reference standards. Taking compounds 98 and 106 as example, both have the same protonated ion [M + H]+ at m/z 1413.6530 and 1413.6530. In the results, they matched 3″-O-acetylpolygalacin D2 and 2″-O-acetylpolygalacin D2, respectively.
Their identical MS fragment pattern were similar. But according to the literature, the C3-glucoside was eluted earlier than the C2-glucoside [26,27,28] in the ESI-BPI chromatogram, so the compound with the earlier RT was identified as the C3-glucoside, 3″-O-acetylpolygalacin D2, and the other one with the later RT was identified as the C2-glucoside, 2″-O-acetylpolygalacin D2.
2.2. Biomarker Discovery for Differentiating Four Parts of PG
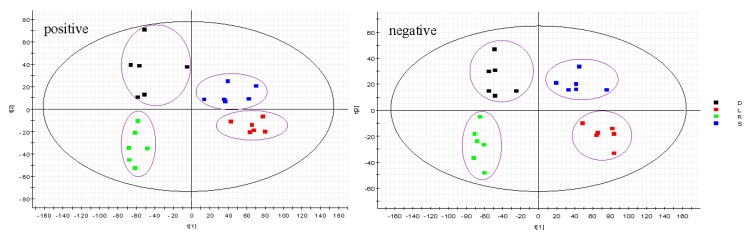
The PCA 2D plots of the samples from the root, stem, leaf and seed groups were classified in four clusters according to their common spectral characteristics (Figure 3). That means the four parts of PG could be easily differentiated.
Figure 3.
PCA of root (R), stem (S), leaf (L) and seed (D) of PG in positive mode and negative mode.
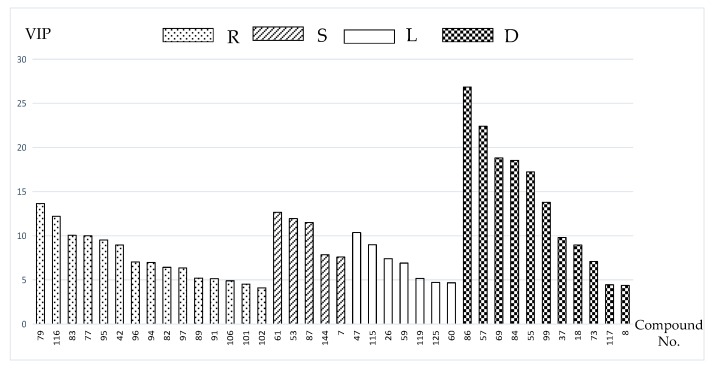
