Abstract
Melanization is an intrinsic characteristic of many fungal species, but details of this process are poorly understood because melanins are notoriously difficult pigments to study. While studying the binding of cell-wall dyes, Eosin Y or Uvitex, to melanized and non-melanized Cryptococcus neoformans cells we noted that melanization leads to reduced fluorescence intensity, suggesting that melanin interfered with dye binding to the cell wall. The growth of C. neoformans in melanizing conditions with either of the cell-wall dyes resulted in an increase in supernatant-associated melanin, consistent with blockage of melanin attachment to the cell wall. This effect provided the opportunity to characterize melanin released into culture supernatants. Released melanin particles appeared mostly as networked structures having dimensions consistent with previously described extracellular vesicles. Hence, dye binding to the cell wall created conditions that resembled the ‘leaky melanin’ phenotype described for certain cell-wall mutants. In agreement with earlier studies on fungal melanins biosynthesis, our observations are supportive of a model whereby C. neoformans melanization proceeds by the attachment of melanin nanoparticles to the cell wall through chitin, chitosan, and various glucans.
Keywords: fungal, melanin, melanin anchor, vesicles, leaky melanin, Uvitex, Eosin Y
Introduction
Melanization is common in fungi and is associated with virulence among many pathogenic species. Melanin production serves a multitude of functions in fungi including increased cell wall strength [1], decreased susceptibility to heat and cold [2], resistance against ultraviolet radiation [3], protection from amoeboid predators [4] and energy transduction (reviewed in [5, 6]). In the context of virulence, melanin reduces the vulnerability of fungal cells to antimicrobial oxidative fluxes and proteins produced by immune cells and increases resistance to certain antifungal drugs [7, 8]. Melanin is a potential drug target, since compounds that interfere with melanization increase microbial clearance in animal models of fungal infection [9]. Hence, dissecting the process of melanization in fungi is essential for understanding critical aspects of fungal pathogenesis. Nonetheless, studies on melanin have been hampered by this pigment’s amorphous structure and insoluble nature, which is notoriously difficult to isolate and analyse with standard techniques.
Cryptococcus neoformans is a human pathogenic basidiomycete that is a primary cause of life-threatening meningoencephalitis in individuals with impaired immunity. Melanization in C. neoformans is considered one of the most important virulence factors together with the polysaccharide capsule [10]. The current model of melanization in C. neoformans posits that it is catalysed by the enzyme laccase in the presence of exogenous phenolic compounds [11]. Melanin and laccase have been shown to co-localize near the C. neoformans cell wall [11, 12] and have been found in extracellular vesicles [13, 14]. Melanization in vesicles helps contain the many cytotoxic intermediates produced during melanin biosynthesis [13], and these structures have been referred to as fungal melanosomes [15]. The mechanism by which the fungal melanosome is exported to the exterior of the cells and attaches to the cell wall is unknown. Solid-state nuclear magnetic resonance (NMR) has provided evidence that C. neoformans melanin has covalent links to cell-wall polysaccharides [16]. When chitin or chitosan synthesis is altered, C. neoformans cells release more melanin into the surrounding medium [17, 18]. Taken together, these data suggest that cell-wall polysaccharides may serve as anchors for melanin into the cell wall.
While studying staining of melanized and non-melanized C. neoformans cells with cell-wall fluorescent dyes, we noted a lessening of fluorescent intensity in the presence of melanin. To explain this observation, we considered two hypotheses: (1) melanization blocked accessibility to the dye-binding site within the cell wall and/or (2) melanin’s broad-spectrum absorbance ‘quenched’ dye fluorescence. Consequently, we studied the effect of melanization in the presence of cell-wall dyes and established that both of the above-mentioned explanations contribute to the observed outcome. Our data demonstrate that cell-wall dyes interfered with melanin incorporation into the cell wall, thus promoting the release of melanin particles to the culture media. These observations give additional support to the view that Cryptococcal melanin structure is formed by melanin nanoparticles, individual or as strings, which are attached to the cell wall through polysaccharide interactions.
Methods
Yeast growth conditions
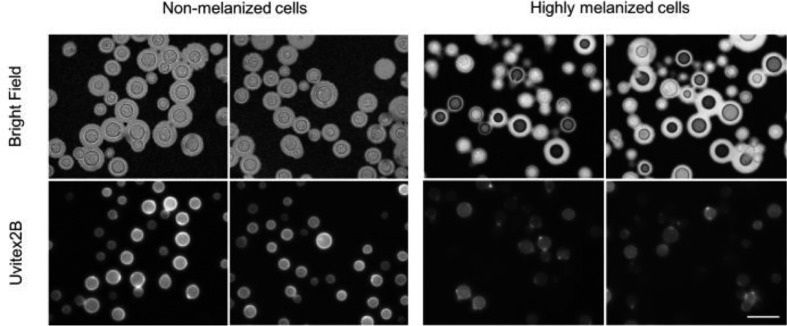
C. neoformans var. grubii serotype A strain H99 was used for all experiments. Fungal cultures were started from frozen yeast stocks and grown for 48 h in 10 ml yeast peptone dextrose (YPD) broth (Becton, Dickinson and Company, Sparks, MD, USA) shaking at 30 °C. This culture was then used to seed a larger volume of YPD, which was then grown for an additional 48 h. For some experiments, heat-killing of yeast cells was done by incubating the cells in a 55 °C water bath for 1 h. The highly melanized cultures described in Fig. 1 were obtained by growing yeast cells in minimal media supplemented with N-acetylglucosamine as described by [19].
Fig. 1.
C. neoformans melanized cells show dimmed Uvitex fluorescent signal. Light microscopy of C. neoformans cells stained with India Ink (top panel) or with 5 µl of Uvitex (stock 20 mg ml−1) in the dark for 10 min at room temperature (bottom panel). Cell images were obtained using 100× oil-immersed objective. Scale bar, 10 µm.
