Abstract
Objective
SSc is an autoimmune disease characterized by progressive fibrosis of the skin and internal organs. IL-6 and related cytokines that signal through STAT3 have been implicated in the pathogenesis of SSc and mouse models of fibrosis. The aim of this study was to investigate the efficacy of inhibiting STAT3 in the development of fibrosis in two mouse models of skin fibrosis.
Methods
Biopsy samples of skin from SSc patients and healthy control subjects were used to determine the expression pattern of phosphotyrosyl (pY705)-STAT3. C188-9, a small molecule inhibitor of STAT3, was used to treat fibrosis in the bleomycin-induced fibrosis model and Tsk-1 mice. In vitro studies were performed to determine the extent to which STAT3 regulates the fibrotic phenotype of dermal fibroblasts.
Results
Increased STAT3 and pY705-STAT3 was observed in SSc skin biopsies and in both mouse models of SSc. STAT3 inhibition with C188-9 resulted in attenuated skin fibrosis, myofibroblast accumulation, pro-fibrotic gene expression and collagen deposition in both mouse models of skin fibrosis. C188-9 decreased in vitro dermal fibroblast production of fibrotic genes induced by IL-6 trans-signalling and TGF-β. Finally, TGF-β induced phosphotyrosylation of STAT3 in a SMAD3-dependent manner.
Conclusion
STAT3 inhibition decreases dermal fibrosis in two models of SSc. STAT3 regulates dermal fibroblasts function in vitro and can be activated by TGF-β. These data suggest that STAT3 is a potential therapeutic target for dermal fibrosis in diseases such as SSc.
Keywords: fibrosis, systemic sclerosis, STAT3
Rheumatology key messages
STAT3 regulates the development of skin fibrosis in multiple mouse models.
STAT3 regulates the fibrotic phenotype in dermal fibroblasts.
STAT3 regulates TGF-β signaling.
Introduction
SSc is an autoimmune disease that is characterized by progressive fibrosis of the skin and internal organs [1]. Although the initial stages of SSc may involve vascular changes and inflammatory infiltrates with monocytes and T cells, the later stages are characterized by tissue fibrosis due to the excessive deposition of extracellular matrix (ECM) components, such as type I collagen [2]. Multiple molecular pathways lead to fibroblast activation and differentiation into myofibroblasts, a key effector cell in fibrosis. In particular, TGF-β is an important cytokine that modulates fibrosis by stimulating fibroblasts and differentiation of myofibroblasts.
Two mouse models commonly used to determine pathways involved in skin fibrosis are the subcutaneous bleomycin (BLM) model and the tight-skin mouse (Tsk-1). The BLM model is an inflammation-driven dermal fibrosis model resembling the early inflammatory stages of SSc [3]. BLM treatment induces increased production of reactive oxygen species, thereby causing cellular damage to endothelial cells and other cell types, leading to the production of important cytokines such as TGF-β and IL-6 [4]. In contrast the Tsk-1 mice have a duplication of the fibrillin-1 gene, resulting in an oversized fibrillin-1 protein that leads to activation of TGF-β pathways, abnormal microfibril deposition and thickening of the hypodermis [5]. Both models complement each other and serve as tools for testing anti-fibrotic agents in an inflammatory-dependent induction of fibrosis and a TGF-β–driven fibrosis absent of significant inflammation.
In addition to TGF-β, there is substantial evidence that IL-6 may be an important cytokine that can promote fibrosis [6–9]. The IL-6 family of cytokines signal through gp130. Upon cytokine-induced oligomerization, gp130-associated Janus kinases trans-activate and phosphotyrosylate gp130. gp130 subsequently recruits STAT3, through its Src-homology (SH) 2 domain, leading to the phosphotyrosylation of STAT3 (pY-STAT3). Phosphorylated STAT3 homodimerizes and translocates to the nucleus, where it induces production of a variety of genes that regulate cellular differentiation, apoptosis and gene transcription [10]. C188-9 is a potent, small-molecule STAT3 inhibitor designed to bind to the SH2 domain of STAT3 [11, 12]. C188-9 prevents STAT3 activation by preventing STAT3 recruitment to gp130, phosphorylation and homodimerization, but does not have activity against other STAT proteins [11, 12]. Toxicolology studies in mice demonstrate that C188-9 is well tolerated and does not cause clinical, laboratory or pathological abnormalities [11, 12]. STAT3 inhibition via C188-9 has been shown to decrease fibrosis in the BLM model of pulmonary fibrosis [13]. Interestingly, these studies also demonstrated that TGF-β itself could activate STAT3, suggesting alternative mechanisms by which STAT3 could be activated to modulate fibrosis [13]. Therefore, it is of interest to determine whether STAT3 can decrease fibrosis in other mouse models, particularly a model with significant TGF-β activation, such as the Tsk-1 mouse. The aim of this study was to investigate the effect of inhibiting STAT3 in the development of dermal fibrosis in two complementary mouse models of SSc, one involving inflammation-dependent pathways (BLM model) and the other involving fibroblast activation devoid of inflammation (Tsk-1 model).
Methods
Mice
Animal care was in accordance with institutional and National Institues of Health guidelines and approved by the Baylor College of Medicine Animal Welfare Committee. Mice were housed in ventilated cages equipped with microisolator lids and maintained under strict containment protocols. Wild type C57BL/6 mice, Tsk-1 mice and pallid (Pa/Pa) controls were purchased from Jackson Laboratories.
Human skin biopsies
SSc patients and healthy control skin biopsies were obtained from the University of Texas Health Science Center at Houston Medical School. SSc patients met the ACR classification criteria for SSc. Healthy control subjects had no personal or family medical history of autoimmune diseases [14]. Biopsy samples were obtained from a similar location on the forearms of affected SSc patients and controls. This study was approved by the University of Texas Health Science Center at Houston Medical School institutional review board. All subjects provided written informed consent.
Skin biopsy microarray analysis
Experimental details for generating STAT3 gene expression levels in human skin samples have been previously published [15]. Briefly, the gene expression data were generated using Illumina HumanHT-12 bead arrays. Raw gene expression data were analysed with BRB ArrayTools. Data were normalized by the quantile method. Genes whose log intensity variance was in the bottom 75th percentile were filtered out, and 11 819 transcripts remained. In the present study, the differential expression of one target gene, STAT3, was investigated however, correction for multiple comparison using the false discovery rate (FDR) was also done. Finally analyses were also adjusted for age, gender and ethnicity. Pearson’s correlation analysis was used to identify correlation of STAT3 transcript levels with the concomitant modified Rodnan Skin Scores (mRSSs).
Subcutaneous BLM dermal fibrosis model
Wild-type C57BL/6 female mice (age 6–8 weeks) were injected with 0.1 ml of BLM (0.01 U/g Monday through Thursday and 0.02 U/g on Fridays) via s.c. route (three sites per mouse) for 4 weeks [16, 17]. Subcutaneous injections with 0.1 ml of PBS were used as a control. C57BL/6 J female mice exposed to s.c. BLM or PBS received daily intraperitoneal injections of either C188-9 [50 mg/kg in dimethyl sulfoxide (DMSO), obtained from StemMed, Ltd] or vehicle (DMSO) starting on day 15 and continuing until day 26. On day 28, mice were sacrificed and lesional skin was harvested using an 8 mm skin biopsy punch. Skin biopsies were used for histology, protein analysis and total RNA isolation. Experiments were performed three times, with four mice per experimental group each time.