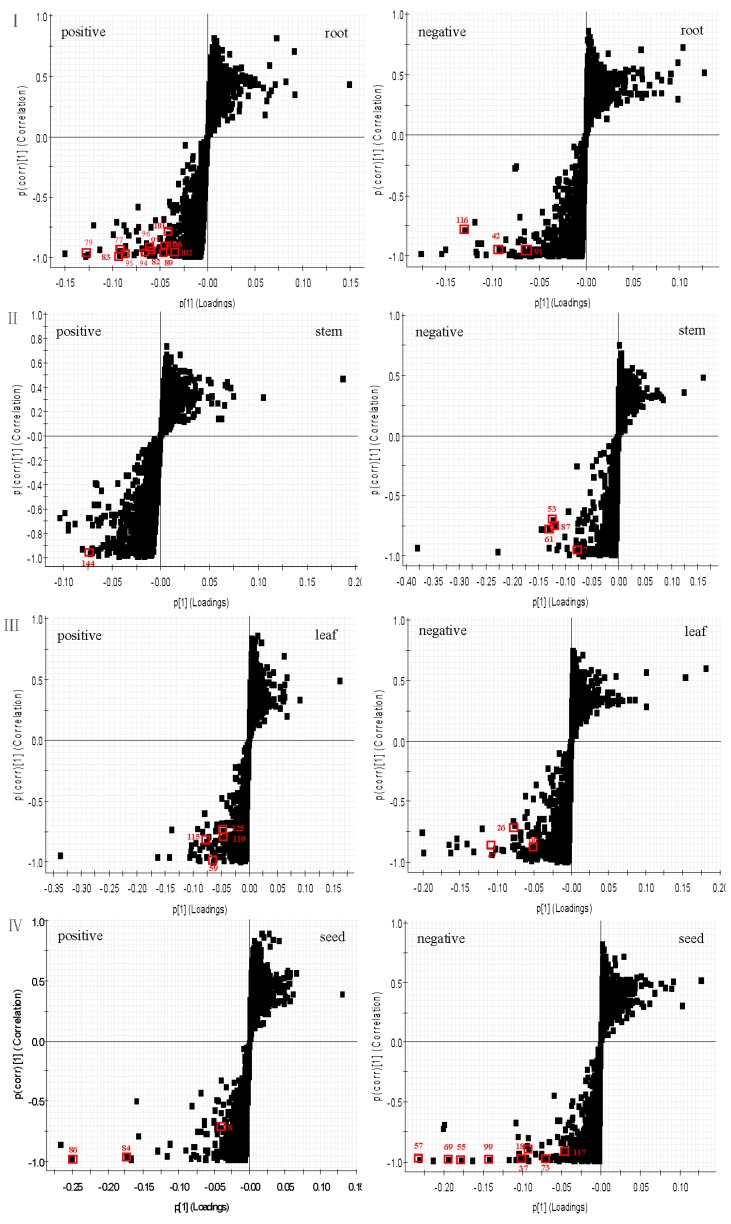
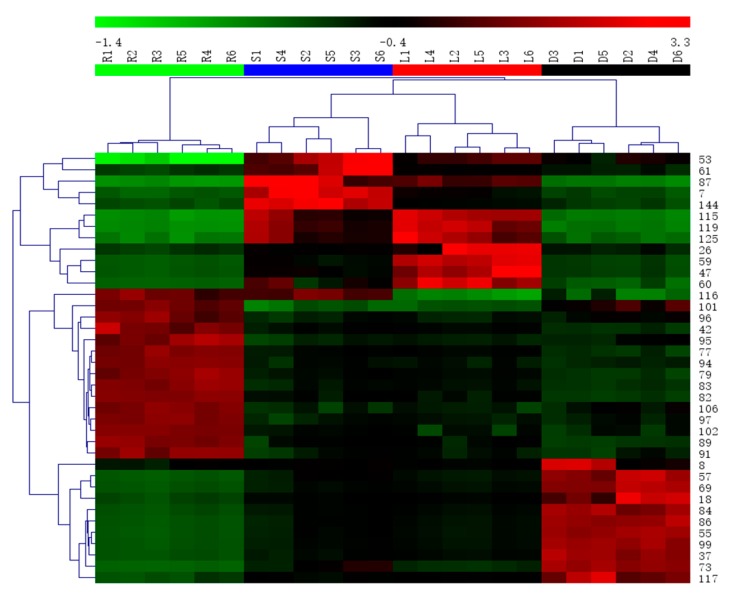
In order to differentiate one part from other three parts, the OPLS-DA models were built in both positive and negative modes. Then, OPLS-DA score plot, S-plot, variable trend and VIP (variable importance in the projection) values were obtained to understand which variables are the responsible for this sample separation [29]. Based on VIP values (VIP > 4) (Figure 4) and p values (p < 0.05) [30] from univariate statistical analysis, 38 robust known biomarkers enabling the differentiation among root, stem, leaf and seed, were discovered and marked in S-plots (Figure 5). In order to systematically evaluate the biomarkers, a heatmap was generated from these biomarkers (shown in Figure 6), which shows distinct segregation among the four parts.
Figure 4.
VIP value obtained from OPLS-DA model of the potential markers in root (R), stem (S), leaf (L) and seed (D) of PG.
Figure 5.
The OPLS-DA/S-plots of root (I), stem (II), leaf (III) and seed (IV) of PG in positive mode and negative mode.