Staining procedure
C. neoformans fluorescent staining was performed by suspending cells at a density of 1×108 cells ml−1 in the presence of cell-wall dyes, Uvitex (Polysciences, Inc., Warrington, PA, USA) or Eosin Y (Chroma Gesellschaft, Münster, Germany), for 1 h at room temperature in 50 ml PBS, shaking and protected from light. Unstained and heat-killed controls were treated with an equal volume of PBS instead of the dye solutions. The pH of fungal solutions was measured immediately after the addition of dye; there was no appreciable difference between stained and unstained control samples (data not shown). Samples were then washed three times with PBS and three times with minimal media without L-DOPA. Yeast cells were then transferred into 150 ml of minimal media (29.4 mM KH2PO4, 10 mM MgSO4 7H2O, 13 mM glycine and 3 µM thiamine) with 1 mM L-DOPA and low dextrose (1.5 mM) to induce overnight melanization. Forty ml cell-free supernatant aliquots were collected at the start of the experiment and every 24 h for 2 days. At each time interval, the culture viability, pH and laccase activity were measured, and cells were fixed and examined microscopically. For each time point, the cell-free supernatant was ultra-centrifuged at 100 000 g for 1 h in an Optima L-90K UltraCentrifuge (Beckman Coulter, USA) using a swinging bucket SW28 rotor (Beckman Coulter, USA). The resulting pellet was subjected to particle size analysis and optical absorbance for melanin quantification (see below).
Viability assay
Culture viability was tested via tadpoling as previously described by [20]. Briefly, 200 µl of each sample aliquot (40 ml) was plated into the first column of a 96-well plate and serially diluted (1 : 10) in Sab broth 11 times. To avoid unaccounted carryover of cells, new tips were used for every dilution. The plate was covered and the cells grown at 30 °C for 2–3 days without shaking. Once colonies grew on the bottom, the number of countable colonies in the most dilute well(s) were multiplied by the dilution factor of the cell to get the number of viable cells in the initial culture.
Culture pH measurements
Cultures were removed from the incubator and onto sterile conditions. A few drops of each culture were used to measure pH by means of intermediate range (5–10) ColorpHast pH strips (Millipore, USA).
Cell morphology analysis
To establish cell morphology in the presence and absence of dyes, a 1 ml volume of cell pellet was washed twice with PBS and incubated with 4 % EM grade paraformaldehyde (VWR, USA) for 15 min, rocking at room temperature. After 15 min, the cells were washed twice with PBS and stored at 4 °C. Fluorescent and bright field images were recorded at 100X on an AX720 microscope (Olympus, USA) equipped with a digital camera (32-0021B-249, QImaging).
Melanin analyses and quantification
Melanin has a unique broadband absorption spectrum [21]. Culture supernatant absorbance has been used as an indirect semi-quantitative approach to ascertaining the amount of extracellular melanin [18, 22]. Combining these two aspects, we performed a semi-quantitative measurement of melanin content. Each sample was analysed in triplicate by loading 200 µl of cell-free supernatant on a 96-well plate. Relative absorbance units from 200 to 900 µm were measured using a SpectraMax M3 microplate reader (Molecular Devices, USA). For analysing the size and heterogeneity of the melanin particles in the supernatant, dynamic light scattering (DLS) was used. This method works best with dilute suspensions and does not destroy the sample. Relative particle sizes were measured using a ZetaPlus Zeta Potential Analyzer (Brookhaven Instruments Corporation, USA).
Ultra-centrifugation to isolate extracellular vesicles/melanin
The protocol used is similar to one previously reported for extracellular vesicle isolation in C. neoformans [23]. Briefly, cell-free supernatant was spun at 100, 000 g for 1.5 h in an Optima L-90K UltraCentrifuge (Beckman Coulter, USA) using a swinging bucket SW28 (Beckman Coulter, USA). This supernatant was collected, and absorbance was recorded. The pellet was resuspended in 3 ml Milli-Q purified water (EMD Millipore, USA) and subjected to another 100, 000 g spin for 1 h in an Optima TL 100 Ultracentrifuge (Beckman Coulter, USA) using a TLA 100.3 fixed angle rotor (Beckman Coulter, USA). The aqueous supernatant was discarded, and the aqueous content was then evaporated using a vacuum concentrator (LABCONCO, USA). Each dried sample was resuspended in exactly 100 µl of Milli-Q water in a UVette (Eppendorf, USA) for comparative analysis.
Transmission electron microscopy (TEM)
The ultra-centrifuged pellets were fixed in 2.5 % (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer for 15 min at room temperature, collected by ultra-centrifugation (100 000 g, 1 h, 4 °C) and stored overnight at 4 °C. The following day, samples were treated with osmium tetraoxide, serially dehydrated, embedded in epoxy resin and ultra-thin sections were obtained. Images were recorded using a Philips/FEI Bio Twin CM120 transmission electron microscope. Average particle size was measured using ImageJ (v 1.48 s).
Cell wall stains binding to polysaccharides
Polysaccharides [Amylose (Sigma # 859656), Cellulose (Sigma # S3504), Chitosan (Sigma # 9012-76-4), Crab Chitin (Sigma # C9752), Corn Starch (Signature Kitchens), Curdlan (Waco Chemicals # 032–09902) and Shrimp Chitin (Sigma # C7170)] were sonicated into a 10 ml aqueous solution. The larger fragments were then pelleted, and 1 ml of the smaller fragments from the supernatant were transferred into a 1.5 ml tube. The solution was then split in half: one aliquot was stained with Uvitex or Eosin Y (1 h at room temperature) while the other remained in DI water. Samples were washed twice with Milli Q water to remove unbound dye. Stained and unstained controls were loaded into black 96-well plates in triplicate and fluorescence was recorded (Uvitex excitation wavelength was 350 nm, emission wavelength was 450 nm; Eosin Y excitation wavelength was 475 nm, emission wavelength was 550 nm).
Laccase activity
Laccase activity was measured using a modified version of the ABTS assay [24]. Briefly, cells collected at various time intervals were washed twice in 50 mM Na2HPO4 (pH 7.0) and concentrated to 1×108 cells in 900 µl of 50 mM Na2HPO4. We then added 100 µl of 10 mM ABTS, a substrate for laccase that yields an optically active product once it is reduced. The cell mixture was incubated overnight at 30 °C and absorbance was recorded at 420 nm. All sets included a control without cells as a blank to account for possible contaminants or degradation products that may interfere with the reaction. Heat-killed samples were used as negative controls and their average value was used for background subtraction. Experiments were carried out in biological triplicate and all measurements were taken in technical triplicate. This method was especially attractive to us because it measures laccase catalysis on the exterior surfaces of whole live cells and omits interference from intracellular enzymes.