Tsk-1 mouse model
Tsk-1 and Pa/Pa control mice (aged 5 weeks) were treated with daily intraperitoneal injections of C188-9 (50 mg/kg in DMSO) or DMSO as vehicle control for 2 weeks. At 7 weeks of age, skin biopsies from identical anatomic areas on the back were obtained and used for histology, protein and RNA analysis. Experiments were performed three times, with four mice per experimental group each time.
Histology
Formalin-fixed paraffin-embedded skin biopsies were stained with haematoxylin and eosin or Masson’s Trichrome. Skin fibrosis was measured by quantifying the thickness of the dermis in the BLM model and the hypodermis (measuring the subcutaneous connective tissue beneath the panniculus carnosus) in the Tsk-1 model. The dermis or hypodermis was measured at 10 randomly selected sites/microscopic field in each animal [17, 18].
Immunohistochemistry
Tissue sections were rehydrated and antigen retrieval was performed (Dako). Endogenous peroxidases were quenched, and endogenous avidin and biotin were blocked with BLOX ALL blocking solution (Vector Labs). Slides were blocked with 2.5% normal horse serum (Vector Labs). Slides were then incubated overnight at 4 °C with primary antibodies for pY-STAT3 (1:100 dilution, Abcam) or α-SMA (1:1000 dilution, Sigma-Aldrich). Secondary antibodies were incubated (20 min) using the ImmPRESS detection kit (Vector Labs). For pY-STAT3 staining, slides were developed using the ImmPACT DAB (Vector Labs). For α-SMA staining, slides were processed with the Mouse on Mouse Kit and the ImmPACT Vector Red substrate kit (Vector Labs). Isotype controls included rabbit IgG for pY-STAT3 and mouse IgG for α-SMA.
Biochemical analysis of skin biopsies
The collagen content was determined from lesional skin by using the Sircol Collagen Assay kit (Biocolor, Newtown Abbey, UK) [17, 18]. The total protein assay (Bio-Rad Laboratories, Hercules, CA, USA) was used as a control to normalize the collagen content of each sample.
RNA analysis
Real-time quantitative PCR (RT-PCR) was used to determine tissue mRNA levels. Total RNA was isolated from skin biopsies frozen in RNA Later (Qiagen Sciences, Maryland, MA, USA) using Trizol Reagent (Invitrogen, Camarillo, CA, USA), purified with an RNA mini kit (Qiagen Sciences, Maryland, MA, USA) and reverse transcribed. RT-PCR was performed using validated TaqMan Gene Expression Assays for Col1α1, αSMA, TGF-β, connective tissue growth factor (CTGF), IL-6, Fibronectin (Fn), cadherin 11 (Cad11) and 18 s RNA (Applied Biosystems) on an Applied Biosystems Step One Plus PCR System. The 18 s RNA gene was used as an endogenous control to normalize transcript levels of mRNA in each sample, presented as mean normalized transcript levels using the comparative Ct method (2ΔΔCt).
Cell culture
Primary dermal fibroblasts were isolated from C57/BL6 mice [9, 19]. Cells were stimulated with: media alone, TGF-β (10 ng/ml) or IL-6 (20 μg/ml) and sIL-6 Rα (20 μg/ml) together (all from R&D Systems, Minneapolis, MN, USA). To block STAT3, SMAD2 or SMAD3, dermal fibroblasts were also cultured with and without the addition of C188-9 (20 μM), a SMAD-2 inhibitor (EMD Millipore 616464, 10 μM) or a SMAD3 inhibitor (SIS3, 10 μM, Millipore 566405), respectively. Cells were harvested at 24 h post treatment for RNA analysis and for western blotting.
Western blot analysis
Cells were lysed with 1X RIPA Lysis Buffer (EMD Millipore) freshly supplemented with 1X protease and 1X phosphatase inhibitor mixture (Roche Diagnostics). A 50 μg lysate of total protein was electrophoresed on 10% SDS–PAGE gels and transferred to polyvinylidene difluoride membrane. For primary antibody incubation (overnight at 4 °C), rabbit polyclonal antibodies were used against phospho-SMAD-2, phospho-SMAD3, SMAD2 and SMAD3 (1:500, Cell Signaling), pY-STAT3 and STAT3 (1:500, Abcam) and a mouse monoclonal anti-mouse against GAPDH (1:1,000, Abcam). Species-specific horseradish peroxidase–conjugated secondary antibodies (Jackson Laboratories) were applied for 1 h at room temperature, and blots were developed using ECL western blotting substrate (Thermo Fischer Scientific, Waltham, MA, USA). ImageJ analysis was used to quantify western blots.
Statistical analysis
Results are expressed as the means (s.d.) or mean (s.e.m.). Mann–Whitney’s U test (in vivo studies) was used for comparison between two groups. P values < 0.05 were considered statistically significant.
Results
STAT3 expression was increased in skin of SSc patients and mouse models of skin fibrosis
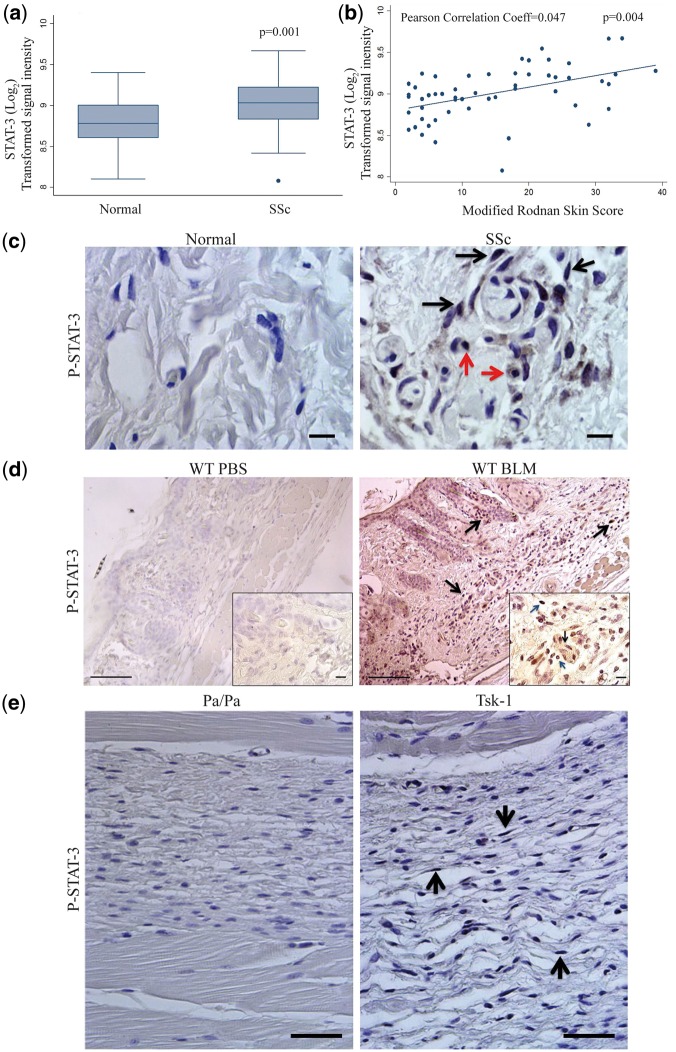
To determine whether levels of STAT3 expression were increased in SSc, we queried the existing published gene expression dataset from 61 SSc patients and 36 controls [15]. STAT3 mRNA expression was increased in SSc skin biopsies compared with control skin biopsies (P = 0.001, Fig. 1A). Similar differences were observed with two other STAT3 transcripts (data not shown.) The increase in STAT3 transcript remained significantly increased in SSc skin biopsies compared with age-, gender- and ethnicity-matched controls in the global analysis (FDR-adjusted P = 0.009). Furthermore, in SSc patients, STAT3 mRNA levels positively were correlated with the extent of skin fibrosis as determined by the mRSS (Pearson correlation coefficient 0.47, P = 0.004; Fig. 1B). STAT3 also correlated significantly with mRSS in the global analysis (FDR-adjusted P = 0.0104). Immunohistochemical analyses of skin biopsies were then used to determine the cellular localization of pY-STAT3 expression. In normal human skin samples there was no pY-STAT3 expression detected. In contrast, nuclear pY-STAT3 expression was detected in the dermis of fibrotic skin from SSc patients, localizing to nuclei of fibroblasts (black arrows) and macrophages (red arrows) (Fig. 1C and supplementary Fig. 1C, available at Rheumatology Online).