Figure 6.
Heatmap visualizing the intensities of potential biomarkers.
3. Discussion
There are 73, 42, 35, 44 compounds that were characterized from the root, stem, leaf and seed, respectively. As the results show, 95 compounds were identified in ESI(−) mode and 64 compounds were identified in ESI(+) mode. According to the BPI chromatograms of the four parts of PG, it seems that ESI(−) ionization mode is better than ESI(+) based on the quantity and the responses of the identified compounds, but it is still necessary to run the ESI(+) mode because some compounds showed better respond than in ESI(−) mode.
Compared with the results from previous studies [2,8,16,31,32], 56 chemical components were identified for the first time in Campanulaceae. The stem, leaf and seed contain more flavonoids but few saponins that can be easily discriminated from the root. In previous study, various metabolites in Korean Platycodon grandiflorum were profiled by UPLC-QTOF/MS [16]. Compared with the root of PG in Korea, there were only nine constituents (compounds 5, 31, 76, 79, 83, 91, 94, 95, 97) in common. Meanwhile, the stems and leaves of PG in Korea and in China are both rich in natural components with various structural patterns, including triterpenoid saponins, flavonoids, organic acids, phenols, alcohols, amino acids, coumarins and amino acids, etc., but there are only two similar chemical components (compounds 99, 104). It is also interesting that there are eleven components (compounds 5, 14, 17, 21, 23, 31, 52, 83, 94, 95, 97) reported in stems and leaves of PG in Korea that were found in the root of PG in China. The reason for this phenomenon may be the different analytical methods and the different growing locations.
In this paper, 38 robust known biomarkers enabling the differentiation among root, stem, leaf and seed, were discovered. For the root part, there are 15 potential biomarkers including triterpenoid saponins (77, 79, 82, 83, 89, 91, 94, 95, 96, 97, 101, 102, 106), an organic acid (116) and a phenyl-propanoid (42). For stem part, there are five potential biomarkers including flavonoids (53, 61, 87), a tannin (7) and a triterpenoid saponin (144). For leaf part, there are seven potential biomarkers including flavonoids (47, 59, 125), sesquiterpenoids (115, 119) and tannins (26, 60). For seed part, there are 11 potential biomarkers including flavonoids (8, 18, 37, 57, 69, 73, 84, 99), quinones (55, 86) and an organic acid (117). These robust biomarkers enabling the differentiation among root, stem, leaf and seed can be used for rapid identification of four different parts of PG grown in northeast China.
Even so, there are still some unresolved issues. Firstly, pharmaceutical effects associated with these robust biomarkers or these identified compounds should be screened in the future. Additionally, as shown in BPI chromatograms, though 159 compounds were identified there are still many unidentified components. Further research should be carried on based on the formula of these unknown compounds [13]. Most importantly, the stems and leaves of PG should be developed and utilized due to the presence of so many different components from the root. This comprehensive and unique phytochemical profile study revealed the structural diversity of secondary metabolites and the different patterns in various parts of PG. The method developed in this study can be used as a standard protocol for discriminating and predicting parts of PG directly.
4. Experimental Section
4.1. Materials and Reagents
All samples were harvested from Jilin Province, China, as listed in Table 2, and identified by Professor Ping-Ya Li (School of Pharmaceutical Sciences, Jilin University, Changchun, China). The voucher specimens (No. 2016121-2016144) had been deposited at the Research Center of Natural Drug, School of Pharmaceutical Sciences, Jilin University, Changchun, China. The cultivation ages of the roots are all 2 years, while the others are all 1 year old.
Table 2.