Statistical methods
Statistical analyses were carried out using Prism version 7.0C (GraphPad Software, San Diego, CA, USA). Uvitex binding differences among melanized and non-melanized cells (Fig. 2c) were analysed using an unpaired two-tailed t-test. Two-way analysis of variance (ANOVA) followed by Turkey’s multiple comparison test was used to analyse supernatant absorbance as a result of staining (Fig. 4b), laccase activity data (Fig. 6) and culture viability (Fig. S1, available in the onversion of this article). TEM particle size data (Fig. 7) was analysed with two-way ANOVA and Sidak’s multiple comparisons test.
Fig. 2.
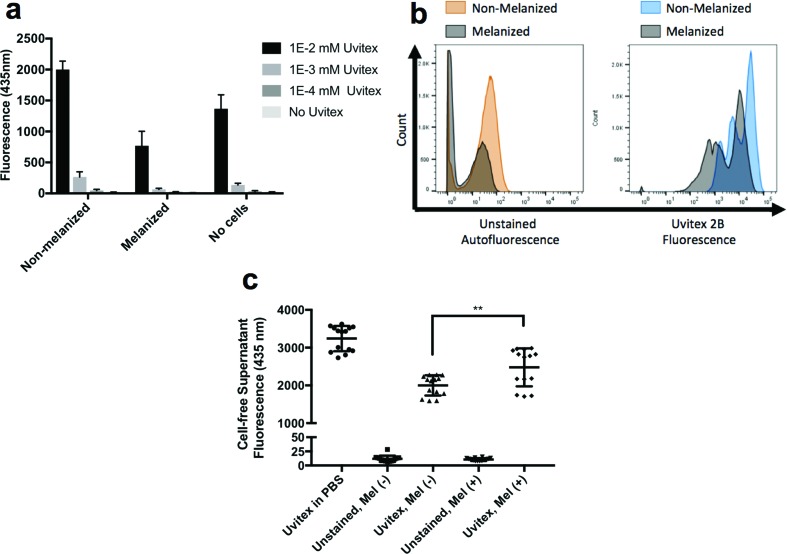
Melanin quenched the Uvitex fluorescence signal and interfered with its binding to the cell wall. (a) Incubation of melanized C. neoformans cells with Uvitex reduced the amount of spectroscopically-measured fluorescence of this cell wall stain. (b) The presence of melanin in the fungal cell wall reduced both auto-fluorescence (left histogram) and Uvitex fluorescence (right histogram) measured using flow cytometry. (c) Melanized cells bound less Uvitex than non-melanized cells, as evidenced by the increased amount of dye in the cell-free supernatant of stained cultures.
Fig. 4.
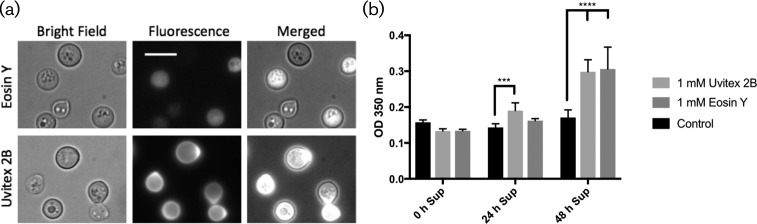
Staining C. neoformans with either Uvitex or Eosin Y affected melanin deposition on the cell wall. (a) C. neoformans cells stained with either Eosin Y or Uvitex exhibited increased fluorescence localized near the fungal cell wall. (b) Cell-free supernatant absorbance was measured at different times for cultures previously stained. At 24 h, only Uvitex showed a darker supernatant while at 48 h both stains were effective at increasing melanin content in the culture supernatant.
Fig. 6.
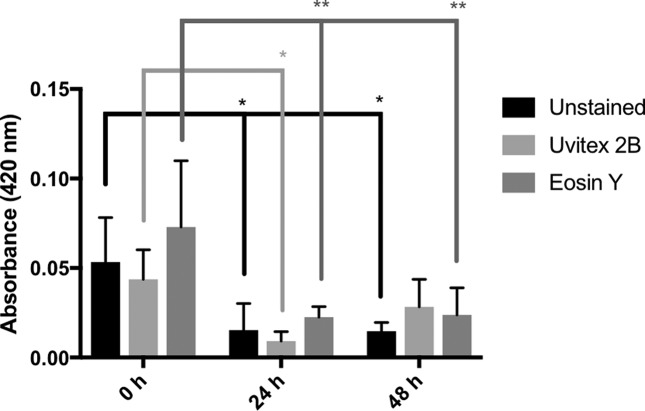
Laccase activity in the presence and absence of cell-wall dye as a function of time. There was no significant difference in laccase activity among the stained samples and unstained control at any time interval. All samples exhibited a statistically significant (P<0.05) drop in laccase activity between the 0 and 24 h time intervals [Unstained (*), Uvitex (*), Eosin Y (**)]. Only the Unstained and Eosin Y samples retained a statistically significant reduction in laccase activity between the 0 and 48 h time intervals.