Fig. 1.
Characterization of STAT-3 expression and activation in SSc patients and the bleomycin and Tsk-1 model
(A) Microarray expression profiling STAT3 expression on SSc patients vs normal controls. Data are given as log2 transformed signal intensity from the microarray, mean + s.e.m. (P = 0.001 normal (n = 36) vs SSc (n = 61)). (B) Correlation of modified Rodnan skin scores with STAT3 mRNA expression from the microarray dataset of SSc skin biopsies. N = 54 SSc patients, P = 0.004. (C) Immunolocalization of pY-STAT-3 expression in skin biopsies from healthy control and SSc patients (dermal fibroblasts: black arrows; macrophages: red arrows). Images are representative of five patients from each group. Scale bars: ×100, 10 μm. (D) Immunolocalization expression of pY-STAT-3 in skin sections from wild-type mice treated with PBS and bleomycin (BLM). Dermal fibroblasts are identified by black arrows; macrophages are identified by red arrows. Images are representative of eight mice from each group. Scale bars: ×20, 100 μm; ×100, 10 μm. (E) Immunolocalization of pY-STAT-3 expression was determined on hypodermal fibroblasts in skin sections from Pa/Pa and Tsk-1 mice (black arrows). Images are representative of 10 mice from each group. Scale bars: ×40, 50 μm.
To determine whether pY-STAT3 expression was also increased in fibrotic skin in murine models of fibrosis, immunohistochemistry was also performed on skin biopsies in the BLM model and Tsk-1 model. As seen in Fig. 1D and E, very low levels of pY-STAT3 expression were observed in PBS-treated mice or pallid control mice. In contrast, increased nuclear pY-STAT3 expression was observed in the fibrotic dermis region of BLM-treated mice (Fig. 1D and supplementary Fig. 1D, available at Rheumatology Online). As seen in the Fig. 1D insert, fibroblasts (black arrows) and macrophages (red arrows) were observed to have nuclear pY-STAT3 detected. In addition, Tsk-1 mice expressed abundant nuclear pY-STAT3 on fibroblasts located in the hypodermis region (black arrows; Fig. 1E and supplemental Fig. 1E, available at Rheumatology Online). These data demonstrate that levels of STAT3 mRNA and pY-STAT3 are increased in fibrotic skin of SSc patients, BLM-treated mice and the Tsk-1 mouse model.
STAT3 inhibition reduced dermal fibrosis in the BLM model
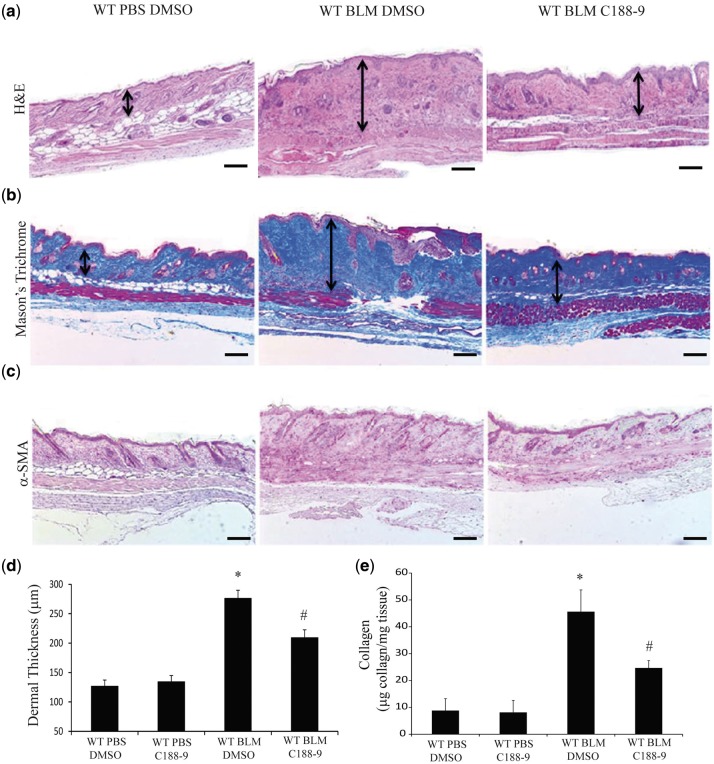
To determine the effect of STAT3 inhibition in dermal fibrosis, mice were injected with subcutaneous BLM for four weeks and treated with C188-9, a small-molecule inhibitor of STAT3, during the last 2 weeks of the model. Mice exposed to C188-9 were phenotypically healthy, displaying no observable signs of toxicity. Relative to saline-injected mice, skin from BLM-injected mice exhibited increased thickness of the dermal layer, with dense bands of collagen and extracellular matrix (Fig. 2a, black arrows). Mice treated with C188-9 had an attenuated response to BLM, with reduced dermal thickness relative to that of control-treated mice. Immunohistochemistry analysis confirmed that C188-9 decreased pY-STAT3 expression in BLM-treated mice (supplementary Fig. 1A and D). Masson’s Trichrome staining of lesional skin confirmed the increase in ECM deposition in BLM-injected mice and the decrease in ECM deposition with C188-9 treatment (Fig. 2B, black arrows). Quantification of dermal thickness demonstrated that BLM injections increased the thickness of the dermis, which was markedly attenuated in mice treated with C188-9 (Fig. 2D). Immunohistochemical staining for α-SMA, a marker of myofibroblasts, established the large numbers of myofibroblasts in BLM-injected mice (Fig. 2C). STAT3 inhibition with C188-9 decreased the number of myofibroblasts in lesional skin (Fig. 2C). Finally, the Sircol assay was used to measure total collagen deposition in lesional skin. As expected, BLM increased collagen levels in the skin, which were decreased with treatment with C188-9 (Fig. 2E).