Information of samples from Jilin Province, China.
| Collection Region | Mark of Samples | Collection Date | Collection Region | Mark of Samples | Collection Date |
|---|---|---|---|---|---|
| Antu County | S1 | 2 October 2016 | Fusong County | S4 | 4 October 2016 |
| L1 | 2 October 2016 | L4 | 4 October 2016 | ||
| R1 | 26 October 2016 | R4 | 30 October 2016 | ||
| D1 | 2 October 2016 | D4 | 4 October 2016 | ||
| Hunchun City | S2 | 1 October 2016 | Tonghua City | S5 | 5 October 2016 |
| L2 | 1 October 2016 | L5 | 5 October 2016 | ||
| R2 | 27 October 2016 | R5 | 28 October 2016 | ||
| D2 | 1 October 2016 | D5 | 5 October 2016 | ||
| Changbai County | S3 | 30 September 2016 | Jiaohe City | S6 | 3 October 2016 |
| L3 | 30 September 2016 | L6 | 3 October 2016 | ||
| R3 | 29 October 2016 | R6 | 25 October 2016 | ||
| D3 | 30 September 2016 | D6 | 3 October 2016 |
S: stem, L: leaf, R: root; D: seed.
Acetonitrile and methanol suitable for UHPLC-MS purchased from Fisher Chemical Company (Geel, Belgium). Formic acid for UPLC was purchased from Sigma-Aldrich (St. Louis, MO, USA). Deionized water was purified using a Millipore water purification system (Millipore, Billerica, MA, USA). All other chemicals were of analytical grade. Fourteen standard compounds including platycodin D (111851-201607), mannitol (100533-201304), citric acid (111679-201602), phenylalanine (140676-201405), tryptophan (140686-201303), chlorogenic acid (110753-201716), caffeic acid (110885-201102), dibutyl sebacate (190102-201501), linolenic acid (111631-201605), sucrose (111507-201303), adenosine (110879-201202), monopalmitin (190011-201302), rutin (100080-201610), quercetin (100081-201610), were purchased from the National Institutes for Food and Drug Control (Beijing, China). Seven standard compounds including gallocatechin (201512013), quinine acid (20150321), brusatol (20150410), stigmasterol (20150111), xanthotoxol (20109376), delphinidin (20159567), and atractylenolide ІІІ (2014712) were purchased from Beijing Putian Genesis Biotechnology Co., Ltd. (Beijing, China). Nine standard compounds including deapioplatycoside E (160712), deapioplatycodin D (160518), -D2 (160407), platycoside E (160112), platycodin D2 (160721), -D3 (160909), platycoside G3 (160921), 2′-O-acetyl-platycodin D2 (160112), 3′-O-acetylplatycodin D2 (160923) were provided by Institute of Frontier Medical Science of Jilin University (Changchun, China).
4.2. Sample Preparation and Extraction
The roots, stems, leaves and seeds of PG from the different sites were respectively air dried, ground and sieved (40 mesh) to give a homogeneous powder. Then 200 mg of the powder was respectively extracted thrice with 80% methanol at 80 °C for 3 h each time. After filtering, the extracts were combined, concentrated and evaporated to dryness. Finally, the desiccated extracts were dissolved and diluted with 80% methanol to 10.0 mL. The solution was filtered through a syringe filter (0.22 µm) and injected directly into the UPLC system. The volume injected was 2 μL for each run.
4.3. UPLC-QTOF-MSE
The UPLC analysis was performed by a Waters ACQUITY UPLC System. The column used was an ACQUITY UPLC BEH C18 (100 mm × 2.1 mm, 1.7 μm) from Waters Corporation (Milford, MA, USA). The mobile phases consisted of eluent A (0.1% formic acid in water, v/v) and eluent B (0.1% formic acid in acetonitrile, v/v) with flow rate of 0.4 mL/min with a liner gradient program: 10% B from 0 to 2 min, 10–90% B from 2 to 26 min, 90% B from 26 to 28 min, 90–10% B from 28 to 28.1 min, 10% B from 28.1 to 30 min. The temperature of the UPLC column and autosampler were set at 30 °C and 15 °C. Mixtures of 10/90 and 90/10 water/acetonitrile were used as the strong wash and the weak wash solvent respectively.