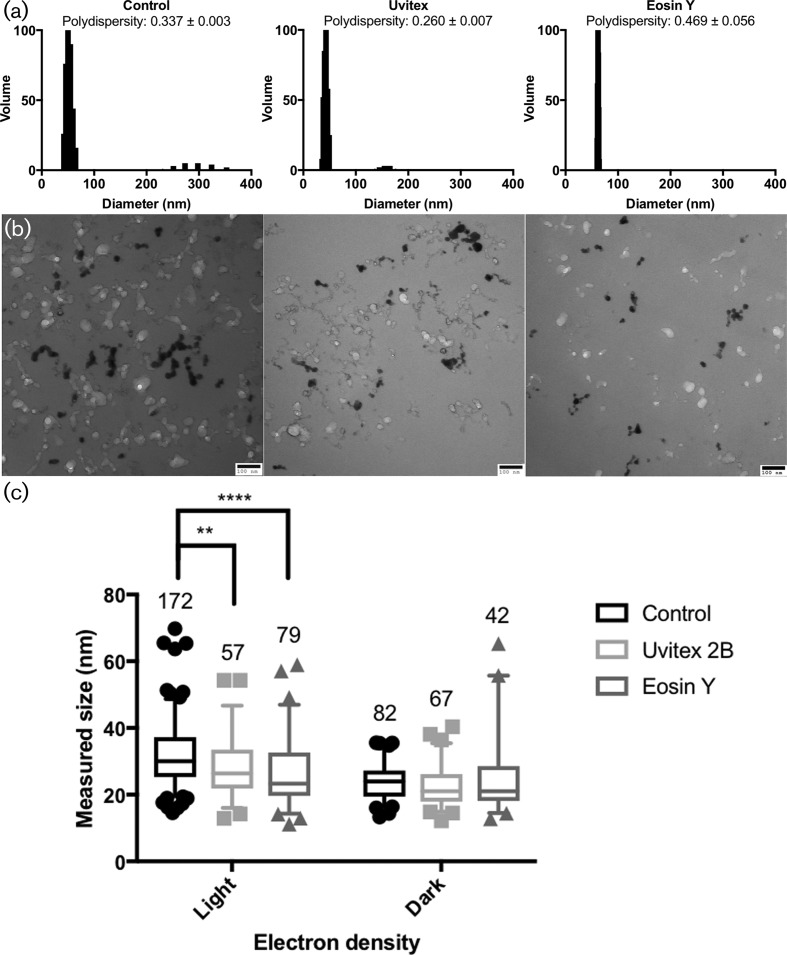
Fig. 7.
DLS and TEM particle size analysis from ultra-centrifuged cell-free supernatant. (a) Dynamic light scattering (DLS) measurements of washed ultra-centrifuged particles in Milli Q water. Polydispersity (mean±std. error) values from DLS confirm the size distributions visualized in the TEM data in panel (b). (b) Transmission electron microscopy of the same particles post fixation and dehydration; scale bar, 100 nm. (c) Size comparison of light and dark particles on TEM images as measured with ImageJ. Among the light (less electron-dense) particles, both Uvitex and Eosin Y were significantly smaller than the unstained control.
Results
Melanin-Uvitex interaction
When staining C. neoformans with Uvitex to study cell-wall composition, it was noted that overall chitin fluorescence intensity on melanized cells was significantly dimmer relative to the brightly fluorescent non-melanized cells (Fig. 1). The budding sites in melanized cells were brightly stained, consistent with the thinning of the melanin coat on these regions [25]. We were intrigued by this observation and considered two hypotheses that could explain this phenomenon: (1) melanin is physically blocking the dye binding sites and/or (2) melanin is quenching Uvitex fluorescence due to its ability to absorb light across the electromagnetic spectrum.
Does melanin quench Uvitex fluorescence or hinder its binding?
To test melanin’s possible contribution towards the quenching of Uvitex fluorescence, we mixed 100 µl (1×107cells ml−1) of melanized or non-melanized C. neoformans cells with 100 µl of various concentrations of Uvitex dissolved in PBS in a black opaque-bottom 96-well plate. Cellular suspensions were mixed vigorously and Uvitex fluorescence emitted at 435 nm was immediately recorded to prevent cells settling on the bottom of the plate. We found that melanized C. neoformans yeast cells show decreased Uvitex fluorescence when compared to controls (Fig. 2a). This suggests that cell-wall melanization contributes to fluorescence quenching (Fig. 2a). Melanin’s ability to quench Uvitex fluorescence was also evident when flow cytometry was used to measure the fluorescence of melanized and non-melanized C. neoformans cells (Fig. 2b). Signals from melanized cells were generally less intense than those of non-melanized cells, regardless of Uvitex staining. Thus, our data demonstrate that due to the wide range absorption spectrum of melanin pigments, Uvitex fluorescence, as well as some of the cell’s autofluorescence, can be quenched.
To test the possibility that melanin could be sterically hindering Uvitex binding to the cell wall, melanized and non-melanized C. neoformans cells were stained with equimolar concentrations of Uvitex in a 4 ml aliquot for 1 h in the dark, at room temperature, with moderate rocking. Cells were then pelleted, and supernatant fluorescence was recorded at 435 nm. The more binding of Uvitex to cells, the less fluorescence remained in the supernatant. Thus, supernatant fluorescence could be used to estimate the amount of Uvitex that bound to the cells. Both melanized and non-melanized C. neoformans cells showed reduced Uvitex fluorescence compared to controls (Fig. 2c), confirming that both cells' phenotypes bind Uvitex. However, the supernatant from melanized cells showed higher average fluorescence signal than the non-melanized ones, suggesting that melanin interfered with Uvitex binding to the cell wall.
Cell-wall stains alter C. neoformans melanin deposition
We considered that if melanin interfered with Uvitex binding, then it was possible that Uvitex could interfere with melanin deposition on the C. neoformans cell wall. To test this possibility, we observed the progression of melanization in cells previously stained or not stained with Uvitex (Fig. 3a). Cells were then pelleted via centrifugation, and supernatant absorbance was used as a proxy for melanin released into the medium. While we did not see any noticeable difference in the pigmentation of the cell pellet, we did notice what appeared to be a direct correlation between the concentration of stain used and the darkness of the cell-free supernatants (Fig. 3b). The trend was quantitatively confirmed through spectroscopic measurements (Fig. 3c).