Fig. 2.
STAT3 inhibition with C188-9 decreased dermal fibrosis in the bleomycin model
Histologicial examination of skin sections stained with H&E (A), Mason’s Trichome (B) and α-SMA antibodies (C), from PBS-treated mice exposed to DMSO (left, similar results were observed in PBS-treated mice exposed to C188-9, data not shown), BLM-treated mice treated with DMSO vehicle control (middle), and BLM-treated mice treated with C188-9 (right). These images revealed that STAT3 inhibition reduces dermal fibrosis. Images are representative of 10–12 mice from each group. Scale bars: ×4, 200 μm. (D) Dermal thickness was measured to determine the degree of dermal fibrosis, demonstrating reduced dermal fibrosis with C188-9 treatment. Data are presented as mean (s.e.m.), n = 10–12 mice per group. (*P ≤ 0.05 PBS DMSO vs BLM DMSO; #P ≤ 0.05 BLM DMSO vs BLM C188-9). (E) Soluble collagen protein levels were measured using Sircol Assay from skin samples obtained from PBS-treated mice exposed to DMSO or C188-9 and BLM-treated mice exposed to DMSO or C188-9. Data presented as mean microgram collagen per milligram tissue ± s.e.m., n = 10–12 mice per group (*P ≤ 0.05 PBS DMSO vs BLM DMSO; #P ≤ 0.05 BLM DMSO vs BLM C188-9).
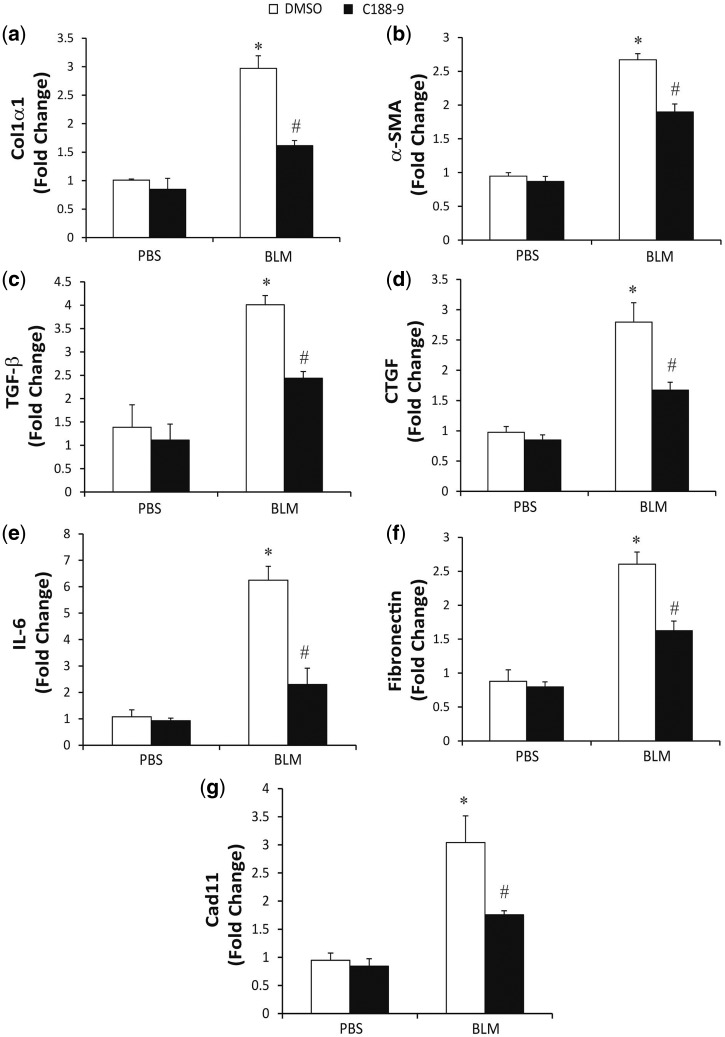
To further quantify the STAT3-dependent changes in BLM-induced dermal fibrosis, total RNA was isolated and relative RT-PCR was used to determine the expression levels of fibrotic mediators (Fig. 3). In comparison with PBS treatment, BLM-treated mice displayed an increased expression of col1α1, α-SMA, TGF-β, CTGF, IL-6, fibronectin and Cad11. Consistent with a decrease in dermal fibrosis, inhibition of STAT3 with C188-9 decreased the BLM-induced expression of all of these fibrotic mediators relative to DMSO treatment as the control. Together, the histological and RNA analyses demonstrate that STAT3 contributes to the development of dermal fibrosis in the BLM model and that targeting STAT3 with C188-9 can attenuate the development of dermal fibrosis.
Fig. 3.
Pro-fibrotic mediators in bleomycin model following STAT3 inhibition
Total mRNA was isolated from skin biopsies from PBS-treated mice exposed to DMSO or C188-9 and bleomycin treated mice exposed to DMSO or C188-9. RNA transcripts were determined for Col1α1 (A), α-SMA (B), TGF-β (C), CTGF (D), IL-6 (E), Fibronectin (F) and Cad11 (G). Transcripts were measured in parallel with 18S rRNA and values are presented as mean of fold change transcripts ± s.e.m., n = 10–12 mice per group (*P ≤ 0.05 PBS DMSO vs BLM DMSO; **P ≤ 0.05 BLM DMSO vs BLM C188-9).