The MS experiments were performed on a Waters Xevo G2-S QTOF mass spectrometer (Waters Co., Milford, MA, USA.) connected to the UPLC system through an electrospray ionization (ESI) interface. The optimized instrumental parameters were as follows: capillary voltage floating at 2.6 kV (ESI+) or 2.2 kV (ESI−); cone voltage at 40 V; source temperature at 120 °C, desolvation temperature at 300 °C and cone gas flow was 50 L/h, desolvation gas flow was 800 L/h. In MSE mode, collision energy of low energy function was set at 6 V, while ramp collision energy of high energy function was set at 20–40 V. To ensure mass accuracy and reproducibility, the mass spectrometer was calibrated over a range of 100–1600 Da with sodium formate. Leucine-enkephalin (m/z 556.2771 in positive ion mode; m/z 554.2615 in negtive ion mode) was used as the lockmass at a concentration of 200 ng/mL and flow rate of 20 μL/min. Data were collected in continuum mode, all the acquisition of data were controlled by the Waters MassLynx v.4.1 software ( waters, Milford, MA, USA).
4.4. Data Analysis
For the screening analysis, the raw data were processed using the streamlined workflow of UNIFI 1.7.0 software (Waters, Manchester, UK) to quickly identify the chemical components [15]. Besides the Waters Traditional Medicine Library in the UNIFI software, a self-built database was created including the information of chemical components from PG based on the literature and on-line databases such as China Full-text Journals Database (CNKI), PubMed, Medline, Web of Science and ChemSpider. Minimum peak area of 200 was set for 2D peak detection.The peak intensity of high energy over 200 counts and over 1000 counts for low energy were the selected parameters in 3D peak detection. A margin of error up to 5 ppm for identified compounds was allowed. Positive adducts containing +H, +Na, and negative adducts including +COOH and −H were selected. The verification of compounds was carried out by comparison with retention time of reference standards and characteristic MS fragmentation patterns reported in literature.
For metabonomics analysis, the raw data were processed by MarkerLynx XS V4.1 software for alignment, deconvolution, data reduction, etc. [33]. As a result, the list of mass and retention time pairs with corresponding intensities for all the detected peaks from each data file. The main parameters were as follows: retention time range 0–28 min, mass range 100–1600 Da, mass tolerance 0.10, minimum intensity 5%, marker intensity threshold 2000 counts, mass window 0.10, retention time window 0.20, and noise elimination level 6. The resulting data were analyzed by principle component analysis (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA). S-plots and VIP-plots were obtained via OPLS-DA analysis to find potential biomarkers that significantly contributed to the difference among the groups.
5. Conclusions
In the present study, UPLC-QTOF-MSE coupled with UNIFI platform and precise multivariate statistical analyses was used to profile the four parts of PG. For the constituent screening under the optimized conditions, a total of 159 chemical compounds (73, 42, 35, 44 compounds characterized from root, stem, leaf and seed, respectively) were identified from PG. The results showed various structural patterns including triterpenoid saponins, organic acids, steroids, phenols, flavonoids, alcohols, amino acids, coumarins, terpenoids, alkaloids and amides. The stem, leaf and seed contain more flavonoids but few saponins that can be easily discriminated from the root.
For the metabolomic analysis, four parts of PG were successfully discriminated into four different clusters. A total of 38 robust biomarkers were discovered. That is to say, 15, 5, 7, and 11 robust biomarkers enabling the differentiation among root, stem, leaf and seed, were characterized. These biomarkers can be suitable for the simultaneous differentiation of four different parts of PG, which is reported for the first time. In a word, these results provided the reliable characterization profiles and the differentiate components among root, leaf, stem and seed of PG grown in northeast China. The method developed in this study can be used as a standard protocol for discriminating and predicting the different parts of PG directly.
Acknowledgments
This work was supported by Talents Team Major Program of Jilin Province of China (JRCBTZ. [2016] No. 3).
Author Contributions
Pingya Li and Jinping Liu conceived and designed the experiments; Cuizhu Wang, Nanqi Zhang and Zhenzhou Wang performed the experiments; Cuizhu Wang, Zeng Qi, Hailin Zhu and Bingzhen Zheng were responsible for data analysis. Cuizhu Wang wrote the paper. Jinping Liu and Pingya Li assisted paper revision.