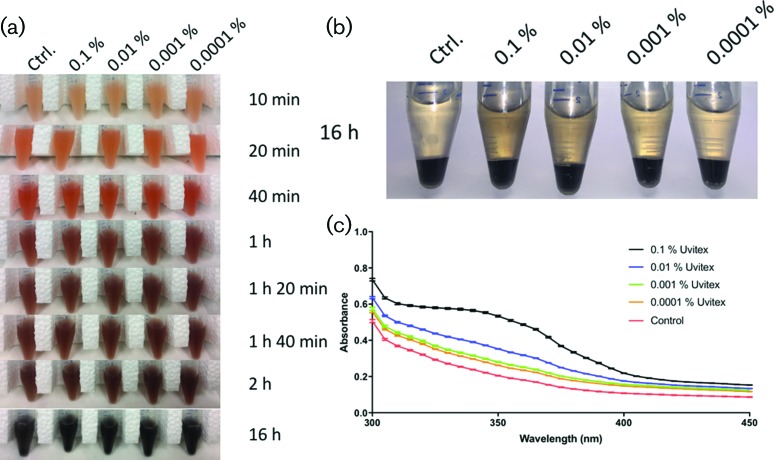
Fig. 3.
C. neoformans stained with Uvitex released more melanin into the culture supernatant. (a) Under the conditions tested, cells rapidly melanized with little visual difference in pigmentation. (b) While the cell pellet did not show differences in pigmentation, the supernatant of highly stained cells looked darker than the control. (c) Visual differences in the supernatant were quantified by absorbance. Cells stained with increasing quantities of Uvitex released more melanin into the extracellular media.
Chitosan plays a role in the binding of melanin to the C. neoformans cell wall
Early studies had shown that Uvitex binds chitin [26], and chitin and chitosan are critical for melanin retention within the cell wall of various fungi (C. neoformans [17, 18, 27], A. nidulans [28], Exophilia dermatitidis [29], Candida albicans [30]). We assessed the role of chitinous cell-wall components in C. neoformans melanin deposition after staining with Uvitex or Eosin Y (Fig. 4a), which has a higher affinity for chitosan [31]. Following 48 h of culture melanization, supernatants from cells stained with either cell dye showed higher absorbance indicative of greater melanin release (Fig. 4b). Melanin content in the extracellular milieu was found to increase over time in all samples. However, as early as 48 h the stained samples showed increased melanin content. These results imply that when we blocked chitosan binding sites with specific stains, the effect was similar to that observed by staining with a more promiscuous stain such as Uvitex. These data are consistent with chitosan playing an important role in melanin deposition on the C. neoformans cell wall.
Analysis of collected particles confirms the presence of melanin
We analysed the size, colour and absorbance of the particles collected in the ultra-centrifuged pellet, which was re-suspended in a known volume to facilitate comparisons between samples (Fig. 5a). Spectroscopic analysis of pelleted material showed melanin’s characteristic broadband absorption spectrum. While all pelleted substances produced a broad absorption spectrum, large differences in absorbance were consistent with the pellet size of melanin particles, where larger pellets absorbed quantitatively more light than their smaller counterparts (Fig. 5b).
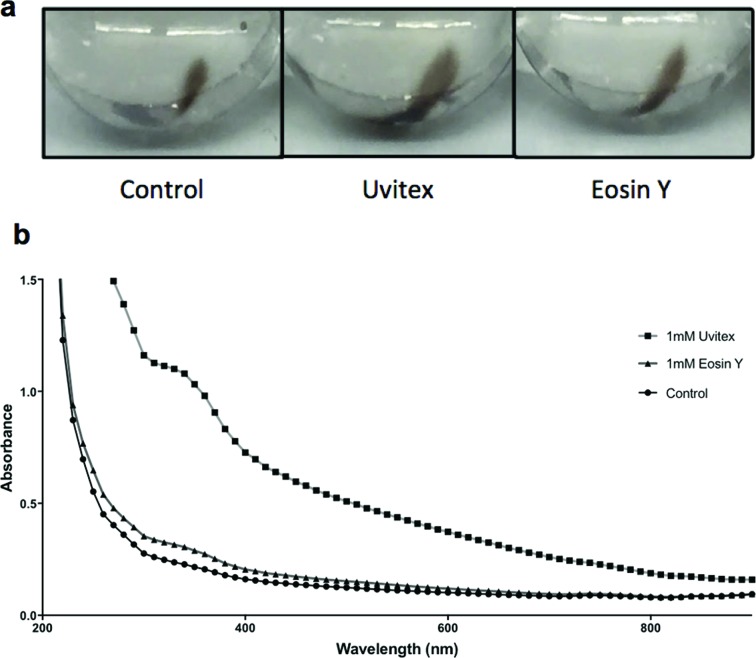
Fig. 5.
Measured melanin content is higher in stained samples vs unstained control. The supernatant of cultures stained with either Uvitex or Eosin Y was ultra-centrifuged. (a) Qualitatively, the pellets of both stained samples are larger than the control. To quantitatively measure these differences, pellets were dried, re-suspended in equal volumes and their absorbance measured in a spectrophotometer. (b) Optical absorbance correlates with pellet sizes observed for each sample.
Changes in culture pH are not responsible for changes in supernatant coloration
Slightly basic conditions (pH >8) can induce l-DOPA auto-polymerization (described in [32]). To verify that the increase in supernatant absorbance was not due to pH variations, we measured individual culture pH at each time point. There was an even increase in all samples from 5.5 to 7 within 24 h. The pH stabilized, and no further pH differences were found between samples at later time points. Heat-killed controls did not alter pH at any time post inoculation (data not shown).
Cell morphology and viability testing
Cell wall stains were previously reported to act as stressors that affect fungal cell viability and morphology. Growing fungi in the presence of stain can lead to growth inhibition and failed separation between mother and daughter cells [33]. While our growth conditions are different from those used in the above-referenced publication, we set out to quantify the effects of our staining protocol on melanizing C. neoformans cells. To examine culture viability we used a tadpoling assay [20], which revealed no difference in growth between stained and control samples (Fig. S1). Our data suggest that under the conditions utilized, these stains are not toxic to C. neoformans cells. To ascertain whether there were changes to cell morphology, we fixed cells collected at various time intervals, analysed cells by microscopy and noted no discernable differences. Cells appeared to grow well as evidenced by the increased number of buds and the appearance of smaller cells at later time intervals (Fig. S2). We then assessed defects in mother-to-daughter cell separation by calculating the average cell number per cell group (total cell number divided by the number of groups, including single cells) as described by [33]. This ratio is equal to one when only single cells are observed and becomes increasingly large when groups of mother-daughter cells are found. Our values ranged between 1.07 and 1.29 (Fig. S2), which were very similar to the control cultures of [33], who calculated 1.1 to 1.4; their stress conditions showed a maximum value of 3.8.