STAT3 inhibition reduced hypodermal fibrosis in the Tsk-1 model
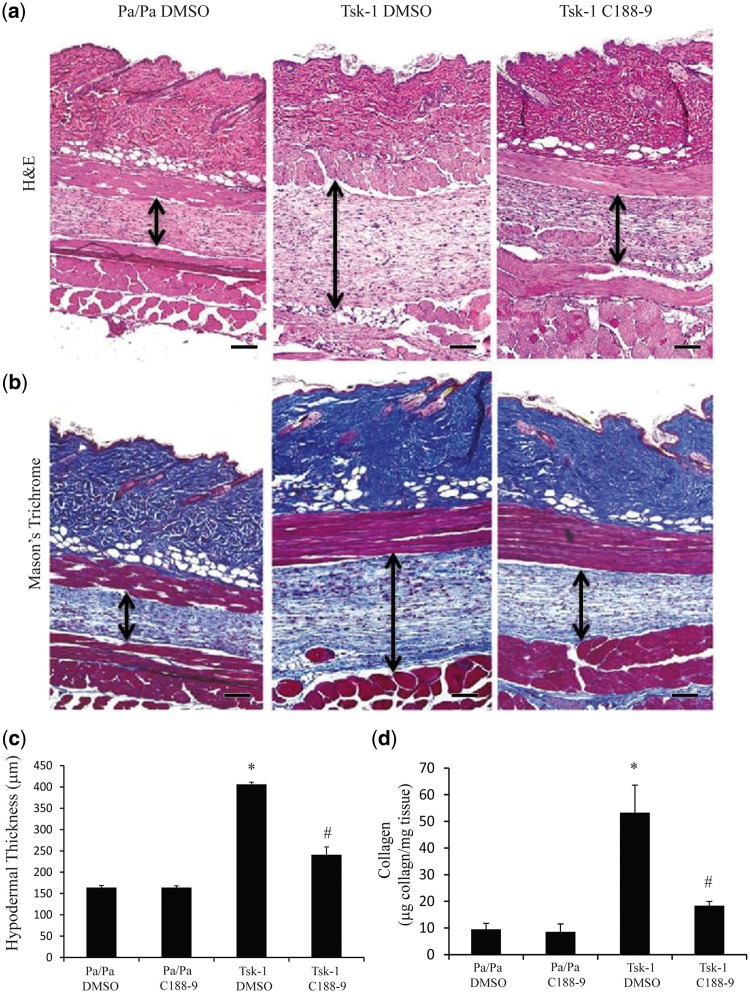
To determine the effect of STAT3 inhibition in a complementary mouse model of skin, Tsk-1 mice were treated with C188-9 or DMSO vehicle control for 2 weeks starting at 5 weeks of age. Mice exposed to C188-9 displayed a healthy phenotype lacking any observable signs of toxicity. As seen in Fig. 4, Tsk-1 mice had increased thickness of the hypodermis relative to Pa/Pa control mice. C188-9 attenuated the hypodermal thickness observed in Tsk-1 mice. Immunostaining confirmed that C188-9 reduced pY-STAT3 expression in Tsk-1 mice (supplementary Fig. 1B and E, available at Rheumatology Online). Similarly, Masson’s Trichrome staining demonstrated increased collagen deposition in the hypodermis in Tsk-1, which was reduced with C188-9 treatment (Fig. 4B). Quantification of hypodermal thickness demonstrated that STAT3 inhibition with C188-9 reduced the hypodermal thickness in Tsk-1 mice compared with in the DMSO vehicle control (Fig. 4C). No changes were seen in Pa/Pa control mice treated with C188-9 vs DMSO. Finally, total collagen deposition, as measured by Sircol assay, also demonstrated that C188-9 significantly reduced collagen accumulation in Tsk-1 mice (Fig. 4D).
Fig. 4.
STAT3 inhibition with C188-9 decreases hypodermal fibrosis in Tsk-1 mice
Histological examination of skin sections stained with H&E (A) and Mason’s Trichome (B) from Pa/Pa mice treated with DMSO (left; similar results were observed in Pa/Pa mice exposed to C188-9), Tsk-1 mice treated with DMSO (middle) and Tsk-1 mice treated with C188-9 (right). These images revealed that STAT inhibition displayed a reduction in hypodermal fibrosis. Images are representative of 10–12 mice per group. Scale bars: ×10, 100 μm. (C) Hypodermal thickness was measured to determine the degree of fibrosis in the hypodermis section. Data are presented as mean (s.e.m.), n = 10–12 mice per group (*P ≤ 0.05 Pa/Pa DMSO vs Tsk-1 DMSO; **P ≤ 0.05 Tsk-1 DMSO vs Tsk-1 C188-9). (D) Soluble collagen protein levels were measured using Sircol Assay from skin samples obtained from Pa/Pa control mice treated with DMSO or C188-9 and Tsk-1 mice treated with DMSO or C188-9. Data presented as mean microgram collagen per milligram tissue (s.e.m.), n = 10–12 mice per group (*P ≤ 0.05 Pa/Pa DMSO vs Tsk-1 DMSO; **P ≤ 0.05 Tsk-1 DMSO vs Tsk-1 C188-9).
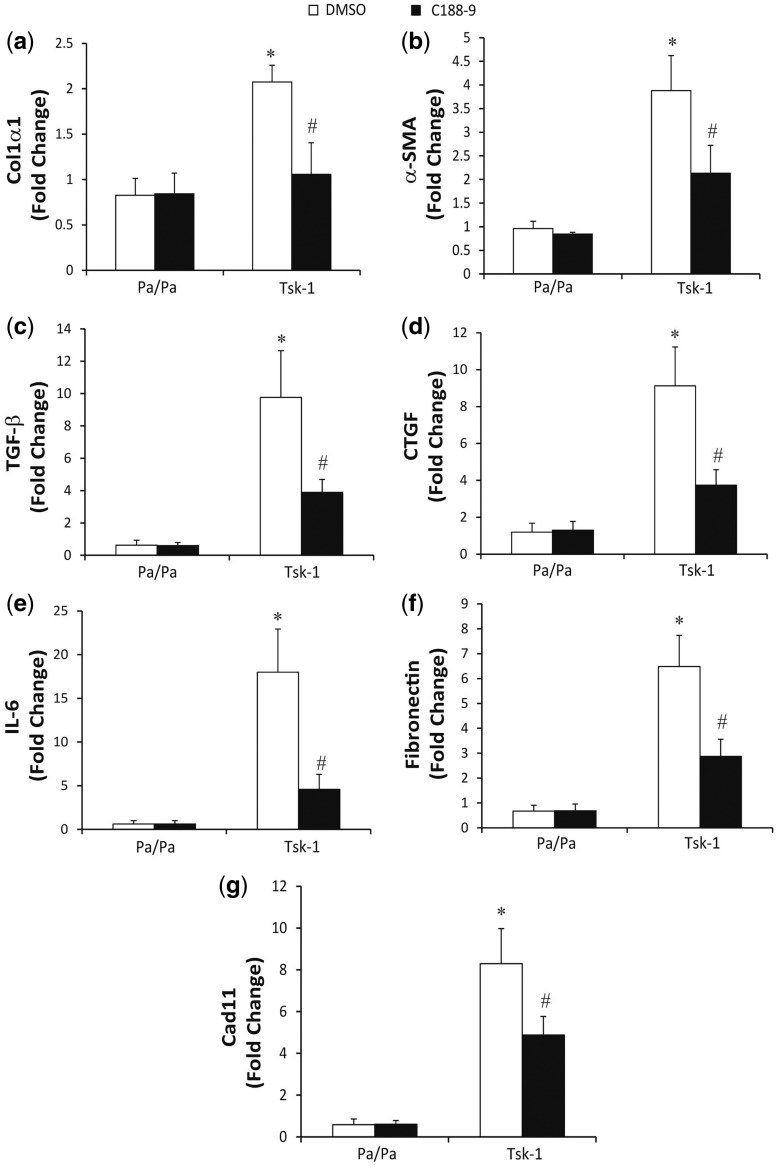
To further quantify the changes in skin fibrosis in Tsk-1 mice treated with C188-9, RT-PCR was used to determine the expression levels of fibrotic mediators (Fig. 5). In comparison with skin from Pa/Pa mice, skin from Tsk-1 mice had increased expression of col1α1, α-SMA, TGF-β, CTGF, IL-6, fibronectin and Cad11. Consistent with a reduction in fibrosis, treatment with C188-9 decreased the expression of these fibrotic mediators in Tsk-1 mice. Together, these data demonstrate that STAT3 contributes to the development of skin fibrosis in the Tsk-1 model and can regulate fibrotic pathways in the absence of significant inflammation. Furthermore, as in the BLM model, targeting STAT3 with C188-9 in Tsk-1 mice can attenuate development of skin fibrosis.