Conflicts of Interest
The authors declare that they have no conflicts of interest concerning this article.
Footnotes
Sample Availability: Samples of the compounds are available from the authors.
References
- 1.Qi Y.F. The functional differences in different parts from the same plant. Inf. Tradit. Chin. Med. 1988;3:40–42. [Google Scholar]
- 2.Zhang L., Wang Y.L., Yang D.W., Zhang C.H., Zhang N., Li M.H., Liu Y.Z. Platycodon grandiflorus—An Ethnopharmacological, phytochemical and pharmacological review. J. Ethnopharmacol. 2015;164:147–161. doi: 10.1016/j.jep.2015.01.052. [DOI] [PubMed] [Google Scholar]
- 3.Choi J.H., Hwang Y.P., Lee H.S., Jeong H.G. Inhibitory effect of Platycodi Radix on ovalbumin-induced airway inflammation in a murine model of asthma. Food Chem. Toxicol. 2009;47:1272–1279. doi: 10.1016/j.fct.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Ahn K.S., Noh E.J., Zhao H.L., Jung S.H., Kang S.S., Kim Y.S. Inhibition of inducible nitric oxide synthase and cyclooxygenase II by Platycodon grandiflorum saponins via suppression of nuclear factor-κB activation in RAW 264.7 cells. Life Sci. 2005;76:2315–2328. doi: 10.1016/j.lfs.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 5.Xie Y., Pan H., Sun H., Li D. A promising balanced Th1 and Th2 directing immunological adjuvant, saponins from the root of Platycodon grandiflorum. Vaccine. 2008;26:3937–3945. doi: 10.1016/j.vaccine.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 6.Jeong C.H., Choi G.N., Kim J.H., Kwak J.H., Kim D.O., Kim Y.J., Heo H.J. Antioxidant activities from the aerial parts of Platycodon grandiflorum. Food Chem. 2010;118:278–282. doi: 10.1016/j.foodchem.2009.04.134. [DOI] [Google Scholar]
- 7.Güvenç A., Akkol E.K., Hürkul M.M., Süntar I., Keles H. Wound healing and anti-inflammatory activities of the Michauxia L’Hérit (Campanulaceae) species native to Turkey. J. Ethnopharmacol. 2012;139:401–408. doi: 10.1016/j.jep.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Jeong C.H., Shim K.H. Chemical Composition and Antioxidative Activities of Platycodon grandiflorum Leaves and Stems. J. Korean Soc. Food Sci. Nutr. 2006;35:685–708. [Google Scholar]
- 9.Liu D., Tan W. Nutritional composition and antioxidant activities of Platycodon grandiflorum flower and leaf. Agric. Food Ind. Hi Tech. 2016;27:44–46. [Google Scholar]
- 10.Choi J.H., Jin S.W., Choi C.Y., Kim H.G., Kim S.J., Lee H.S., Chung Y.C., Kim E.J., Lee Y.C., Jeong H.G. Saponins from the roots of Platycodon grandiflorum ameliorate high fat diet-induced non-alcoholic steatohepatitis. Biomed. Pharmacother. 2017;86:205–212. doi: 10.1016/j.biopha.2016.11.107. [DOI] [PubMed] [Google Scholar]
- 11.Mazol I., Gleńsk M., Cisowski W. Polyphenolic compounds from Platycodon grandiflorum A. DC. Acta Pol. Pharm. 2004;61:203–208. [PubMed] [Google Scholar]
- 12.Inada A., Yamada M., Murata H., Kobayashi M., Toya H., Kato Y., Nakanishi T. Phytochemical studies of seeds of medicinal plants. I. Two sulfated triterpenoid glycosides, sulfapatrinosides I and II, from seeds of Patrinia scabiosaefolia FISCHER. Chem. Pharm. Bull. 1988;36:4269–4274. doi: 10.1248/cpb.36.4269. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F.X., Li M., Qiao L.R., Yao Z.H., Li C., Shen X.Y., Wang Y., Yu K., Yao X.S., Dai Y. Rapid characterization of Ziziphi Spinosae Semen by UPLC/Qtof MS with novel informatics platform and its application in evaluation of two seeds from Ziziphus species. J. Pharm. Biomed. Anal. 2016:59–80. doi: 10.1016/j.jpba.2016.01.047. [DOI] [PubMed] [Google Scholar]