Laccase activity
Recently, it was reported that some industrial synthetic dyes could affect laccase activity [34]. This prompted us to test the possibility that the cell-wall stains used in this study were interacting with laccase, the protein that catalyses melanin polymerization. There was no significant difference between stained and unstained samples using the ABTS assay (Fig. 6), suggesting that our results are not due to laccase inhibition by dye. It was interesting that laccase activity was maximal during the early hours of the experiment and dropped significantly with time to remain low from 24 to 48 h.
Particle size analysis
Extracellular melanin-containing particles were collected by ultra-centrifugation and analysed by both dynamic light scattering (DLS) and transmission electron microscopy (TEM). The coloration of the supernatant post-ultra-centrifugation suggests that melanin material remains dispersed in the supernatant following ultra-centrifugation. DLS measurements of pelleted particles predicted a population with hydrodynamic radii ranging from between 40–60 nm across all samples (Fig. 7a). Both the control and Uvitex-treated samples also showed a second population of particles in the 300 and 150 nm hydrodynamic diameter, respectively (Fig. 7a).
The pellet was then fixed, dehydrated and visualized by TEM. Images revealed a mix of spherical particles ranging in diameter from 10 to 70 nm, with averages around 20–30 nm (Fig. 7b, c). Particle electron density varied and ranged across the grey scale, from dark (electron-rich) to light (electron-poor). There was no observable correlation between electron density and particle size of. Many appeared strung together or connected like a string of particles (Fig. 7b). Very few single spherical particles were observed in the samples, possibly due to aggregation during fixation and dehydration. Both dark and light particles ranged in their measured diameter, with the lighter population being slightly bigger than the darker populations. When comparing the ratio of light to dark particles present, control samples had the highest value and Uvitex the lowest (data not shown), which is consistent with our results indicating increased supernatant absorbance in the stained samples versus controls.
Cell-wall targets for Uvitex and Eosin Y
The C. neoformans cell wall is composed of chitin, chitosan and various glucans. Eosin Y specifically binds chitosan on the cryptococcal cell wall [18]. However, the molecular structure of the Uvitex binding site in chitin is unknown. We used spectroscopic techniques to probe for the interaction between cell-wall components and Uvitex. Chitin and chitosan are beta-1,4-linked polymers of N-acetyl-d-glucosamine (GlcNAc), while the others are a mixture of alpha- and beta-linked, branched and unbranched polymers of glucose. Although the methods used in this analysis do not allow for comparisons of fluorescence intensity and polysaccharide concentration between samples, a positive signal implies that the stain has bound to the polysaccharide. These data demonstrate that Uvitex and Eosin bind to a number of polysaccharides (Fig. S3). All but one unstained control had absorbance curves parallel to the x-axis. The only exception was the chitosan unstained control, which rose higher than others but still much below the stained sample. While on this assay cellulose did not give a strong signal, microscopic analysis showed a very strong signal suggesting that Uvitex binds cellulose in vitro (data not shown).
Discussion
Understanding the structure of the fungal cell wall is one of the most difficult problems in biology [35]. The fungal cell wall is composed of an intricate network of polysaccharides that harbors many macromolecules including proteins, lipids and sometimes melanin. This structure is sufficiently rigid to protect the fungal cell against a variety of insults and yet is sufficiently flexible to allow changes in cellular morphology such as hyphae formation and daughter cell budding. Recently, the fungal cell wall was shown to manifest viscoelastic properties that allow the transition of amphotericin-laden liposomes indicating that it must have sufficient porosity to allow the transit of these structures [36]. Earlier vesicular structures were visualized in the cell wall, which were interpreted to be extracellular vesicles transiting this structure [23]. In this regard, current models of cryptococcal cell-wall melanization involve the transit and assembly of melanin-containing vesicles into a melanized coat [13].
One of the major obstacles to achieving a better understanding of the cell wall in its melanized and non-melanized states is the relative paucity of techniques to probe this structure. While studying C. neoformans melanization, we made the serendipitous observation that melanized cells stained with Uvitex manifested much dimmer fluorescence that non-melanized cells. We investigated this phenomenon and found that the presence of melanin interfered with Uvitex fluorescence in two ways. First, melanin's broad absorption quenched Uvitex fluorescence, making darker cells appear to have a dimmer fluorescent signature. This observation raises the interesting possibility that if fluorescence intensity correlates with pigment concentration in a consistent fashion it could be developed to estimate the amount of cell wall-associated melanin per cell. Second, we found that melanized cells bound less Uvitex than non-melanized cells.
The observation that cell-wall melanin interfered with Uvitex binding was intriguing because it suggested that both melanin and the dye bound to the same place. Specifically, we considered that if Uvitex and melanin shared binding sites, then studying Uvitex binding to the cell wall could shed light on the mechanism by with melanin is anchored in that structure. Chitin [30, 37], chitosan [18], and other cell-wall structures [15] have been implicated as being important for melanin deposition on the cell wall. Early studies aiming to assess binding specificity of Calcofluor White and Uvitex, both fluorescents stilbene derivatives, reported that Uvitex binds to several polysaccharides, mainly fungi-derived [26, 38, 39]. In fungal cells, Uvitex localizes to the cell wall, but little is known about its interaction at molecular level with cell-wall components.