Fig. 5.
Pro-fibrotic mediators in Tsk-1 model following STAT3 inhibition
Total RNA was isolated from skin biopsies from Pa/Pa control mice treated with DMSO or C188-9 and Tsk-1 mice treated with DMSO or C188-9. RNA transcripts were determined for Col1α1 (A), α-SMA (B), TGF-β (C), CTGF (D), IL-6 (E), Fibronectin (F) and Cad11 (G). Transcripts were measured in parallel with 18S rRNA and values are presented as mean of fold change transcripts ±s.e.m., n = 10–12 mice per group (*P ≤ 0.05 Pa/Pa DMSO vs Tsk-1 DMSO; **P ≤ 0.05 Tsk-1 DMSO vs Tsk-1 C188-9).
STAT3 regulated fibroblast activation and myofibroblasts
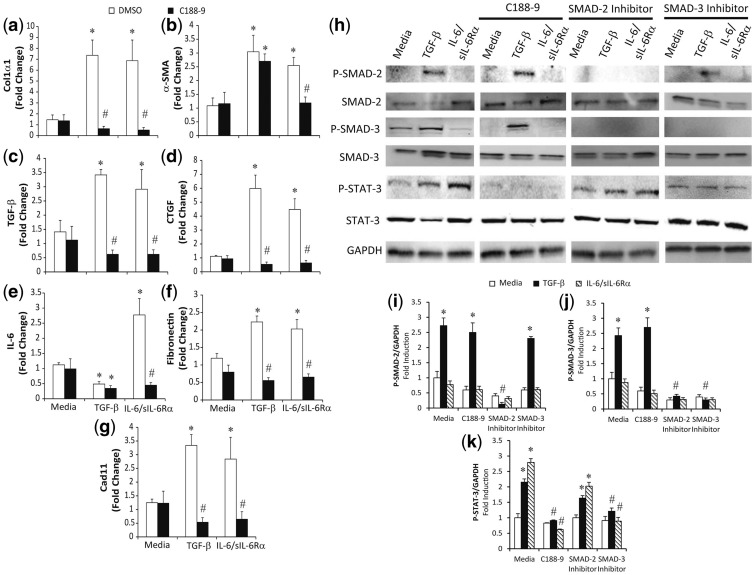
Fibroblasts and myofibroblasts are key drivers of fibrosis [20]. Given the decrease in fibrosis with inhibition of STAT3 in the mouse models, we hypothesized that STAT3 regulates fibrosis pathways in dermal fibroblasts. Murine dermal fibroblasts were activated and induced to differentiate into myofibroblast by treatment with TGF-β or the IL-6/sIL-6 Rα complex [8]. Total mRNA was isolated to quantify expression of the fibrotic mediators by RT-PCR. As seen in Fig. 6, both TGF-β and the IL-6/sIL-6 Rα complex increased expression of col1α1, α-SMA, TGF-β, CTGF, fibronectin and Cad11 transcripts. However, IL-6 transcripts were reduced with stimulation with TGF-β, but increased with IL-6 trans-signalling (Fig. 6E). As expected, STAT3 inhibition with C188-9 attenuated the upregulation of the fibrotic mediators induced by IL-6 trans-signalling. Interestingly, STAT3 inhibition with C188-9 also attenuated TGF-β–induced expression of col1α1, TGF-β, CTGF, fibronectin and Cad11. Finally, C188-9 treatment was able to reduce the increased α-SMA levels induced via IL-6 trans-signalling, but had no effect on the increased α-SMA levels following TGF-β treatment. These data indicate that STAT3 regulates both IL-6–mediated and TGF-β–mediated fibroblast activation and IL-6–mediated myofibroblast differentiation in murine dermal fibroblasts.
Fig. 6.
STAT3 regulation of cultured fibroblasts and myofibroblasts
Primary dermal fibroblasts were isolated, cultured and subsequently stimulated with TGF-β (10 ng/ml), IL-6/sIL-6Rα complex (20 μg/ml) and C188-9 inhibitor (20 μM). Twenty-four hours later, RNA was isolated and transcripts were determined for Col1α1 (A), α-SMA (B), TGF-β (C), CTGF (D), IL-6 (E), Fibronectin (F) and Cad11 (G). Transcripts were measured in parallel with 18S rRNA, and values are presented as mean of fold change transcripts ± s.e.m., n ≥ 4 (*P ≤ 0.05 Media vs TGF-β, IL-6/sIL-6Rα; **P ≤ 0.05 TGF-β vs TGF-β C188-9; and IL-6/sIL-6Rα vs IL-6/sIL-6Rα C188-9). (H–J) Lysates were obtained from dermal fibroblasts treated for 1 h with different inhibitors (C188-9, 20 μM; SMAD-2 inhibitor, 10 μM; SMAD-3 inhibitor, 10 μM), and subsequently stimulated with TGF-β and IL-6/sIL-6Rα complex. Western blots were performed for total and phosphorylated forms of STAT3, SMAD2 and SMAD3. GAPDH levels were used as a loading control. Phospho-SMAD2 (I), phosph0SMAD3 (J) and phospho-STAT3 (K) band intensities were quantified using Image J analysis. Values are presented as the percentage of GAPDH ± s.e.m., n ≥ 4 (*P ≤ 0.05 Media vs TGF-β or IL-6/sIL-6Rα; **P ≤ 0.05 TGF-β vs TGF-β with either C188-9, SMAD2 or SMAD3 inhibitors or IL-6/sIL-6Rα vs IL-6/sIL-6Rα C188-9).
The observation that STAT3 can regulate TGF-β signalling is novel. Therefore, we wanted to characterize the pathways used by TGF-β to activate STAT3. Western blotting was performed on dermal fibroblasts stimulated with TGF-β or the IL-6/sIL-6 Rα complex (Fig. 6H–J). TGF-β, but not IL-6 trans-signalling, increased phosphorylation of SMAD2 and SMAD3. As expected, stimulation of dermal fibroblasts with the IL-6/sIL-6 Rα complex increased STAT3 phosphorylation at Y705. Interestingly, levels of pY-STAT3 were increased in dermal fibroblasts by TGF-β, consistent with the ability of STAT3 inhibition to block TGF-β activation of fibrotic genes in Fig. 6. The phosphorylation of STAT3 by TGF-β was observed as early as at 3 h (supplementary Fig. 2, available at Rheumatology Online) and persisted until up to 24 h (Fig. 6).
To determine the signalling pathways that contribute to STAT3 phosphorylation, dermal fibroblasts were treated with TGF-β and IL-6 trans-signalling in the presence and absence of the STAT3 inhibitor (C188-9), a SMAD2 inhibitor or a SMAD3. C188-9 blocked STAT3 phosphorylation induced by both TGF-β and IL-6 trans-signalling. C188-9 did not block TGF-β–induced phosphorylation of SMAD2 or SMAD3. As expected, inhibitors of SMAD2 and SMAD3 blocked TGF-β–induced phosphorylation of SMAD2 and SMAD3, respectively. With regards to STAT3 phosphorylation, inhibition of SMAD2 had a non-significant decrease in STAT3 activation. In contrast, inhibition of SMAD3 resulted in a larger and significant decrease in STAT3 phosphorylation induced by TGFβ. Finally, inhibitors of c-jun N-terminal kinase and extracellular signal-related kinase signalling, did not block TGF-β activation of STAT3 (data not shown). These data demonstrate TGF-β–induced STAT3 phosphorylation is dependent on SMAD3.