- 14.Deng L., Shi A.M., Liu H.Z., Meruva N., Liu L., Hu H., Yang Y., Huang C., Li P., Wang Q. Identification of Chemical Ingredients of Peanut Stems and Leaves Extracts using UPLC-QTOF-MS Coupled With Novel Informatics UNIFI Platform. J. Mass Spectrom. 2016;51:1157–1167. doi: 10.1002/jms.3887. [DOI] [PubMed] [Google Scholar]
- 15.Tang J.F., Li W.X., Tan X.J., Li P., Xiao X.H., Wang J.B., Zhu M.J., Li X.L., Meng F. A novel and improved UHPLC-QTOF/MS method for the rapid analysis of the chemical constituents of Danhong injection. Anal. Method. 2016;8:2904–2914. doi: 10.1039/C5AY03173G. [DOI] [Google Scholar]
- 16.Lee J.W., Ji S.H., Kim G.S., Song K.S., Um Y., Kim O.T., Lee Y., Hong C.P., Shin D.H., Kim C.K., et al. Global Profiling of Various Metabolites in Platycodon grandiflorum by UPLC-QTOF/MS. Int. J. Mol. Sci. 2015;16:26786–26796. doi: 10.3390/ijms161125993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H.P., Zhang Y.B., Yang X.W., Yang X.B., Xu W., Xu F., Cai S.Q., Wang Y.P., Xu Y.H., Zhang L.X. High-Performance Liquid Chromatography with Diode Array Detector and Electrospray Ionization Ion Trap Time-of-Flight Tandem Mass Spectrometry to Evaluate Ginseng Roots and Rhizomes from Different Regions. Molecules. 2016;2:603. doi: 10.3390/molecules21050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee B.J., Jeon S.H., Lee S.W., Chun H.S., Cho Y.S. Soil Physico-Chemistry and Saponins Content of Platycodon grandiflorum Radix Cultured from Different Sites in Gyeongnam Province. Korean J. Med. Crop. Sci. 2014;22:463–468. doi: 10.7783/KJMCS.2014.22.6.463. [DOI] [Google Scholar]
- 19.Nguyen H., Lee D.K., Choi Y.G., Min J.E., Yoon S.J., Yu Y.H., Lim J., Lee J., Kwon S.W., Park J.H. A 1H-NMR-based metabolomics approach to evaluate the geographical authenticity of herbal medicine and its application in building a model effectively assessing the mixing proportion of iIntentional admixtures: A case study of panax ginseng metabolomics for the authenticity of herbal medicine. J. Pharm. Biomed. Anal. 2016;124:120–128. doi: 10.1016/j.jpba.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Xiao J.F., Zhou B., Ressom H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC Trends Anal. Chem. 2012;32:1–14. doi: 10.1016/j.trac.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J.R., Yau L.F., Gao W.N., Liu Y., Yick P.W., Liu L., Jiang Z.H. Quantitative comparison and metabolite profiling of saponins in different parts of the root of Panax notoginseng. J. Agric. Food Chem. 2014;62:9024–9034. doi: 10.1021/jf502214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubert J., Righetti L., Stranska-Zachariasova M., Dzuman Z., Chrpova J., Dall’Asta C., Hajslova J. Untargeted metabolomics based on ultra-high-performance liquid chromatography–high-resolution mass spectrometry merged with chemometrics: A new predictable tool for an early detection of mycotoxins. Food Chem. 2016;224:423. doi: 10.1016/j.foodchem.2016.11.132. [DOI] [PubMed] [Google Scholar]