To further explore targets for Uvitex in the cell wall we tested its affinity for a series of compounds that could be found in fungal and plant cell walls, including various polysaccharides. Uvitex bound with high affinity to three major components of fungal cell wall; namely 1–3, 1–6-β-glucans, chitin and chitosan. Thus, we considered that the diminished fluorescence resulting from Uvitex binding was a result of melanin interference. To test this hypothesis, we stained C. neoformans cells with Uvitex and observed the process of melanization. While we did not see any visually apparent difference in melanization of the cells, we noticed that the culture supernatant of C. neoformans cells grown with Uvitex were darker. We also assessed the effect of Eosin Y, a chitosan-specific dye [31], on the cell-wall melanization. While Uvitex was generally more effective than Eosin Y at promoting darker culture supernatants, both stains produced a similar effect. Since Eosin Y is a more specific stain than Uvitex, it is possible that Uvitex produced darker supernatants because it was more effective at interfering with melanin deposition resulting in less retained pigment. Taken together, while chitosan is an important melanin anchor, the current study provides additional evidence to suggest that other cell-wall structures also play a key role in C. neoformans melanin deposition. Melanization of C. albicans also depends on cell-wall chitin [30]. We note the similarity between the effects of cell-wall dyes on C. neoformans and the ‘leaky melanin’ phenotype described for cell-wall mutants of C. neoformans [18, 27, 40] and for methlyxanthine treated cells [41].
The finding that these dyes promoted darkening of supernatants implied release of melanin and suggested that we could use this technique to isolate melanin granules with sizes consistent to those described in previously published literature [14, 42]. Analysis of these released melanin particles is also consistent with the idea that C. neoformans melanin has been shown to be arranged in concentric layers surrounding in the cell wall [25, 43]. Curiously, we observed that released melanin tended to aggregate forming structures that resembled electron dense beads on a string. While we cannot ascertain from our images the presence of a membrane surrounding the electron dense particles, they appear with similarly sized particles with a broad range in electron density. Thus, it may be possible that the more electron dense particles depict the progression of melanization. Given that our samples were collected from the supernatant of cells a few days after transferring the cells from rich media to minimal media, it is possible that the linear structures of melanin are fragments of cell-wall layers that detached during cell division events, or during the process of capsule growth. Alternatively, it is possible that they are released melanin particles that are undergoing some form of extracellular self-assembly.
In summary, melanin on the cell wall of C. neoformans reduces Uvitex fluorescence. This finding has the potential for development as a method for quantifying melanin in melanized cells based in the absence of fluorescence as a quantitative measure of melanin content. The observation that Uvitex could interfere with melanin deposition on the cell wall allowed us to characterize these elusive particles released into the culture supernatant, a mixture of pigmented particles. Overall, the results are consistent and supportive of current models of cell-wall melanization whereby melanin is attached to chitin, chitosan and glucans in the cell wall.
Funding information
This research was supported by a grant from the National Institute of Allergy and Infectious Diseases (R01-A1052733). R. P. D. was supported by a grant from the U.S. Health Resources and Services Administration (D18HP29037). E. C. was supported by a Postdoctoral Fellowship from the Johns Hopkins Malaria Research Institute.
Acknowledgements
We thank Barbara Smith and the collective staff at Johns Hopkins School of Medicine’s Microscopy Facility for their technical support in sample preparation and TEM imaging.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Three supplementary figures are available with the online version of this article.
Abbreviations: ABTS, 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid); L-DOPA, L-3,4-dihydroxyphenylalanine; DLS, dynamic light scattering; GlcNAc, N-acetyl-D-glucosamine; Mel (−), Not melanized; Mel (+), Melanized; ANOVA, analysis of variance; TEM, transmission electron microscopy.
Edited by: A. Alastruey-Izquierdo and V. J. Cid
References
- 1.Chen Z, Nunes MA, Silva MC, Rodrigues CJ. Appressorium turgor pressure of Colletotrichum kahawae might have a role in coffee cuticle penetration. Mycologia. 2004;96:1199–1208. doi: 10.1080/15572536.2005.11832868. [DOI] [PubMed] [Google Scholar]
- 2.Ál R, Casadevall A. Melanization affects susceptibility of Cryptococcus neoformans to heat and cold1. FEMS Microbiol Lett. 2006;153:265–272. doi: 10.1111/j.1574-6968.1997.tb12584.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Casadevall A. Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Appl Environ Microbiol. 1994;60:3864–3866. doi: 10.1128/aem.60.10.3864-3866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA. 2001;98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A, Cordero RJB, Bryan R, Nosanchuk J, Dadachova E. Melanin, radiation, and energy transduction in fungi. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0037-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobson ES. Pathogenic roles for fungal melanins. Clin Microbiol Rev. 2000;13:708–717. doi: 10.1128/CMR.13.4.708-717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda R, Sugita T, Jacobson ES, Shinoda T. Effects of melanin upon susceptibility of Cryptococcus to antifungals. Microbiol Immunol. 2003;47:271–277. doi: 10.1111/j.1348-0421.2003.tb03395.x. [DOI] [PubMed] [Google Scholar]
- 8.van Duin D, Casadevall A, Nosanchuk JD. Melanization of Cryptococcus neoformans and Histoplasma capsulatum reduces their susceptibilities to amphotericin B and caspofungin. Antimicrob Agents Chemother. 2002;46:3394–3400. doi: 10.1128/AAC.46.11.3394-3400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nosanchuk JD, Ovalle R, Casadevall A. Glyphosate inhibits melanization of Cryptococcus neoformans and prolongs survival of mice after systemic infection. J Infect Dis. 2001;183:1093–1099. doi: 10.1086/319272. [DOI] [PubMed] [Google Scholar]