Discussion
In the current study, it was demonstrated that STAT3 is activated in fibrotic skin tissue from SSc patients and mouse models of skin fibrosis. It was also demonstrated that inhibition of STAT3 phosphorylation, using C188-9, reduces skin fibrosis in two different mouse models. Finally, STAT3 not only modulates IL-6–stimulated expression of fibrotic genes, but also regulates TGF-β–stimulated expression of fibrotic genes in dermal fibroblasts.
Multiple molecular pathways are involved in SSc and tissue fibrosis, including multiple cytokines such as CCL2, type I interferons, TGF-β and the IL-6 family of cytokines. These cytokines ultimately lead to the accumulation of activated fibroblasts and myofibroblasts, key producers of ECM [21]. STAT3 is a latent cytoplasmic transcription factor that is activated predominantly by the gp130 receptor via the IL-6 family of cytokines, which includes IL-6, IL-11, leukemia inhibitory factor (LIF), oncostatin M (OSM), ciliary neutotrophic factor (CNTF), IL-27, IL-31 and neuropoietin [10]. IL-6 signals via the ‘classical’ pathway by binding to cell-surface IL-6 Rα or via the ‘trans-signalling’ pathway mediated by soluble IL-6 and soluble IL-6 Rα complex binding to cell-surface gp130 [22]. STAT3 is activated via other pathways, including those downstream of IL-10 signalling and growth factors, such as epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) [23]. Indeed multiple members of the IL-6 family have been shown to be involved in the development of fibrosis [8, 24, 25]. In the recent trial of anti–IL-6 receptor for the treatment of SSc, microarray analyses indicated that the STAT3 pathway remained elevated despite blocking the IL-6 pathway [26]. Therefore, targeting downstream mediators such as STAT3, other STAT proteins or Janus kinases may be a better strategy.
The current study utilized two distinct and complementary models of skin fibrosis to advance our understanding of how STAT3 may regulate the development of fibrosis. These models allow for investigation of the different phases or pathways involved in SSc. The BLM model triggers cellular damage and inflammation that lead to activation of TGF-β pathways and fibrosis [4, 27]. In contrast, the Tsk-1 model is dependent on the activation of TGF-β, independent of a significant inflammatory component [28, 29]. The current results demonstrate that STAT3 inhibition decreases fibrosis in both models, which suggests that STAT3 is, at least in part, acting through modulation of the TGF-β and fibrosis pathways. Consistent with a role for STAT3 in regulating the dermal fibroblasts is the observation of nuclear localization of pY-STAT3 in fibroblasts in SSc skin biopsies and fibrotic skin from mice. STAT3 expression and phosphorylation has also been reported in keloid fibroblasts and keloid scar tissue [30]. Furthermore, the in vitro studies with dermal fibroblasts clearly demonstrate that STAT3 activation can regulate the fibrotic phenotype of dermal fibroblasts triggered by either IL-6 or TGF-β. Together the in vivo and in vitro studies strongly suggest that the fibroblast would be one of the potential targets of STAT3 inhibition in fibrotic tissue. However, we do not rule out the potential for STAT3 to regulate macrophages during the development of fibrosis, as has been previously reported [31]. It would definitely be appealing to be able to target multiple cells involved in the development of tissue fibrosis, which STAT3 inhibition potentially could do.
As expected, in vitro stimulation of dermal fibroblasts with IL-6 activated STAT3. However, it is notable that IL-6 triggered fibrotic pathways in dermal fibroblasts, consistent with its pleiotropic role in regulating cell behaviour. Previous reports have demonstrated that STAT3 inhibition by siRNA in keloid fibroblasts resulted in decreased collagen production [30]. Also, hypertrophic scar fibroblasts treated with a cell-permeable STAT3 peptide inhibitor resulted in decreased collagen and fibronectin production [32]. Indeed, in vitro STAT3 inhibition of dermal fibroblasts with C188-9 also decreased expression of collagen and other fibrotic genes.
More notable was the ability of the TGF-β pathway to activate STAT3 and for STAT3 inhibition to block fibrotic gene expression in dermal fibroblasts induced by TGF-β. These data are similar to observations in alveolar epithelial cells [13]. The interaction between TGF-β and STAT3 pathways has been previously suggested during the development of liver fibrosis, in which TGF-β required STAT3 activation to induce CTGF expression [33]. The signalling pathways by which TGF-β activates STAT3 remain unknown. Our data support that the activation of STAT3 by TGF-β is more dependent on SMAD3 than SMAD2 activation. It remains unclear why the activation of STAT3 by TGF-b is more dependent on SMAD3 than SMAD2. Prior studies have emphasized differences in SMAD2 and SMAD3 with regards to downstream responses as well as associations with different intracellular proteins [34–36]. It is intriguing to hypothesize that STAT3 may form a molecular complex with SMAD3 in dermal fibroblasts stimulated with TGF-β. This complex may involve p300 as has been suggested in Hep3B cells [37]. Alternatively, it is also possible that TGF-β indirectly activates STAT3 by inducing IL-6 or IL-6 family members, which then subsequently trigger STAT3 phosphorylation [38, 39]. These potential mechanisms are being investigated.
In conclusion, these findings demonstrate the importance of STAT3 signalling in regulating dermal fibroblasts during the development of dermal fibrosis. Based on these data, it is hypothesized that STAT3 activation in dermal fibroblasts promotes the pro-fibrotic environment that leads to ECM deposition by fibroblasts and myofibroblasts. The current report adds to our understanding of the role of STAT3 in dermal fibrosis and suggests that STAT3 may be a therapeutic strategy for fibrotic diseases such as SSc.
Funding: This study was funded by the National Institutes of Health (NIH) R01AR062056 (S.K.A., S.T.), the Scleroderma Foundation Established Investigator Award (S.K.A.), NIH 2T32AI053831-11A1 (M.P.) and the Ford Foundation Postdoctoral Award Program (M.P.).