- 23.Andersen M.B., Rinnan Å., Manach C., Poulsen S.K., Pujos-Guillot E., Larsen T.M., Astrup A., Dragsted L.O. Untargeted metabolomics as a screening tool for estimating compliance to a dietary pattern. J. Proteome Res. 2014;13:1405–1418. doi: 10.1021/pr400964s. [DOI] [PubMed] [Google Scholar]
- 24.Garcíavillalba R., Tomásbarberán F.A., Fançaberthon P., Roller M., Zafrilla P., Issaly N., García-Conesa M.T. Targeted and Untargeted Metabolomics to Explore the Bioavailability of the Secoiridoids from a Seed/Fruit Extract (Fraxinus angustifolia Vahl) in Human Healthy Volunteers: A Preliminary Study. Molecules. 2015;20:22202. doi: 10.3390/molecules201219845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P., Zhu W., Wang D., Yan J., Wang Y., He L. Enantioselective Effects of Metalaxyl Enantiomers on Breast Cancer Cells Metabolic Profiling Using HPLC-QTOF-Based Metabolomics. Int. J. Mol. Sci. 2017;18:142. doi: 10.3390/ijms18010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng L.F., Kong H.J., Zhu M., Yan W.D. A facile method to evaluate the quality of Platycodon grandiflorum, A. De Candolle using reference standard extract. J. Funct. Foods. 2016;26:48–56. doi: 10.1016/j.jff.2016.07.008. [DOI] [Google Scholar]
- 27.Yoo D.S., Choi Y.H., Cha M.R., Lee B.H., Kim S.J., Yon G.H., Hong K.S., Jang Y.S., Lee H.S., Kim Y.S., et al. HPLC-ELSD analysis of 18 platycosides from balloon flower roots (Platycodi Radix) sourced from various regions in Korea and geographical clustering of the cultivation areas. Food Chem. 2011;129:645–651. doi: 10.1016/j.foodchem.2011.04.106. [DOI] [PubMed] [Google Scholar]
- 28.Ha Y.W., Na Y.C., Seo J.J., Kim S.N., Linhardt R.J., Kim Y.S. Qualitative and quantitative determination of ten major saponins in Platycodi Radix by high performance liquid chromatography with evaporative light scattering detection and mass spectrometry. J. Chromatogr. A. 2006;1135:27–35. doi: 10.1016/j.chroma.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira A.C.S., Monforte A.R., Teixeira C.S., Martins R., Fairbairn S., Bauer F.F. Monitoring Alcoholic Fermentation: An Untargeted Approach. J. Agric. Food Chem. 2014;62:6784–6793. doi: 10.1021/jf502082z. [DOI] [PubMed] [Google Scholar]
- 30.Zou Z.J., Liu Z.H., Gong M.J., Han B., Wang S.M., Liang S.W. Intervention effects of puerarin on blood stasis in rats revealed by a 1H-NMR-based metabonomic approach. Phytomedicine. 2015;22:333–343. doi: 10.1016/j.phymed.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 31.He J.Y., Ma N., Zhu S., Komastsu K., Li Z.Y., Fu W.M. The genus Codonopsis (Campanulaceae): A review of phytochemistry, bioactivity and quality control. J. Nat. Med. 2015;69:1–21. doi: 10.1007/s11418-014-0861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H.J., Kang S.H. Ethnobotany, Phytochemistry, Pharmacology of the Korean Campanulaceae: A Comprehensive Review. Korean J. Plant Res. 2017;30:240–264. doi: 10.7732/kjpr.2017.30.2.240. [DOI] [Google Scholar]
- 33.Zhao Y.Y., Cheng X.L., Wei F., Xiao X.Y., Sun W.J., Zhang Y., Lin R.C. Serum metabonomics study of adenine-induced chronic renal failure in rats by ultra performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Biomarkers. 2012;17:48–55. doi: 10.3109/1354750X.2011.637180. [DOI] [PubMed] [Google Scholar]