- 10.Coelho C, Bocca AL, Casadevall A. The tools for virulence of Cryptococcus neoformans. Adv Appl Microbiol. 2014;87:1–41. doi: 10.1016/B978-0-12-800261-2.00001-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhu X, Gibbons J, Garcia-Rivera J, Casadevall A, Williamson PR. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect Immun. 2001;69:5589–5596. doi: 10.1128/IAI.69.9.5589-5596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. Vesicle-associated melanization in Cryptococcus neoformans. Microbiology. 2009;155:3860–3867. doi: 10.1099/mic.0.032854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, et al. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franzen AJ, Cunha MM, Miranda K, Hentschel J, Plattner H, et al. Ultrastructural characterization of melanosomes of the human pathogenic fungus Fonsecaea pedrosoi. J Struct Biol. 2008;162:75–84. doi: 10.1016/j.jsb.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhong J, Frases S, Wang H, Casadevall A, Stark RE. Following fungal melanin biosynthesis with solid-state NMR: biopolymer molecular structures and possible connections to cell-wall polysaccharides. Biochemistry. 2008;47:4701–4710. doi: 10.1021/bi702093r. [DOI] [PubMed] [Google Scholar]
- 17.Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, et al. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2005;4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho E, Chrissian C, Cordero RJB, Liporagi-Lopes L, Stark RE, et al. N-acetylglucosamine affects Cryptococcus neoformans cell-wall composition and melanin architecture. Microbiology. 2017;163:1540–1556. doi: 10.1099/mic.0.000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welch AZ, Koshland DE. A simple colony-formation assay in liquid medium, termed 'tadpoling', provides a sensitive measure of Saccharomyces cerevisiae culture viability. Yeast. 2013;30:501–509. doi: 10.1002/yea.2989. [DOI] [PubMed] [Google Scholar]
- 21.Riesz JJ. PhD Thesi. University of Queensland, Australia: 2007. The Spectroscopic Properties of Melanin. [Google Scholar]
- 22.Eisenman HC, Mues M, Weber SE, Frases S, Chaskes S, et al. Cryptococcus neoformans laccase catalyses melanin synthesis from both D- and L-DOPA. Microbiology. 2007;153:3954–3962. doi: 10.1099/mic.0.2007/011049-0. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourbonnais R, Leech D, Paice MG. Electrochemical analysis of the interactions of laccase mediators with lignin model compounds. Biochim Biophys Acta. 1998;1379:381–390. doi: 10.1016/S0304-4165(97)00117-7. [DOI] [PubMed] [Google Scholar]
- 25.Eisenman HC, Nosanchuk JD, Webber JB, Emerson RJ, Camesano TA, et al. Microstructure of cell wall-associated melanin in the human pathogenic fungus Cryptococcus neoformans. Biochemistry. 2005;44:3683–3693. doi: 10.1021/bi047731m. [DOI] [PubMed] [Google Scholar]
- 26.Coleman T, Madassery JV, Kobayashi GS, Nahm MH, Little JR. New fluorescence assay for the quantitation of fungi. J Clin Microbiol. 1989;27:2003–2007. doi: 10.1128/jcm.27.9.2003-2007.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- 28.Bull AT. Chemical composition of wild-type and mutant Aspergillus nidulans cell walls. The nature of polysaccharide and melanin constituents. J Gen Microbiol. 1970;63:75–94. doi: 10.1099/00221287-63-1-75. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Szaniszlo PJ. WdCHS3, a gene that encodes a class III chitin synthase in Wangiella (Exophiala) dermatitidis, is expressed differentially under stress conditions. J Bacteriol. 2000;182:874–881. doi: 10.1128/JB.182.4.874-881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker CA, Gómez BL, Mora-Montes HM, MacKenzie KS, Munro CA, et al. Melanin externalization in Candida albicans depends on cell wall chitin structures. Eukaryot Cell. 2010;9:1329–1342. doi: 10.1128/EC.00051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heustis RJ, Ng HK, Brand KJ, Rogers MC, Le LT, et al. Pharyngeal polysaccharide deacetylases affect development in the nematode C. elegans and deacetylate chitin in vitro. PLoS One. 2012;7:e40426. doi: 10.1371/journal.pone.0040426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng W, Fan H, Wang L, Jin Z. Oxidative self-polymerization of dopamine in an acidic environment. Langmuir. 2015;31:11671–11677. doi: 10.1021/acs.langmuir.5b02757. [DOI] [PubMed] [Google Scholar]
- 33.Roncero C, Durán A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernández C, Farnet da Silva AM, Ziarelli F, Perraud-Gaime I, Gutiérrez-Rivera B, et al. Laccase induction by synthetic dyes in Pycnoporus sanguineus and their possible use for sugar cane bagasse delignification. Appl Microbiol Biotechnol. 2017;101:1189–1201. doi: 10.1007/s00253-016-7890-0. [DOI] [PubMed] [Google Scholar]
- 35.Nar G, Latge J-P, Munro CA. The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.funk-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker L, Sood P, Lenardon MD, Milne G, Olson J, et al. The viscoelastic properties of the fungal cell wall allow traffic of AmBisome as intact liposome vesicles. MBio. 2018;9:e02383-17. doi: 10.1128/mBio.02383-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Zheng L, Hauser M, Becker JM, Szaniszlo PJ. WdChs4p, a homolog of chitin synthase 3 in Saccharomyces cerevisiae, alone cannot support growth of Wangiella (Exophiala) dermatitidis at the temperature of infection. Infect Immun. 1999;67:6619–6630. doi: 10.1128/iai.67.12.6619-6630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda H, Ishida N. Specificity of binding of hexopyranosyl polysaccharides with fluorescent brightener. J Biochem. 1967;62:276–278. doi: 10.1093/oxfordjournals.jbchem.a128660. [DOI] [PubMed] [Google Scholar]
- 39.Wachsmuth ED. A comparison of the highly selective fluorescence staining of fungi in tissue sections with Uvitex 2B and Calcofluor White M2R. Histochem J. 1988;20:215–221. doi: 10.1007/BF01747466. [DOI] [PubMed] [Google Scholar]
- 40.Zhang P, Wei D, Li Z, Sun Z, Pan J, et al. Cryptococcal phosphoglucose isomerase is required for virulence factor production, cell wall integrity and stress resistance. FEMS Yeast Res. 2015;15:fov072. doi: 10.1093/femsyr/fov072. [DOI] [PubMed] [Google Scholar]
- 41.Tsirilakis K, Kim C, Vicencio AG, Andrade C, Casadevall A, et al. Methylxanthine inhibit fungal chitinases and exhibit antifungal activity. Mycopathologia. 2012;173:83–91. doi: 10.1007/s11046-011-9483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf JM, Espadas-Moreno J, Luque-Garcia JL, Casadevall A. Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryot Cell. 2014;13:1484–1493. doi: 10.1128/EC.00111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandal P, Roy TS, das TK, Banerjee U, Xess I, et al. Differences in the cell wall architecture of melanin lacking and melanin producing Cryptococcus neoformans clinical isolates from India: an electron microscopic study. Braz J Microbiol. 2007;38:662–666. doi: 10.1590/S1517-83822007000400015. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.