Disclosure statement: D.J.T. holds patents covering the chemical composition and uses of C188-9 that have been filed by Baylor College of Medicine, which has issued an exclusive licence to StemMed, Ltd and is an owner, President and CEO of StemMed. S.K.A. has a patent related to the use of C188-9 in fibrosis. All other authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Charles C, Clements P, Furst DE.. Systemic sclerosis: hypothesis-driven treatment strategies. Lancet 2006;367:1683–91. [DOI] [PubMed] [Google Scholar]
- 2. Gabrielli A, Avvedimento EV, Krieg T.. Scleroderma. New Engl J Med 2009;360:1989–2003. [DOI] [PubMed] [Google Scholar]
- 3. Avouac J. Mouse model of experimental dermal fibrosis: the bleomycin-induced dermal fibrosis. Methods Mol Biol 2014;1142:91–8. [DOI] [PubMed] [Google Scholar]
- 4. Yamamoto T, Nishioka K.. Cellular and molecular mechanisms of bleomycin-induced murine scleroderma: current update and future perspective. Exp Dermatol 2005;14:81–95. [DOI] [PubMed] [Google Scholar]
- 5. Pablos JL, Everett ET, Norris JS.. The tight skin mouse: an animal model of systemic sclerosis. Clin Exp Rheumatol 2004;22:S81–5. [PubMed] [Google Scholar]
- 6. O’Reilly S, Cant R, Ciechomska M, van Laar JM.. Interleukin-6: a new therapeutic target in systemic sclerosis? Clin Transl Immunol 2013;2:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fielding CA, Jones GW, McLoughlin RM. et al. Interleukin-6 signaling drives fibrosis in unresolved inflammation. Immunity 2014;40:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pedroza M, Schneider DJ, Karmouty-Quintana H. et al. Interleukin-6 contributes to inflammation and remodeling in a model of adenosine mediated lung injury. PloS One 2011;6:e22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seluanov A, Vaidya A, Gorbunova V.. Establishing primary adult fibroblast cultures from rodents. J Vis Exp 2010;44:2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knight D, Mutsaers SE, Prele CM.. STAT3 in tissue fibrosis: is there a role in the lung? Pulmonary Pharmacol Therapeutics 2011;24:193–8. [DOI] [PubMed] [Google Scholar]
- 11. Bharadwaj U, Eckols TK, Xu X. et al. Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget 2016;7:26307–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu X, Kasembeli MM, Jiang X, Tweardy BJ, Tweardy DJ.. Chemical probes that competitively and selectively inhibit Stat3 activation. PloS One 2009;4:e4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pedroza M, Le TT, Lewis K. et al. STAT-3 contributes to pulmonary fibrosis through epithelial injury and fibroblast–myofibroblast differentiation. FASEB J 2016;30:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subcommittee for scleroderma criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Preliminary criteria for the classification of systemic sclerosis (scleroderma). Arthritis Rheum 1980;23:581–90. [DOI] [PubMed] [Google Scholar]
- 15. Assassi S, Swindell WR, Wu M. et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol 2015;67:3016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takagawa S, Lakos G, Mori Y. et al. Sustained activation of fibroblast transforming growth factor-beta/Smad signaling in a murine model of scleroderma. J Invest Dermatol 2003;121:41–50. [DOI] [PubMed] [Google Scholar]
- 17. Wu M, Pedroza M, Lafyatis R. et al. Identification of cadherin 11 as a mediator of dermal fibrosis and possible role in systemic sclerosis. Arthritis Rheumatol 2014;66:1010–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu M, Schneider DJ, Mayes MD. et al. Osteopontin in systemic sclerosis and its role in dermal fibrosis. J Invest Dermatol 2012;132:1605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Corti M, Brody AR, Harrison JH.. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol 1996;14:309–15. [DOI] [PubMed] [Google Scholar]
- 20. Kendall RT, Feghali-Bostwick CA.. Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol 2014;5:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abraham DJ, Varga J.. Scleroderma: from cell and molecular mechanisms to disease models. Trends Immunol 2005;26:587–95. [DOI] [PubMed] [Google Scholar]
- 22. Schaper F, Rose-John S.. Interleukin-6: biology, signaling and strategies of blockade. Cytokine Growth Factor Rev 2015;26:475–87. [DOI] [PubMed] [Google Scholar]
- 23. Darnell JE Jr, Kerr IM, Stark GR.. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994;264:1415–21. [DOI] [PubMed] [Google Scholar]
- 24. Bamber B, Reife RA, Haugen HS, Clegg CH.. Oncostatin M stimulates excessive extracellular matrix accumulation in a transgenic mouse model of connective tissue disease. J Mol Med 1998;76:61–9. [DOI] [PubMed] [Google Scholar]
- 25. Tang W, Geba GP, Zheng T. et al. Targeted expression of IL-11 in the murine airway causes lymphocytic inflammation, bronchial remodeling, and airways obstruction. J Clin Invest 1996;98:2845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khanna D, Denton CP, Jahreis A. et al. Safety and efficacy of subcutaneous tocilizumab in adults with systemic sclerosis (faSScinate): a phase 2, randomised, controlled trial. Lancet 2016;387:2630–40. [DOI] [PubMed] [Google Scholar]
- 27. Giacomelli R, Liakouli V, Berardicurti O. et al. Interstitial lung disease in systemic sclerosis: current and future treatment. Rheumatol Int 2017;37:853–63. [DOI] [PubMed] [Google Scholar]
- 28. Saito E, Fujimoto M, Hasegawa M. et al. CD19-dependent B lymphocyte signaling thresholds influence skin fibrosis and autoimmunity in the tight-skin mouse. J Clin Invest 2002;109:1453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sgonc R. The vascular perspective of systemic sclerosis: of chickens, mice and men. Int Archives Allergy Immunol 1999;120:169–76. [DOI] [PubMed] [Google Scholar]
- 30. Lim CP, Phan TT, Lim IJ, Cao X.. Stat3 contributes to keloid pathogenesis via promoting collagen production, cell proliferation and migration. Oncogene 2006;25:5416–25. [DOI] [PubMed] [Google Scholar]
- 31. Le TT, Karmouty-Quintana H, Melicoff E. et al. Blockade of IL-6 Trans signaling attenuates pulmonary fibrosis. J Immunol 2014;193:3755–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ray S, Ju X, Sun H. et al. The IL-6 Trans-signaling-STAT3 pathway mediates ECM and cellular proliferation in fibroblasts from hypertrophic scar. J Invest Dermatol 2013;133:1212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y, Liu H, Meyer C. et al. Transforming growth factor-β (TGF-β)-mediated connective tissue growth factor (CTGF) expression in hepatic stellate cells requires Stat3 signaling activation. J Biol Chem 2013;288:30708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown KA, Pietenpol JA, Moses HL.. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-β signaling. J Cell Biochem 2007;101:9–33. [DOI] [PubMed] [Google Scholar]
- 35. Brown KA, Ham AJ, Clark CN. et al. Identification of novel Smad2 and Smad3 associated proteins in response to TGF-β1. J Cell Biochem 2008;105:596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Budi EH, Duan D, Derynck R.. Transforming growth factor-β receptors and Smads: regulatory complexity and functional versatility. Trends Cell Biol 2017;27:658–72. [DOI] [PubMed] [Google Scholar]
- 37. Yamamoto T, Matsuda T, Muraguchi A, Miyazono K, Kawabata M.. Cross-talk between IL-6 and TGF-β signaling in hepatoma cells. FEBS Lett 2001;492:247–53. [DOI] [PubMed] [Google Scholar]
- 38. Elias JA, Lentz V, Cummings PJ.. Transforming growth factor-β regulation of IL-6 production by unstimulated and IL-1–stimulated human fibroblasts. J Immunol 1991;146:3437–43. [PubMed] [Google Scholar]
- 39. Rochester CL, Ackerman SJ, Zheng T, Elias JA.. Eosinophil–fibroblast interactions. Granule major basic protein interacts with IL-1 and transforming growth factor-β in the stimulation of lung fibroblast IL-6-type cytokine production. J Immunol 1996;156:4449–56. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.